Abstract
Caused by porcine epidemic diarrhea virus (PEDV), porcine epidemic diarrhea (PED) is an acute infectious disease which causes damage to the intestine including intestinal villus atrophy and shedding, leading to serious economic losses to the pig industry worldwide. In order to obtain detailed information about the pathogenesis and host immune response in a PEDV-infected host for first In vivo study we used high-throughput sequencing to analyze the gene expression differences of the small intestinal mucosa after infection with PEDV. Transcripts obtained were over 65,525,000 clean reads after reassembly were 22,605 genes detected, of which 22,248 were known genes and 371 new genes were predicted. Moreover, 3168 genes expression was up-regulated and 3876 genes down-regulated. (Gene Ontology) GO annotation and functional enrichment analysis indicated that all of the DEGs (differentially expressed genes) were annotated into biological process, cellular component and molecular function. Most of these unigenes are annotated in cellular processes, the cell and binding. KEGG analysis of the DEGs showed that a total of 7044 DEGs unigenes were annotated into 323 pathways classified into 6 main categories. Most of these unigenes are annotated were related to immune system response to the infectious diseases pathways. In addition, 20 DEGs were verified by quantitative real-time PCR. As the first, in vivo, RNAseq analysis of piglets and PEDV infection, our study provides knowledge about the transcriptomics of intestinal mucosa in PEDV-infected piglets, from which a complex molecular pathways and pathogenesis-related biological processes are involved in PEDV interaction with piglet intestinal mucosa.
Keywords: PEDV, RNAseq, Piglet intestinal mucosa, Gene expression, Transcriptome analysis
Highlights
-
•
Within vivo RNAseq analysis, the mapping rate of PEDV infected piglets was 93.55%, 22,248 known genes and 371 novel genes.
-
•
3168 genes were up-regulated and 3876 genes were down-regulated compare to uninfected piglets.
-
•
Most of expressed genes were related to immune system followed by endocrine system.
-
•
TLRs expression were elevated in the PEDV infected piglets and induced subsequent downstream signaling pathways.
1. Introduction
Porcine epidemic diarrhea (PED) is a highly contagious acute infectious disease caused by porcine epidemic diarrhea virus (PEDV) which belongs to the genus Alphacoronavirus in the family Coronaviridae. It causes acute enteritis and lethal watery diarrhea in piglets resulting in dehydration and ultimately death. The main pathological changes in the intestinal villi is shrinkage and shedding [1]. PED is a major issue that has severely affected the pig industry in Asian countries in recent decades. Effective prevention and control of PED is still a worldwide problem at present [2].
Proteomic analysis by Zeng et al. [3] showed that many changes in protein expression were present in PEDV infected Vero cells. These proteins are involved in the regulation of apoptosis, signal transduction and the stress response. Moreover, Kim et al. [4] showed that the apoptosis induced by PEDV infection mainly activates the intracellular caspase-dependent mitochondrial apoptosis-inducing factor (AIF) signaling pathway. PEDV infection can also cause intracellular MAPK signaling including ERK (extracellular signaling-regulated kinase), p38 MAPK and JNK (c-Jun N-terminal kinase) activation [5,6]. Therefore, the proliferation of PEDV will stimulate a variety of signaling pathways, such as apoptosis [4], MAPK signaling pathway, endoplasmic reticulum stress response, making cells provide a microenvironment conducive to PEDV proliferation, and the induction of cellular pathological changes [6]. Most research on PED has focused on pathogen isolation and identification, genomic and structural protein analysis, diagnosis and vaccine development [7]; however, the detailed molecular mechanisms of viral pathogenesis remain unclear. The results of a virus infection are mainly determined by the interaction between the virus and the innate immune system [8]. Therefore, understanding how the virus interacts with the innate immune system of the host is essential to understand the molecular mechanisms of viral pathogenesis. In addition, PEDV infection not only causes ER stress and activates NF-κB [6,9]. Furthermore, it has been reported that stable expression of PEDV N and E proteins in porcine intestinal epithelial cells (IECs) can activate NF-κB and up-regulate the expression of IL-8 [10]. Ding et al. [11] reported that transient expression of PEDV N protein could inhibit NF-κB activation in HEK-293T cells. However, the exact mechanism that regulates NF-κB activity during PEDV infection remains to be elucidated.
In recent years, with the continuous development of high-throughput sequencing technology, high-throughput sequencing has been widely used by researchers studying crops, livestock and poultry diseases. The technique provides a powerful tool for finding candidate genes responsible for diseases. In the present study, BGISEQ-500 was used for RNA sequencing (RNAseq) analysis to compare gene expression differences of the small intestine mucosa after host infection with PEDV. Furthermore, aim of the study was to reveal the host-specific mechanisms associated with immune responses and the intestinal mucosa in piglets after PEDV infection.
2. Materials and methods
2.1. Ethics statement
The Animal Ethics Committee of Anhui Agricultural University approved the study (No. 20170517).
