Graphical abstract
Highlights
-
•
Structures have been determined for natural and recombinant polyproteins.
-
•
Native HIV Gag polyprotein architecture was revealed by cryo-EM of immature capsids.
-
•
Recombinant polyprotein technology has resolved sample preparation bottlenecks.
-
•
The high-resolution structure of influenza polymerase has been solved.
-
•
Single-molecule analysis of polyproteins revealed their folding characteristics.
Abstract
Polyproteins are chains of covalently conjoined smaller proteins that occur in nature as versatile means to organize the proteome of viruses including HIV. During maturation, viral polyproteins are typically cleaved into the constituent proteins with different biological functions by highly specific proteases, and structural analyses at defined stages of this maturation process can provide clues for antiviral intervention strategies. Recombinant polyproteins that use similar mechanisms are emerging as powerful tools for producing hitherto inaccessible protein targets such as the influenza polymerase, for high-resolution structure determination by X-ray crystallography. Conversely, covalent linking of individual protein subunits into single polypeptide chains are exploited to overcome sample preparation bottlenecks. Moreover, synthetic polyproteins provide a promising tool to dissect dynamic folding of polypeptide chains into three-dimensional architectures in single-molecule structure analysis by atomic force microscopy (AFM). The recent use of natural and synthetic polyproteins in structural biology and major achievements are highlighted in this contribution.
Current Opinion in Structural Biology 2015, 32:139–146
This review comes from a themed issue on New constructs and expressions of proteins
Edited by Imre Berger and Roslyn M Bill
For a complete overview see the Issue and the Editorial
Available online 18th May 2015
http://dx.doi.org/10.1016/j.sbi.2015.04.007
0959-440X/Crown Copyright © 2015 Published by Elsevier Ltd. All rights reserved.
Introduction
Polyproteins composed of covalently linked individual proteins with different biological functions are prevalent in nature. For instance SARS coronavirus, the agent that causes severe acute respiratory syndrome, realizes its entire proteome from two large polyproteins, each encoded by a long single open reading frame (ORF) [1]. The expressed SARS polyproteins are then processed into the individual functional protein subunits by the action of highly specific proteases also encoded by the ORFs [1, 2]. A further example is human immune deficiency virus (HIV) which causes acquired immune deficiency syndrome (AIDS). The RNA genome of this retrovirus is organized in three major genes gag, pol and env, which encode for polyproteins and undergo proteolytic processing at defined stages during maturation [3]. Recombinant polyprotein approaches mimicking viral polyproteins have recently emerged as a powerful means to express high-value protein complexes for structure determination [4, 5•]. Recently, the long elusive influenza polymerase has been produced successfully from a self-processing synthetic polyprotein, enabling high-resolution structure determination [6••, 7]. Polyprotein fusions which are not processed by protease, but remain covalently conjoined by engineered linkers have been instrumental to obtain important insight into numerous essential physiological processes including multidrug efflux, co-translational protein targeting or enzymatic processing of chromatin, among others [8••, 9, 11•, 12••]. Moreover, single-chain engineering approaches linking protein domains into novel, artificial polyproteins have resulted in new classes of high-affinity binder molecules as potential protein therapeutics [13] and accelerated elucidation of mechanisms governing protein folding by single-molecule techniques [14]. Thus, polyprotein technologies have recently gained prominence as particularly useful tools for unlocking previously often inaccessible protein samples to detailed structural and mechanistic studies, as illustrated in the following. In this contribution, the use of polyproteins in structural biology is discussed, by highlighting recently determined structures of naturally occurring polyproteins on one hand and by reviewing recent structural studies where recombinant polyprotein constructions were utilized on the other.
Natural polyproteins
Polyproteins are used in nature by many viruses to structure their proteome. As a consequence, viral polyproteins are an intense focus of research efforts for numerous reasons [15, 16, 17]. For instance, inhibition by small molecules of proteases that process polyproteins during viral maturation can provide a powerful handle to combat viral disease [2, 18, 19, 20]. In HIV, the physical infrastructure of the virus is provided by the gag gene which gives rise to the precursor Gag (group specific antigen) polyprotein [21]. Viral protease processes Gag during maturation into several proteins and spacer peptides dictating immature and mature viral capsid structure [22]. Maturation is a two-stage process. Precursor Gag polyprotein first forms a hexameric lattice at the plasma membrane of an infected cell. This induces budding and release of immature viral particles. Proteolytic processing of Gag then rearranges the viral structure to the mature form [23]. Inhibition of Gag-processing protease enabled preparation of immature retroviral capsids suitable for structure analysis by electron cryo-tomography and subtomogram averaging methods (Figure 1 ) [24, 25, 26, 27••]. Comparison of the structures of HIV Gag protein in reconstituted tubular arrays on one hand and in intact virus particles on the other, provided unprecedented insights into the conformational plasticity of this precursor polyprotein [26, 27••]. The studies also revealed that retroviral capsid proteins can adopt different quaternary arrangements during virus assembly, notwithstanding conserved tertiary structures [27••].