2.2. Piglets and virus infection
The healthy piglets used in the present study were procured from HeFei Huajie livestock and Poultry Breeding Co., Ltd., Anhui Province, China. The experimental piglets (3 days old) were housed individually in the laboratory animal room at a temperature of 28 °C and fed with milkiwean yoghurt (Nutreco N.V.) at the optimum concentration of 25% every 2–3 h at 45 °C during study. PEDV was isolated from the previously infected piglets in Anhui Province, and subsequently achieved in the laboratory. Piglets were divided into two groups (n = 6); the first group were infected with 10 mL of 106.62 copies/μL of PEDV using 9 mm French tube with syringe at the end. Second group was the controls (n = 4), which were fed with 10 mL of culture medium. After infection, we monitored the time of onset of diarrhea and other symptoms of PED and data were recorded. After 6 h of PED symptoms were detected, the pigs were euthanized, and the inner wall of the small intestine was flushed with sterile PBS solution and sections of the small intestinal mucosa were scraped with a sterile glass slide for RNA sequencing.
2.3. RNA extraction, cDNA library construction and sequencing
Total RNA was extracted from the intestinal tissues of the infected and healthy control piglets using TRlzol reagent (Invitrogen, USA) according to the manufacturer's instructions. We used NanoDrop™ to determine the purity of the RNA samples. Subsequently, we determined the total RNA sample QC to ensure selection of samples with: RNA 7.0 RIN value and 28S/18S > 1.8 using Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit). The total mRNA was enriched using magnetic beads containing Oligo dT that binds (QIAGEN, Germany) mRNA polyA tail and DNase followed by amplification using MagJET mRNA Enrichment Kit (Thermo Scientific, USA). Subsequently, Enriched mRNA subjected to reverse transcription using a random N6 primers to synthesize cDNA. Messenger RNA purification, fragmentation, construction of sequencing libraries and sequencing were performed using the Illumina Pipeline Sequencing by BGISEQ-500 Genome Sequencing Platform (BGI, China; http://www.seq50.com/en/).
2.4. RNAseq data analyses
First, we filtered the low quality reads which were more than 20% of the bases qualities are <10 using internal software SOAPnuke. Reads with adaptors and reads with unknown bases (N bases > 5%) to get the clean reads, starting with an average of 74,222,000 raw reads we obtained 65,525,000 clean reads. Clean reads quality metrics are shown in (Table 1 ). We used StringTie [12] to reconstruct transcripts, then we used Cuffmerge (Cufflinks tools [13]) to bring together the refactoring information for all our samples, and used Cuffcompare to compare reconstructed transcripts to reference annotation. Subsequently, we mapped clean reads to a reference using Bowtie 2 [14], followed by the gene expression level calculation using RSEM [15]. RSEM is a software package for estimating gene and isoform expression levels from RNA-Seq data. Moreover, we calculated the Pearson correlation between all samples, carried out hierarchical clustering between all samples using hclust (http://bioinformatics.org.au/tools/hclust/). Furthermore, we performed principal component analysis PCA analysis for all samples using Princomp and drew the diagrams using ggplot2 with d of R. To determine the number of novel genes and known genes, we calculated the gene numbers. We identified DEGs with DEseq2 by screening for differential genes with a fold change ≥2.00 and an adjusted P-value ≤ 0.05 was based on method described previously by Ref. [16].
Table 1.
Clean reads quality metrics.
| Sample | Toatal Raw Reads (Mb) | Total Clean Reads (Mb) | Total Clean Bases (Mb) | Clean Reads Q20 (%) | Clean Reads Q20 (%) | Clean Reads Ratio (%) |
|---|---|---|---|---|---|---|
| CPD1 | 74.58 | 65.66 | 6.57 | 96.70 | 87.91 | 88.04 |
| CPD2 | 72.12 | 65.65 | 6.57 | 97.18 | 89.16 | 91.03 |
| CPD3 | 74.55 | 65.99 | 6.60 | 96.81 | 88.19 | 88.52 |
| CPD4 | 74.59 | 66.26 | 6.63 | 96.81 | 88.21 | 88.82 |
| TPD1 | 74.58 | 65.50 | 6.55 | 97.03 | 88.91 | 87.83 |
| TPD2 | 72.08 | 65.16 | 6.52 | 97.24 | 89.52 | 90.40 |
| TPD3 | 68.50 | 60.96 | 6.10 | 96.82 | 88.33 | 88.99 |
| TPD4 | 74.61 | 67.25 | 6.73 | 97.18 | 89.35 | 90.14 |
| TPD5 | 77.04 | 65.93 | 6.59 | 96.85 | 88.44 | 85.58 |
| TPD6 | 79.57 | 66.89 | 6.69 | 96.83 | 88.52 | 84.07 |
To predict the major biological and molecular functions of these DGEs, we used Gene Ontology (GO) classification and functional description. GO has three aspects; molecular functions, cellular components and biological process (http://www.geneontology.org/). KEGG database one of the main public database that are sources for signaling pathways information. After using KEGG (Kyoto Encyclopedia of Genes and Genomes) NCBI BLAST tool for pig domestic gene annotation, we performed functional pathway analysis using phyper a function of R. The level of significance of all GO and KEGG terms was corrected by controlling the false discovery rate (FDR) of multiple paired comparisons, and the terms with significant values were targeted. P-value ≤ 0.05, FDR ≤0.01, |log2 Ratio| ≥ 1 gene were considered to be significant differentially expressed genes.
2.5. Genes expression analysis by qRT-PCR
To validate the accuracy of the DGE result, 20 of biologically relevant genes from the same RNA used for DGEs sequencing were randomly selected. Primer premier 5.0 was used to design the targeted genes primers (Supplementary Table 1) and the internal reference gene (GAPDH) was selected based on the published reference gene GAPDH (Genbank: 396823) [12].
3. Results
3.1. DEGs sequencing annotation
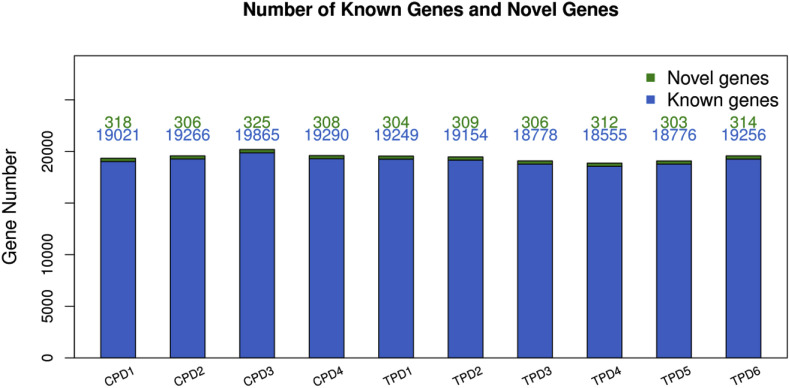
Our data analysis shown that the average genome mapping rate was 93.55% and the average gene mapping rate 75.76%. 22,605 genes were identified in which 22,248 were known genes and 371 were novel genes. 20,894 novel transcripts were identified in which 17,864 were previously unknown splicing event for known genes, 371 of them being novel coding transcripts without any known function and the remaining 2659 were identified as a long noncoding RNA. To show the number of novel genes and known genes, we calculated the gene quantity (Fig. 1 ). Overall, our transcriptomic analysis to the small intestines of PEDV infected piglets shown that the mapping rate was 93.55% and we identified 22,248 known genes and 371 novel genes.
Fig. 1.
Bar graph represents the number of novel genes and known genes in piglets group.
3.2. DEGs analyses reveals that most of expressed genes were related to immune response
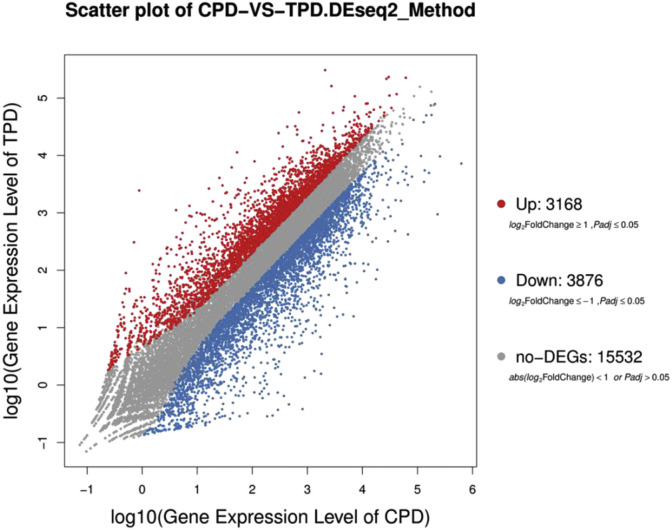
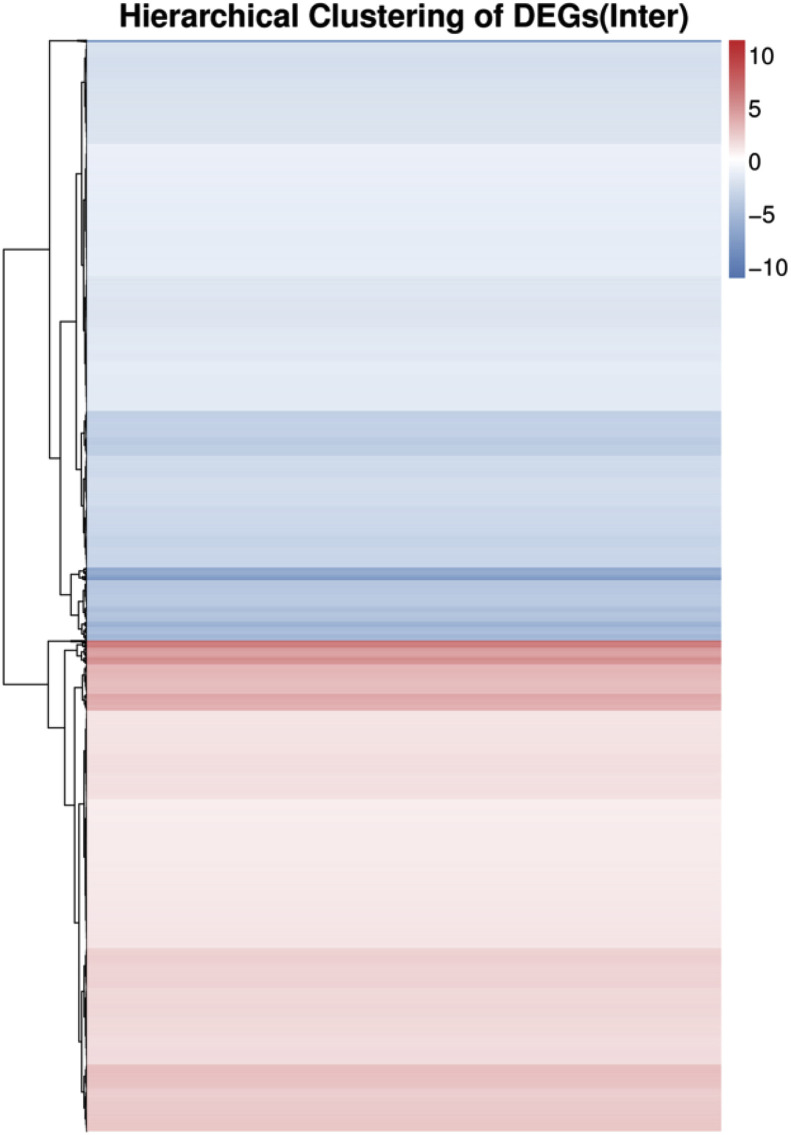
To identify the wide genes involved in response to PEDV infection in piglets. We analyzed the genes that responded to the PEDV infection. We observed that a total of 3168 genes were up-regulated and 3876 genes that were down-regulated in PEDV infected piglets as compared to uninfected piglets (Supplementary Table 2) and (Fig. 2 ). Similarly, to illustrate the differences between the infected and uninfected piglets we performed hierarchical clustering for DEGs and a heat-map was generated to visualize the quantitative differences in the expression levels of 7044 differentially expressed genes between the infected and uninfected groups (Fig. 3 ) and the results were in concordance with previous analysis.
Fig. 2.
Scatter-plot to show the distribution of DEGs. The X-Y axis represents log 10 transformed gene expression level, the red color represents up-regulated genes, the blue color down-regulated genes and the gray color non-DEGs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Quantitative differences in the expression levels of 7044 differentially expressed genes between the infected and Uninfected groups.
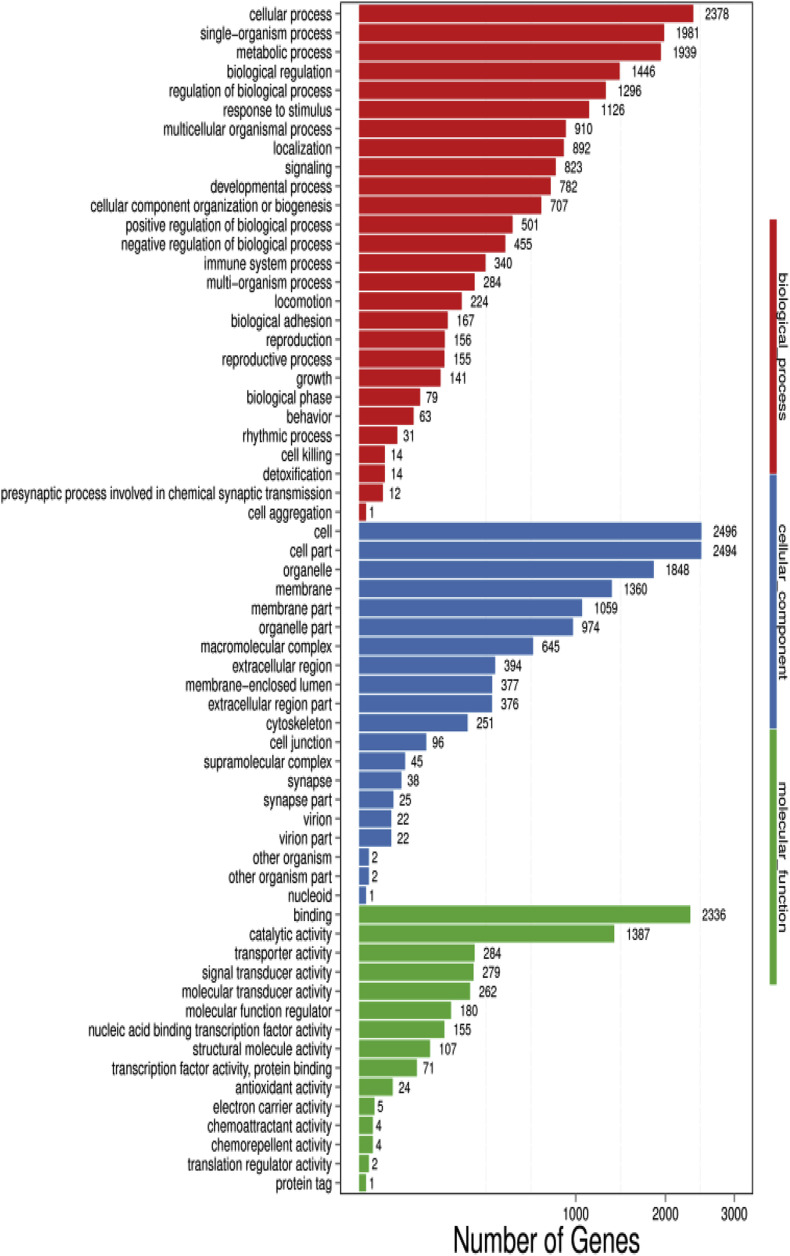
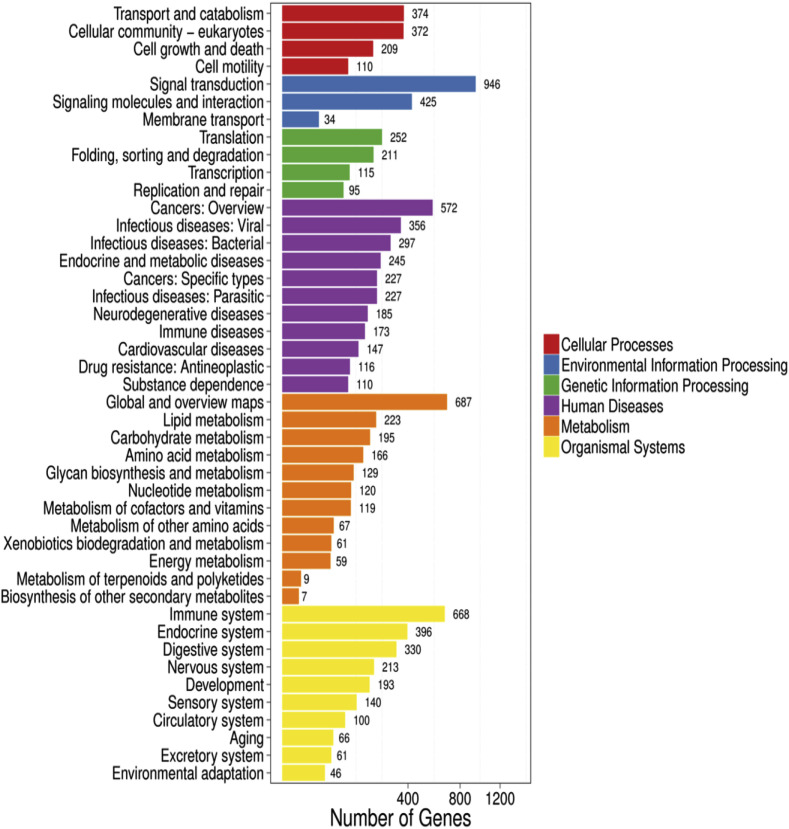
Furthermore, to classify and characterize DEGs functions and pathways, we performed a Gene Ontology (GO) classification and functional annotation of molecular biological function, cellular components and biological process. (Fig. 4 ). Most annotated genes in biological processes category were related to the cellular process with 2378 genes and mainly involved in the single-organism process and metabolic process while the least annotated genes were related to the cell aggregation. Moreover, the most annotated genes related to cellular components were involved in cells, cell part and organelles synthesis while the most annotated genes involved in molecular functions were related to binding catalytic activity and transducer activity and the least were related to translation regulator activity and protein tag. Moreover, we performed KEGG pathway