Figure 1.
Natural polyprotein structures. Polyproteins, prevalent in biology, are illustrated here by the structures of the human immune deficiency virus (HIV) immature capsid determined by electron cryo-tomography revealing molecular details of hexameric Gag [24, 26, 27••], the structure of a repeated unit of the ABA-1 nematode polyprotein allergen derived from nuclear magnetic resonance (NMR) spectroscopy [31] and the crystal structure of a single-chain, multi-domain long-chain acyl-CoA carboxylase, LCC [34]. CT stands for carboxyltransferase, BCCP for biotin carboxyl carrier protein, and BC for biotin carboxylase components.
A different, less frequently encountered type of natural, non-viral polyproteins are so-called tandemly repetitive polyproteins (TRPs). TRPs are also produced as large precursor proteins and then processed by proteases into several copies of proteins with similar function. TRPs are made of consecutively arranged repeats of amino-acid stretches. Examples of known TRPs include polyprotein lipid binding proteins of nematodes [28] and filaggrins, which are keratinocyte produced TRPs crucial to health and appearance of skin [29, 30]. The solution structure of a mature, post-translationally processed repeat unit of a TRP, ABA-1A from the nematode polyprotein allergen of Ascaris, was determined, representing the first structure of this class of proteins [31]. ABA-1A adopts a novel fold comprising two juxtaposed four-helical bundles that share a long central alpha-helix (Figure 1). Nematode polyprotein allergens have no known counterpart in humans. The Ascaris ABA-1A structure therefore may serve as a starting point for the development of new drugs and therapeutic intervention strategies against disease states caused by these intestinal parasites.
An inverse ‘polyprotein’ concept of covalently linking functional protein units into long modular polypeptide chains characterizes mega-enzymes that functionally arrange multiple domains into ordered assembly lines for the production of a wide variety of bioactive molecules. Modular polyketide synthases (PKS) and their metazoan homologs, fatty acid synthases (FAS) belong to this class of catalysts [32, 33]. Recently, the structure and function of a modular multi-domain long-chain acyl-CoA carboxylase from Mycobacterium avium was elucidated [34]. The crystal structure revealed extensive swapping of functional domains in the holo-enzyme which is a homo-hexamer (Figure 1). Thus, four intertwined protomers are involved in completing one catalytic reaction cycle [34].
Synthetic polyproteins
Influenza polymerase produced from a recombinant polyprotein
The highly successful strategy of viruses to utilize polyproteins to their advantage has found its equivalent in recombinant technology. A synthetic polyprotein, expressed recombinantly in baculovirus-infected insect cells, has enabled the structure determination of influenza polymerase. Despite its common appearance, a detailed understanding of the molecular mechanisms of the virus that causes influenza has remained elusive. More than 40 years ago, the influenza polymerase was discovered, a key protein complex that replicates the genetic material of the virus [35, 36]. Atomic resolution information on the structure and function of this protein machine is essential, as it may open up important avenues for drug discovery. However, the influenza polymerase remained inaccessible for decades — to produce this valuable protein complex for detailed analysis, proved to be a seemingly insurmountable technical challenge. This has now changed dramatically with structures of influenza polymerase complex determined by X-ray crystallography [6••, 7]. These break-through studies provide unprecedented insight into the inner workings of this viral protein machine. This revolution in understanding influenza was brought about by applying a polyprotein strategy to produce influenza polymerase recombinantly, in the quality and quantity required for high-resolution structural and functional analysis (Figure 2 ). The strategy applied recapitulates the mechanism adopted by SARS coronavirus. A single ORF was constructed encoding for a highly specific protease, NIa, from tobacco etch virus (TEV), fused in frame with consecutively arranged PA, PB1 and PB2 subunits of the polymerase. At the far end of the polyprotein, a reporter protein was inserted to track protein production ‘in real-time’ during expression, by recording fluorescence [5•]. All protein units within the polyprotein were spaced apart by customized linkers containing the specific site for cleavage by the TEV protease, purification tag sequences and spacer residues (Figure 2). The polyproteins encoding for influenza polymerases were successfully expressed with the MultiBac baculovirus insect cell expression system developed for producing multiprotein complexes [37].
Figure 2.