classification and functional enrichment for the predicted DEGs (Fig. 5 ). We observed that the DGEs (7,044) were found in 323 of KEGG annotated results after PEDV infection in piglets. However, the organismal system annotated more unigenes (2,213), most of which annotated are related to the immune system (668 unigenes). The next order of the KEGG pathway annotation was the endocrine system, digestive system, nervous system and development, respectively.
Fig. 4.
Gene Ontology (GO) classification and functional annotation of molecular biological function, cellular components and biological process.
Fig. 5.
KEGG pathway classification and functional enrichment for the predicted DEGs.
According to the results of the KEGG annotation, a total of 946 unigenes were annotated in signal transduction pathway terms and in 29 categories. DEGs (668 unigenes) were most abundant in terms of the immune system and were annotated in 20 categories (Supplementary Table 3). All together, the wide genes expression in the PEDV infected piglets revealed that 3168 genes were up-regulated and 3876 genes that were down-regulated compare to uninfected piglets and the most of expressed genes were related to immune system followed by endocrine system.
3.3. Gene expression levels are consistent in both qRT-PCR and RNAseq
A total of 20 pairs of DEGs in the RNAseq results were further verified by qRT-PCR. Our results shown that the mRNA expression level of the 20 randomly selected genes were consistent with the expression of RNAseq (Table 2 ) which indicate that the RNAseq data was reliable. Furthermore, the relative mRNA expression of these genes was represented in the (supplementary figures). Overall, our qRT-PCR demonstrated that the RNAseq results were reliable.
Table 2.
Verification of different genes by qRT-PCR.
| Gene | GeneBank ID | RNAseq log2 Fold Change (TPD/CPD) | qRT-PCR (fold change) |
|---|---|---|---|
| FOSL1 | 100525205 | +7.515249739 | 26.51 |
| MAPK9 | 396609 | −1.59479924 | 0.20 |
| IL12B | 397076 | +4.638974482 | 6.49 |
| CXCL10 | 494019 | +5.430593329 | 26.99 |
| IL1B | 397122 | +5.345935977 | 32.95 |
| CD14 | 100037938 | +4.439269932 | 8.96 |
| PLA2G4E | 100514811 | −4.864839375 | 0.13 |
| IL1A | 397094 | +5.163024204 | 26.75 |
| CXCL11 | 100169744 | +7.344878866 | 71.43 |
| CXCL9 | 100135681 | +7.330083072 | 80.77 |
| SIGLEC1 | 397623 | +4.942243902 | 33.51 |
| OSM | 100152038 | +5.362375749 | 14.17 |
| SOCS3 | 493186 | +4.585475261 | 13.50 |
| CXCL2 | 414904 | +5.599152645 | 63.69 |
| GADD45B | 100621090 | +3.077752915 | 4.94 |
| IL1R1 | 100626904 | −3.716191676 | 0.06 |
| IL10 | 397106 | +2.539618277 | 4.78 |
| IL22 | 595104 | +6.096958733 | 14.74 |
| CXCL8 | 396880 | +2.993399022 | 6.71 |
| ISG15 | 100145895 | +4.233992314 | 1.48 |
| GAPDH | 396823 | 0.911362822 | 1 |
4. Discussion
PEDV infection still remain one of the most burden of swine vaccination and immunology. Moreover, PEDV has a strong tendency to infect a specific cell where it can efficiently replicate, particularly villous and intestinal epithelial cells [17]. Interaction between the host and PEDV is not completely deciphered and a comprehensive are needed to unravel the host-pathogen regulation and the PEDV target genes and the host immune response to the infection.
In our study, we used RNAseq technology to analyze the differential expression profiles of small intestinal mucosal genes of PEDV-infected piglets for the first time and verified the accuracy and reliability of transcriptome sequencing data by analyzing various genes mRNA relative expression. Furthermore, we analyzed the functions and pathways of the annotated genes. We observed that some of the genes involved in critical process of the intestinal mucosal immune response in PEDV-infected piglets were significantly up-regulated or down-regulated. Particularly, those genes which are involved in signal transduction and immune system-related signaling pathways were upregulated. In the fact, innate immune response is the first defense line against pathogens which recognizes the pathogens associated molecular pattern (PAMP) [18].
Moreover, Toll-like receptors (TLRs) recognizes the pathogen's PAMPs, the TIR region in the cytoplasm binds downstream linker molecules, triggering a series of signals that activate transcription factors such as NF-κB, AP-1 and IRF-3 and these transcription factors induce inflammatory changes by the production of cytokines and chemokines, thereby inhibiting the entry of pathogens. Cao et al. [19], carried out gene silencing of the TLR family genes and found that PEDV-infected cells with TLR2, TLR3 and TLR9 silenced genes can significantly inhibit the expression of NF-κB and prevent nuclear translocation of NF-κB. Recent studies have shown that TLR4 is also involved in recognizing the envelope proteins and fusion proteins of certain viruses, but the interaction with these ligand structures is unclear. In the present study, the host target organ of PEDV infection significantly up-regulated TLR4. Our data suggest that the expression of TLRs was elevated in the PEDV infected piglets and it induced a subsequent downstream signaling pathways such as NF-κB signaling pathway.
Furthermore, we analyzed the interactions between pathways that are involved in NF-κB signaling such as TNF which can activate the NF-κB signaling pathway may be activated by tumor necrosis factor (TNF) and subsequently, up-regulation of IL-6, CXCL2, IL-1β, IL-8 and another cytokine expression. NF-κB is a transcription factor which regulates the expression of various genes, which are involved in innate and adaptive immunity responses and cell survival [20,21]. NF-κB regulates the transcription of many factors that involve in the immune system stimulation including a variety of proinflammatory cytokines, chemokines, adhesion molecules and induce expression of enzymes and cell proliferation [22]. Reports have shown that Bcl-2 expression is also by NF-κB [23]. As an anti-apoptotic protein, Bcl-xl/Bcl-2l1 protein can directly inhibit channel activity by binding to VDAC on the mitochondrial membrane to stabilize it, inhibit cytochrome C release and block activation of the caspase family of proteins, and thus achieve the purpose of anti-apoptotic action; it can also inhibit Fas and Fas-mediated apoptosis [24]. The results of this study are consistent with the above findings wherein we demonstrated that PEDV infects the host and activated the NF-κB signaling pathway and subsequently, up-regulated the expression of Bcl-2L1 pathway which play a role in resisting apoptosis after PEDV infection.
In our study, interferon (IFN) response was significantly high and is considered to be the first line of defense against all vertebrate-borne viral infections [25,26]. Interferon activates NK and dendritic cells (DCs) to activate the adaptive immune system [27]. Moreover, interferons exerts its antiviral activity by inducing the expression of IFN-inducible antiviral effectors [28], such as IGS15 and MX1. Among them IGS15 inhibits virus replication by enhancing the activity of the JAK/STAT1 signaling pathway [29] or directly modifying the viral proteins [30]. Due to the wide distribution of IFNGR1/2 most cell types respond to IFN-γ, thereby IFN-γ produces pleiotropic effects on immune cells and limited direct antiviral activity [31]. In this study, the high expression of IFN-γobserved in the intestinal mucosa of piglets infected with PEDV may indicate that IFN-γplays an important defensive role against PEDV infection, this is consistent with above finding. Hence, our data suggest that infected piglets have shown to produce antiviral responses through multiple pathways.