Influenza polymerase. (a) A self-processing recombinant polyprotein was used to express influenza polymerase complex in high quality and quantity in insect cells by using the MultiBac system [37] as illustrated (left). The polymerase was expressed from a single open reading frame encoding for tobacco etch virus NIa protease (TEV), the polymerase subunits PA, PB1 and PB2 and a fluorescent protein (CFP). A second, yellow fluorescent protein (YFP) was inserted elsewhere into the MultiBac baculoviral genome as an expression performance marker. The resulting polyprotein is processed into the individual subunits by highly specific proteolysis mediated by TEV. The polyprotein construct is shown schematically (right) illustrating tag placement and design of spacers in between the subunits (adapted from [6••]). The C-terminal part of PB1 and the N-terminal part of PB2 co-fold into a helical bundle necessitating careful linker design (boxed). (b) Crystal structure of influenza polymerase bound to cognate viral RNA (vRNA) is shown in a ribbon representation. PA is colored in blue, PB1 in green and PB2 in red. RNA substrate is colored in orange. Cap-binding domain and endonuclease domain are indicated. The structure motif formed by PB1 C-terminal domain and PB2 N-terminal domain is shown in a magnification (right). PB2 N-terminus and PB1 C-terminus are marked.
Sample preparation bottlenecks resolved by synthetic polyproteins that remain uncleaved
The polyprotein encoding for influenza polymerase was proteolytically cleaved into the constituent protein subunits to yield the sample that crystallized. Conversely, engineering of individual proteins into covalently linked polypeptide chains can also accelerate structure determination considerably. These ‘polyproteins’ remain conjoined as single-chains during sample preparation and structure determination. Particularly prominent examples for such single-chain engineering include the insertion of T4 lysozyme into the primary sequence of G-protein coupled receptors (GPCRs) to facilitate crystallogenesis [38]. In a variation of this approach, the catalytic domain of Pyrococcus abysii glycogen synthase was recently used to stabilize an intracellular loop of the human OX2 orexin receptor to determine its crystal structure bound to an insomnia drug [39].
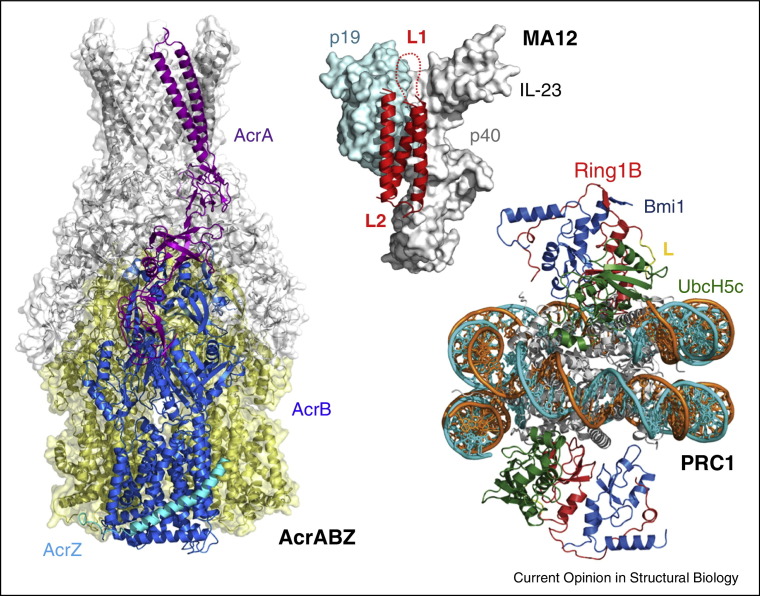
Elaborate single-chain engineering into polyproteins was applied to determine the architecture of the bacterial multidrug efflux pump AcrABZ-TolC [8••]. In this study, the efflux pump was assembled by preparing two single-chain polypeptide fusions, AcrB-AcrA-AcrB and AcrA-AcrZ, respectively, each conjoined by extended glycine/serine rich linkers. TolC was co-expressed with the AcrA-AcrZ fusion. This strategy resulted in a reconstituted complex with the correct stoichiometry enabling structure determination by hybrid methods. Fitting of crystal coordinates into EM densities allowed to model AcrBZ/AcrA interactions (Figure 3 ) as well as the holo-complex containing TolC.
Figure 3.
Linking individual proteins into polypeptide chains. Proteins engineered into single polypeptide chains were used to obtain suitable sample for structure determination of AcrABZ complex [8••], an alphabody (MA12) to neutralize human interleukin Il-23 (p19 and p40) [13] and the first structure of a histone-modifying enzyme, the Polycomb Repressive Complex (PRC) 1 ubiquitylation module, bound to a nucleosome [12••]. Linker amino acid segments are marked (L, L1, L2).