Consequently, Interleukin (IL) members were found to be up-regulated to varying degrees, including IL-1A, IL-1B, IL-6, IL-8, IL-10, IL-12B, IL-18, IL-22 and, IL-27. IL-1 (α/β) is mainly produced by macrophages and all nucleated cells which plays a role in transmitting activation signals. When cells are stimulated by IL-1, PI3 forms a complex with IL-1 and IL-1R1 to regulate the inflammatory response through intracellular signal transduction and activation of the NF-κB pathway. The results of our study showed that the increased expression of IL-1 (α/β) might inhibit the expression of IL-1R1 by activating certain pathways, this observation makes it more important to study the role of PEDV infection in modulation of immune system in piglets.
IL-8, also known as CXCL8, is a proinflammatory cytokine chemokine, which plays an important role in promoting cell-distress signaling and antagonizing the antiviral activity of interferon. Luppi et al. [32] and Waugh et al. [33] found that IL-8 plays a vital role in promoting cell survival signaling. Our data, shown that the expression level of IL-8 was up-regulated after PEDV infection of the host which may be beneficial for leukocyte accumulation during the inflammatory process triggered by PEDV infection. Xue et al. [34] found that recombinant mature pIL-22 (mpIL-22) inhibited alpha coronavirus replication such as PEDV, transmissible gastroenteritis virus (TGEV) and porcine rotavirus (PoRV). In our study, the expression of IL-22 in porcine intestinal mucosa after PEDV infection was increased by 6.1 times compared with the mock infection. Thus, when PEDV infects piglets, the body's immune system expresses a large quantity of IL-22 against the virus.
IL-18 is a known cytokine that improves T cell proliferation and enhances NK and CTL activity [46–49] and induces IFN production [35]. Foss et al. [36] showed that IL-18 might induce immune response against pathogens in porcine mucosa. Therefore, IL-18 up-regulation in infected piglets is likely to be very important in defense against PEDV in piglets.
5. Conclusions
This is the first report of host target tissue gene expression analysis after PEDV-infection by high-throughput RNA-sequencing. Results of this study elucidates the transcriptome of intestinal mucosa in PEDV-infected piglets. Our observations suggest that PEDV infection in piglets induces a strong immune response, particularly activates the antiviral defense mechanisms through the expression of immune genes and the changes of the signaling pathways were identified. Results of this study will further enhance the understanding of PEDV infection in piglets and will guide in devising clinical intervention strategies against PEDV infections.
Authors' contributions
PS, QF and YL performed experiments. PS, QF YL, YZ, YF, XX, GZ, YJ, QX, YL and ZSH analyzed the data. QF and PS conceived experiments. QF, PS, and YL wrote the manuscript. FQ and MAQ interpreted, discussed, edited the draft and final versions of the manuscript. All authors have read and approved the manuscript.
Acknowledgements
This research was supported by Anhui Province Natural Science Fund Project (Grant No.1708085MC83), State Key Laboratory of Veterinary Etiological Biology (Grant No. SKLVEB2016KFKT003), and Anhui Pig Industry Technology System.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micpath.2019.04.033.
Contributor Information
Pei Sun, Email: sunpei1979@126.com.
Qarih Fahd, Email: 1524858261@qq.com.
Yezhen Li, Email: 1050292686@qq.com.
Yao Sun, Email: 282765724@qq.com.
Jie Li, Email: 837367153@qq.com.
Majjid A. Qaria, Email: majidqaria@gmail.com.
Zhan Song He, Email: songhezhan@163.com.
Yuzhen Fan, Email: 1552869864@qq.com.
Qiang Zhang, Email: 13685518887@126.com.
Qianming Xu, Email: xuqianming2006@163.com.
Zongjun Yin, Email: yinzongjun@ahau.edu.cn.
Xingang Xu, Email: tiger2003@nwsuaf.edu.cn.
Yu Li, Email: liyu@ahau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figures: Histogram to verify differential gene expression during PEDV infection. The selected 20 DEGs were validated by the quantitative real-time PCR. Fold changes were normalized against the GAPDH.
Supplementary tables: Tables of the primers used for qRT-PCR, List of differential expression of genes. This file provides the list of part of genes identified in the differential gene expression analysis between PEDV infected and uninfected piglets. File contains, gene ID, gene ontology names, logFC (Fold Change) and P-values. Also table of predicted signaling pathways of immune response and cell transduction.
References
- 1.Pensaert M.B., De Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng S., Zhang H., Ding Z., Luo R., An K., Liu L. Proteome analysis of porcine epidemic diarrhea virus (PEDV)‐infected Vero cells. Proteomics. 2015;15:1819–1828. doi: 10.1002/pmic.201400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim Y., Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. 2014;460:180–193. doi: 10.1016/j.virol.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y., Lee C. Extracellular signal-regulated kinase (ERK) activation is required for porcine epidemic diarrhea virus replication. Virology. 2015;484:181–193. doi: 10.1016/j.virol.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X., Zhang H., Zhang Q., Dong J., Liang Y., Huang Y. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Gene. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frieman M., Heise M., Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133:101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L. Porcine epidemic diarrhea virus (PEDV) nucleocapsid protein antagonizes IFN-β production by sequestering the interaction between IRF3 and TBK1. J. Virol. August 2014;88(16):8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cinar M.U., Islam M.A., Pröll M., Kocamis H., Tholen E., Tesfaye D. Evaluation of suitable reference genes for gene expression studies in porcine PBMCs in response to LPS and LTA. BMC Res. Notes. 2013;6:56. doi: 10.1186/1756-0500-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reguera J., Mudgal G., Santiago C., Casasnovas J.M. A structural view of coronavirus–receptor interactions. Virus Res. 2014;194:3–15. doi: 10.1016/j.virusres.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H., Liu Q., Su W., Wang J., Sun Y., Zhang J. Genome-wide analysis of differentially expressed genes and the modulation of PEDV infection in Vero E6 cells. Microb. Pathog. 2018;117 doi: 10.1016/j.micpath.2018.02.004. S0882401017314584-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mogensen T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao L., Ge X., Gao Y., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus infection induces NF-κB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 2015;96:1757–1767. doi: 10.1099/vir.0.000133. [DOI] [PubMed] [Google Scholar]
- 20.Geng H., Wittwer T., Dittrich‐Breiholz O., Kracht M., Schmitz M.L. Phosphorylation of NF‐κB p65 at Ser 468 controls its COMMD1‐dependent ubiquitination and target gene‐specific proteasomal elimination. EMBO Rep. 2009;10:381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wietek C., O'Neill L.A.J. Diversity and regulation in the NF-κB system. Trends Biochem. Sci. 2007;32:311–319. doi: 10.1016/j.tibs.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Moynagh P.N. 2005. The NF-kappaB Pathway. [DOI] [PubMed] [Google Scholar]
- 23.Fahy B.N., Schlieman M.G., Mortenson M.M., Virudachalam S., Bold R.J. Targeting BCL-2 overexpression in various human malignancies through NF-κB inhibition by the proteasome inhibitor bortezomib. Cancer Chemother. Pharmacol. 2005;56:46–54. doi: 10.1007/s00280-004-0944-5. [DOI] [PubMed] [Google Scholar]
- 24.Donovan M., Cotter T.G. Control of mitochondrial integrity by Bcl-2 family members and caspase-independent cell death. Biochim. Biophys. Acta Mol. Cell Res. 2004;1644:133–147. doi: 10.1016/j.bbamcr.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Robertsen B. The interferon system of teleost fish. Fish Shellfish Immunol. 2006;20:172–191. doi: 10.1016/j.fsi.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Zou J., Secombes C.J. Teleost fish interferons and their role in immunity. Dev. Comp. Immunol. 2011;35:1376–1387. doi: 10.1016/j.dci.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Longhi M.P., Trumpfheller C., Idoyaga J., Caskey M., Matos I., Kluger C. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 2009;206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao C., Denison C., Huibregtse J.M., Gygi S., Krug R.M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. Unit. States Am. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao C., Hsiang T.-Y., Kuo R.-L., Krug R.M. ISG15 conjugation system targets the viral NS1 protein in influenza A virus–infected cells. Proc. Natl. Acad. Sci. Unit. States Am. 2010;107:2253–2258. doi: 10.1073/pnas.0909144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valente G., Ozmen L., Novelli F., Geuna M., Palestro G., Forni G. Distribution of interferon‐γ receptor in human tissues. Eur. J. Immunol. 1992;22:2403–2412. doi: 10.1002/eji.1830220933. [DOI] [PubMed] [Google Scholar]
- 32.Luppi F., Longo A.M., De Boer W.I., Rabe K.F., Hiemstra P.S. Interleukin-8 stimulates cell proliferation in non-small cell lung cancer through epidermal growth factor receptor transactivation. Lung Canc. 2007;56:25–33. doi: 10.1016/j.lungcan.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Waugh D.J.J., Wilson C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 2008;14:6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 34.Xue M., Zhao J., Ying L., Fu F., Li L., Ma Y. IL-22 suppresses the infection of porcine enteric coronaviruses and rotavirus by activating STAT3 signal pathway. Antivir. Res. 2017;142:68–75. doi: 10.1016/j.antiviral.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muneta Y., Mori Y., Shimoji Y., Yokomizo Y. Porcine interleukin 18: cloning, characterization of the cDNA and expression with the baculovirus system. Cytokine. 2000;12:566–572. doi: 10.1006/cyto.1999.0648. [DOI] [PubMed] [Google Scholar]
- 36.Foss D.L., Zilliox M.J., Murtaugh M.P. Bacterially induced activation of interleukin-18 in porcine intestinal mucosa. Vet. Immunol. Immunopathol. 2001;78:263–277. doi: 10.1016/s0165-2427(00)00266-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures: Histogram to verify differential gene expression during PEDV infection. The selected 20 DEGs were validated by the quantitative real-time PCR. Fold changes were normalized against the GAPDH.
Supplementary tables: Tables of the primers used for qRT-PCR, List of differential expression of genes. This file provides the list of part of genes identified in the differential gene expression analysis between PEDV infected and uninfected piglets. File contains, gene ID, gene ontology names, logFC (Fold Change) and P-values. Also table of predicted signaling pathways of immune response and cell transduction.