Single-chain engineering likewise enabled the development of alphabodies, a novel scaffold representing a promising alternative to antibodies for various biomedical and biotechnological applications [13]. Alphabodies comprise in silico designed short individual alpha-helical protein segments that are then conjoined by covalent linkers into single-chain antiparallel coiled-coils that are highly stable and suitable for affinity maturation. The crystal structure of a complex with human interleukin (IL-23) revealed the structural basis of IL-23 antagonism by the alphabody, MA12 (Figure 3).
Covalent linking of subunits of a protein complex was required to obtain the first crystal structure of a histone-modifying enzyme complex bound to a nucleosome core particle [12••]. Polycomb repressor complex (PRC) 1, an essential regulator of cell fate, comprises an activity to ubiquitylate nucleosomal histone H2A at residue K119. PRC1 uses its E3 ubiquitin ligase subunits, Ring1B and Bmi1, together with an E2 ubiquitin-conjugating enzyme, UbcH5c for this purpose. The E2–E3 complex was produced using an engineered single-chain fusion of Ring1B to UbcH5c to overcome the low affinity and salt-sensitive E2–E3 interaction. The resulting single-chain construct was co-expressed with Bmi1 to produce the trimeric complex which was purified and reconstituted with nucleosome core particles, and crystallized [12••]. The structure showed two copies of PRC1 E2–E3 complex bound to one nucleosome, revealing intricate interactions (Figure 3).
SRP pathway revealed by single-chain protein engineering
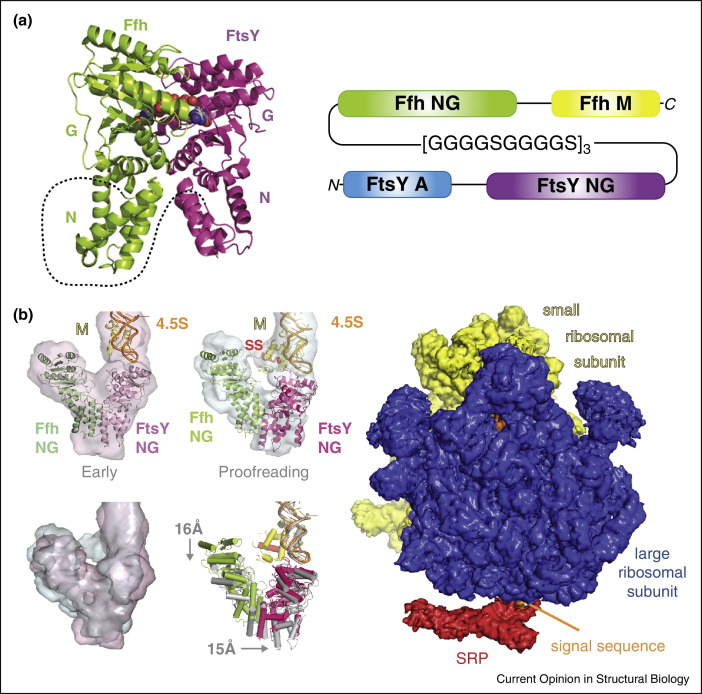
Around one third of the proteins in living cells are delivered to the plasma membrane. This is carried out by a universally conserved, complex mechanism involving ribosomes that are translating mRNA into membrane-bound nascent polypeptide chains, the signal recognition particle, SRP and the SRP receptor, FtsY [9, 40, 41]. Snapshots of this elaborate process were obtained by single particle electron cryo-microscopy and biochemical analysis [9, 10•, 11•, 42, 43, 44]. These studies revealed SRP binding to ribosome nascent chain complexes [42], followed by the ‘early’ [44], ‘proofreading’ [10•] and ‘closed’ [11•] states upon FtsY binding and GTP hydrolysis. Successful structure determination critically relied on stabilizing SRP binding to FtsY. This was achieved by covalently linking the SRP subunit Ffh with FtsY into a single polypeptide chain with virtually wild-type activity (Figure 4 ).
Figure 4.
SRP pathway revealed by single-chain protein engineering. (a)E. coli signal recognition particle (SRP) protein subunit Ffh (green) and the SRP receptor FtsY (purple) were covalently linked into a single polypeptide chain (left). NG domains are marked. The dashed line indicates the extended polypeptide linker. Bound nucleotide is shown (spheres). The single polypeptide construct is illustrated on the right, detailing the arrangement of FtsY A (blue) and NG (purple) domains, the 30 amino acid glycine/serine rich linker connecting FtsY with Ffh, and the Ffh NG (green) and M (yellow) domains. N and C-termini are marked. (b) Structures of SRP/FtsY bound to a translating ribosome were elucidated by electron cryo-microscopy (color coding as in panel a). 4.5S RNA is colored in orange. EM density and fitted models of the ‘early’ and ‘proofreading’ stages are shown (left, top). Substantial rearrangements are observed as illustrated by the overlay of the EM densities and the model coordinates (left, bottom) (adapted from [10•]). The cryo-EM structure of the ribosome-SRP-FtsY co-translational targeting complex in the closed state is shown on the right (adapted from [11•]).
Polyprotein single-molecule structural biology
Transient unfolding and refolding of proteins can be an essential feature of protein structure space in living organisms, for example in the translocation of proteins into and across cellular membranes or the muscle stroke. The use of atomic force microscopy (AFM) has emerged as a powerful technique to probe protein structure, enabling analysis of the mechanical stability and folding pathway of protein specimens at the single-molecule level. By means of a stretching force, applied through a microscopic cantilever to a biological target fixed to a support and recorded by the deflection of a laser beam, the analyzed protein is unfolded to an extended state (Figure 5 ).
Figure 5.
Polyprotein single-molecule structural biology. The setup of atomic force microscopy of polyproteins is shown in a schematic representation (adapted from [14]). The polyprotein is tethered to the gold support resting on the piezoelectronic positioning stage (bottom) on one end, and the tip of a cantilever made of silicon nitride on the other. A laser beam is focused on the back of the cantilever (top). The cantilever is displaced by the force that acts on the polyprotein chain resulting in change of the deflection of the laser beam, recorded by a photodetector. Increasing force causes each domain of the polyprotein to unfold, resulting in characteristic spikes in a force/extension diagram (inset).
Historically, polyproteins were used in single-molecule AFM to measure unique mechanical fingerprint profiles of a protein response to the applied mechanical force [14, 45•]. The muscle protein titin is a prominent example [46, 47, 48]. Titin consists of several hundred repeated protein domains including fibronectin and immunoglobulin-like folds. The use of polyproteins in single-molecule AFM results in considerable improvement of the statistical evaluation of singular domains within the polyprotein chain [14]. This can be exploited in the analysis of homomeric polyproteins that are constructed genetically or by chemical fusion reaction from identical copies of the same protein species of choice. Moreover, in addition to providing a clear fingerprint, polyproteins also have the advantage that a larger number of events can be recorded per experiment as compared to only one event if monomeric proteins are used.
For these reasons polyproteins emerge as work-horses of single-molecule structural biology by AFM. Numerous polyproteins have been analyzed for their mechanical properties by using this technique, including poly-I27, derived from the I-band region of Titin, oligo-calmodulin, poly-ubiquitin, polyproteins made of the virulence factor GB1 of Peptostreptococcus magnus [46, 47, 48, 49, 50, 51, 52, 53, 54] and others, providing unique insights into biological folding/unfolding mechanisms. The availability of a large and growing number of well characterized homomeric specimens furthermore enables now the construction of chimeric polyproteins as a tool to study mechanically uncharacterized proteins, by using the unique fingerprints of the known protein unit as a reference [14, 45•].
Conclusions
Natural and synthetic polyproteins are at the core of contemporary structural biology. Analysis of viral polyproteins and the architectural consequences of their processing during maturation not only furthers our understanding of viral mechanisms but provides important clues for drug design to combat viral diseases. Synthetic polyproteins are emerging as invaluable tools to accelerate research by unlocking hitherto inaccessible proteins for high resolution structure determination. Artificial polyproteins obtained by singe-chain protein engineering approaches are instrumental to overcome sample production bottlenecks and provide novel means to illuminate biological mechanisms, including folding and unfolding properties at the single-molecule level. We anticipate a major increase in the use of polyproteins in structural biology as valuable tools to tackle large and complex biological systems in the future.
Conflict of interest statement
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We thank all members of our laboratories for their assistance. We thank Timothy J. Richmond, Darren Hart, Christiane Schaffitzel, Alexander Pflug and Stefan Reich for their contributions and especially Rob Ruigrok for discussions and helpful suggestions. We are grateful to the Alliance Grenobloise pour la Biologie Structurale et Cellulaire Intégrées (GRAL) (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB) for generous support. Ben Luisi (Cambridge, UK) and John Briggs kindly provided the AcrABZ model coordinates and the HIV immature capsid image, respectively. TC, CS and AM are supported by the Centre National pour la Recherche Scientifique (CNRS), the Agence Nationale de Recherche (ANR) (contract nr. ANR-14-CE09-0017) and the European Commission (EC) Framework Program 7 (FP7) project FluPHARM (contract nr. 259751). CS acknowledges support from the Hofmann-La Roche pRED external collaboration program. IB, MC and FG are supported by the Agence Nationale de Recherche (ANR) through the Investissement d’Avenir program, and the EC through FP7 projects BioSTRUCT-X (contract nr. 283570) and ComplexINC (contract nr. 279039).
Contributor Information
Thibaut Crépin, Email: crepin@embl.fr.
Imre Berger, Email: iberger@embl.fr.
References
- 1.Al-Tawfiq J.A., Zumla A., Memish Z.A. Coronaviruses: severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus in travelers. Emerg Infect Dis. 2014;20:1562–1564. doi: 10.1097/QCO.0000000000000089. [DOI] [PubMed] [Google Scholar]
- 2.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas S., Nelson A.M. HIV and the spectrum of human disease. J Pathol. 2015;235:229–241. doi: 10.1002/path.4449. [DOI] [PubMed] [Google Scholar]
- 4.Vijayachandran L.S., Viola C., Garzoni F., Trowitzsch S., Bieniossek C., Chaillet M., Schaffitzel C., Busso D., Romier C., Poterszman A. Robots, pipelines, polyproteins: enabling multiprotein expression in prokaryotic and eukaryotic cells. J Struct Biol. 2011;175:198–208. doi: 10.1016/j.jsb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Nie Y., Bellon-Echeverria I., Trowitzsch S., Bieniossek C., Berger I. Multiprotein complex production in insect cells by using polyproteins. Methods Mol Biol. 2014;1091:131–141. doi: 10.1007/978-1-62703-691-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed description of the methodology and protocols for expressing protein complexes from synthetic polyproteins in the MultiBac baculovirus insect cell expression system.
- 6••.Reich S., Guilligay D., Pflug A., Malet H., Berger I., Crépin T., Hart D., Lunardi T., Nanao M., Ruigrok R.W., Cusack S. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]; First crystal structure of the influenza polymerase bound to cognate viral RNA substrate, providing unprecedented insights into cap-snatching and RNA synthesis. Influenza polymerase, inaccessible to structural analysis for decades since its discovery, was produced here for the first time successfully from a synthetic polyprotein construct.
- 7.Pflug A., Guilligay D., Reich S., Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 8••.Du D., Wang Z., James N.R., Voss J.E., Klimont E., Ohene-Agyei T., Venter H., Chiu W., Luisi B.F. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structure determination of the bacterial AcrABZ-TolC multidrug efflux pump by hybrid methods combining X-ray crystallography and electron cryo-microscopy. Elaborate single-chain protein engineering was a prerequisite for successful sample preparation of the complete efflux pump.
- 9.Knoops K., Schoehn G., Schaffitzel C. Cryo-electron microscopy of ribosomal complexes in cotranslational folding, targeting, and translocation. Wiley Interdiscip Rev RNA. 2012;3:429–441. doi: 10.1002/wrna.119. [DOI] [PubMed] [Google Scholar]
- 10•.von Loeffelholz O., Knoops K., Ariosa A., Zhang X., Karuppasamy M., Huard K., Schoehn G., Berger I., Shan S.O., Schaffitzel C. Structural basis of signal sequence surveillance and selection by the SRP-FtsY complex. Nat Struct Mol Biol. 2013;20:604–610. doi: 10.1038/nsmb.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structure of the ‘proof-reading’ step in co-translational targeting, revealed by electron cryo-microscopy and biochemical analysis of a reconstituted assembly comprising ribosome nascent chain complex, SRP and SRP receptor, FtsY. The SRP protein subunit Ffh and FtsY were provided as an engineered protein fusion conjoined by an extended glycine/serine rich linker. The resulting protein fusion displayed wild-type activity.
- 11•.von Loeffelholz O., Jiang Q., Ariosa A., Karuppasamy M., Huard K., Berger I., Shan S.O., Schaffitzel C. Ribosome-SRP-FtsY cotranslational targeting complex in the closed state. Proc Natl Acad Sci U S A. 2015;112:3943–3948. doi: 10.1073/pnas.1424453112. [DOI] [PMC free article] [PubMed] [Google Scholar]; High-resolution electron cryo-microscopy study revealing the structure and mechanism of the ‘closed’ step in bacterial co-translational targeting. The emerging signal sequence bound to the Ffh M-domain was observed in near-atomic detail.
- 12••.McGinty R.K., Henrici R.C., Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]; 3.0 Å crystal structure determination of the first histone-modifying enzyme complex, the PRC1 deubiquitylation module, bound to a nucleosome. Single-chain engineering was required to stabilize the enzyme complex for crystallization.
- 13.Desmet J., Verstraete K., Bloch Y., Lorent E., Wen Y., Devreese B., Vandenbroucke K., Loverix S., Hettmann T., Deroo S. Structural basis of IL-23 antagonism by an Alphabody protein scaffold. Nat Commun. 2014;5:5237. doi: 10.1038/ncomms6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann T., Dougan L. Single molecule force spectroscopy using polyproteins. Chem Soc Rev. 2012;41:4781–4796. doi: 10.1039/c2cs35033e. [DOI] [PubMed] [Google Scholar]
- 15.Shin G., Yost S.A., Miller M.T., Elrod E.J., Grakoui A., Marcotrigiano J. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci U S A. 2012;109:16534–16539. doi: 10.1073/pnas.1210418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yost S.A., Marcotrigiano J. Viral precursor polyproteins: keys of regulation from replication to maturation. Curr Opin Virol. 2013;3:137–142. doi: 10.1016/j.coviro.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostyuchenko V.A., Zhang Q., Tan J.L., Ng T.S., Lok S.M. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J Virol. 2013;87:7700–7707. doi: 10.1128/JVI.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller M. The early years of retroviral protease crystal structures. Biopolymers. 2010;94:521–529. doi: 10.1002/bip.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble C.G., Shi P.Y. Structural biology of dengue virus enzymes: towards rational design of therapeutics. Antiviral Res. 2012;96:115–126. doi: 10.1016/j.antiviral.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Konvalinka J., Kräusslich H.G., Müller B. Retroviral proteases and their roles in virion maturation. Virology. 2015 doi: 10.1016/j.virol.2015.03.021. pii: S0042-6822(15)00157-9. [DOI] [PubMed] [Google Scholar]
- 21.Bell N.M., Lever A.M. HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol. 2013;21:136–144. doi: 10.1016/j.tim.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Briggs J.A., Kräusslich H.G. The molecular architecture of HIV. J Mol Biol. 2011;410:491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 23.Sundquist W.I., Kräusslich H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bharat T.A., Davey N.E., Ulbrich P., Riches J.D., de Marco A., Rumlova M., Sachse C., Ruml T., Briggs J.A. Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy. Nature. 2012;487:385–389. doi: 10.1038/nature11169. [DOI] [PubMed] [Google Scholar]
- 25.de Marco A., Heuser A.M., Glass B., Kräusslich H.G., Müller B., Briggs J.A. Role of the SP2 domain and its proteolytic cleavage in HIV-1 structural maturation and infectivity. J Virol. 2012;86:13708–13716. doi: 10.1128/JVI.01704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bharat T.A., Castillo Menendez L.R., Hagen W.J., Lux V., Igonet S., Schorb M., Schur F.K., Kräusslich H.G., Briggs J.A. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc Natl Acad Sci U S A. 2014;111:8233–8238. doi: 10.1073/pnas.1401455111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Schur F.K., Hagen W.J., Rumlová M., Ruml T., Müller B., Kräusslich H.G., Briggs J.A. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature. 2015;517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]; The structure of the immature HIV-1 capsid in intact virus particles formed by HIV Gag precursor polyprotein was elucidated by electron cryo-tomography and sub-tomogram averaging methods, revealing unexpected structural plasticity.
- 28.Kennedy M.W. The polyprotein lipid binding proteins of nematodes. Biochim Biophys Acta. 2000;1476:149–164. doi: 10.1016/s0167-4838(99)00249-6. [DOI] [PubMed] [Google Scholar]
- 29.McAleer M.A., Irvine A.D. The multifunctional role of filaggrin in allergic skin disease. J Allergy Clin Immunol. 2013;131:280–291. doi: 10.1016/j.jaci.2012.12.668. [DOI] [PubMed] [Google Scholar]
- 30.Harding C.R., Aho S., Bosko C.A. Filaggrin — revisited. Int J Cosmet Sci. 2013;35:412–423. doi: 10.1111/ics.12049. [DOI] [PubMed] [Google Scholar]
- 31.Meenan N.A., Ball G., Bromek K., Uhrín D., Cooper A., Kennedy M.W., Smith B.O. Solution structure of a repeated unit of the ABA-1 nematode polyprotein allergen of Ascaris reveals a novel fold and two discrete lipid-binding sites. PLoS Negl Trop Dis. 2011;5:e1040. doi: 10.1371/journal.pntd.0001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith J.L., Skiniotis G., Sherman D.H. Architecture of the polyketide synthase module: surprises from electron cryo-microscopy. Curr Opin Struct Biol. 2015;31:9–19. doi: 10.1016/j.sbi.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bukhari H.S., Jakob R.P., Maier T. Evolutionary origins of the multienzyme architecture of giant fungal fatty acid synthase. Structure. 2014;22:1775–1785. doi: 10.1016/j.str.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 34.Tran T.H., Hsiao Y.S., Jo J., Chou C.Y., Dietrich L.E., Walz T., Tong L. Structure and function of a single-chain, multi-domain long-chain acyl-CoA carboxylase. Nature. 2015;518:120–124. doi: 10.1038/nature13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisfeld A.J., Neumann G., Kawaoka Y. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol. 2015;13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortín J., Martín-Benito J. The RNA synthesis machinery of negative-stranded RNA viruses. Virology. 2015 doi: 10.1016/j.virol.2015.03.018. pii: S0042-6822(15)00154-3. [DOI] [PubMed] [Google Scholar]
- 37.Bieniossek C., Imasaki T., Takagi Y., Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem Sci. 2012;37:49–57. doi: 10.1016/j.tibs.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorsen T.S., Matt R., Weis W.I., Kobilka B.K. Modified T4 lysozyme fusion proteins facilitate G protein-coupled receptor crystallogenesis. Structure. 2014;22:1657–1664. doi: 10.1016/j.str.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin J., Mobarec J.C., Kolb P., Rosenbaum D.M. Crystal structure of the human OX2 orexin receptor bound to the insomnia drug suvorexant. Nature. 2015;519:247–250. doi: 10.1038/nature14035. [DOI] [PubMed] [Google Scholar]
- 40.Saraogi I., Shan S.O. Co-translational protein targeting to the bacterial membrane. Biochim Biophys Acta. 2014;1843:1433–1441. doi: 10.1016/j.bbamcr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X., Shan S.O. Fidelity of cotranslational protein targeting by the signal recognition particle. Annu Rev Biophys. 2014;43:381–408. doi: 10.1146/annurev-biophys-051013-022653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaffitzel C., Oswald M., Berger I., Ishikawa T., Abrahams J.P., Koerten H.K., Koning R.I., Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X., Schaffitzel C., Ban N., Shan S.O. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci U S A. 2009;106:1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estrozi L.F., Boehringer D., Shan S.O., Ban N., Schaffitzel C. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat Struct Mol Biol. 2011;18:88–90. doi: 10.1038/nsmb.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Scholl Z.N., Li Q., Marszalek P.E. Single molecule mechanical manipulation for studying biological properties of proteins, DNA, and sugars. Wiley Interdiscip Rev Nanomed Biotechnol. 2014;6:211–229. doi: 10.1002/wnan.1253. [DOI] [PubMed] [Google Scholar]; Authoritative overview article presenting the state-of-the-art of single-molecule structure analysis of biological samples (proteins, nucleic acid, sugars) by using atomic force microscopy.
- 46.Krüger M., Linke W.A. The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J Biol Chem. 2011;286:9905–9912. doi: 10.1074/jbc.R110.173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bang M.L., Centner T., Fornoff F., Geach A.J., Gotthardt M., McNabb M., Witt C.C., Labeit D., Gregorio C.C., Granzier H., Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 48.Williams P.M., Fowler S.B., Best R.B., Toca-Herrera J.L., Scott K.A., Steward A., Clarke J. Hidden complexity in the mechanical properties of titin. Nature. 2003;422:446–449. doi: 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]
- 49.Valbuena A., Oroz J., Vera A.M., Gimeno A., Gómez-Herrero J., Carrión-Vázquez M. Quasi-simultaneous imaging/pulling analysis of single polyprotein molecules by atomic force microscopy. Rev Sci Instrum. 2007;78:113707. doi: 10.1063/1.2794732. [DOI] [PubMed] [Google Scholar]
- 50.Carrion-Vazquez M., Oberhauser A.F., Fisher T.E., Marszalek P.E., Li H., Fernandez J.M. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog Biophys Mol Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 51.Carrion-Vazquez M., Li H., Lu H., Marszalek P.E., Oberhauser A.F., Fernandez J.M. The mechanical stability of ubiquitin is linkage dependent. Nat Struct Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 52.He C., Genchev G.Z., Lu H., Li H. Mechanically untying a protein slipknot: multiple pathways revealed by force spectroscopy and steered molecular dynamics simulations. J Am Chem Soc. 2012;134:10428–10435. doi: 10.1021/ja3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao Y., Li H. Polyprotein of GB1 is an ideal artificial elastomeric protein. Nat Mater. 2007;6:109–114. doi: 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- 54.Li Y.D., Lamour G., Gsponer J., Zheng P., Li H. The molecular mechanism underlying mechanical anisotropy of the protein GB1. Biophys J. 2012;103:2361–2368. doi: 10.1016/j.bpj.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]