Abstract
In this investigation we report on the influence of volumetric flow rate, flow velocity, complementary DNA concentration, height of a microfluidic flow channel and time on DNA hybridization kinetics. A syringe pump was used to drive Cy3-labeled target DNA through a polydimethylsiloxane (PDMS) microfluidic flow channel to hybridize with immobilized DNA from the West Nile Virus. We demonstrate that a reduction of channel height, while keeping a fixed volumetric flow rate or a fixed flow velocity, enhances mass transport of target DNA to the capture probes. Compared to a passive hybridization, the DNA hybridization in the microfluidic flow channel generates higher fluorescence intensities for lower concentration of target DNA during the same fixed period of time. Within a fixed 2 min time period the fastest DNA hybridization at a 50 pM concentration of target DNA is achieved with a continuous flow of target DNA at the highest flow rate and the lowest channel height.
Keywords: DNA hybridization kinetics, Microfluidic flow channel, PDMS, Pressure-driven flow
1. Introduction
Since the completion of the human genome mapping, interest has shifted to the study of the molecular functions of genes and how they might lead to a diseased state [1]. Consequently, DNA chips have rapidly been applied to fields such as gene identification, genetic expression analysis, DNA sequencing and clinical diagnostics [2]. The detection and analysis of point mutations and single nucleotide polymorphisms (SNP) is one of the main applications of DNA micro-arrays [3]. The majority of DNA micro-array technologies, including the Gene Chip of Affymetrix Inc., are based on passive hybridization, i.e. the binding event depends upon diffusion of target DNA molecules to the capture probes [4]. A passive DNA hybridization approach may take several hours, since target DNAs with a typical diffusion coefficient of 9.943 × 10−7 cm2/s (for 18 base pair oligonucleotides) [5] reach the capture probes only via Brownian random motion in order to hybridize. Because of this dependency on diffusion transport of target DNA, large amounts of target DNA and long hybridization times are often required to achieve sufficient hybridization signals and repeatability of signals. Accordingly, a large number of researchers are seeking alternative methods to make DNA sensing faster and more sensitive in low concentration target DNA solutions [6]. One of the earlier alternative methods was to utilize electric fields to accelerate the rate of interaction between capture probes and target DNA molecules. Nanogen, for example, has overcome the slow diffusion obstacle by transporting negatively charged target DNA via electrophoresis [7]. In diffusion based transport of DNA, the time (τ) it takes a DNA molecule to travel over a distance x is given by [1]:
| (1) |
In Eq. (1), D is the diffusion coefficient and τ is the time required for a molecule to diffuse over distance x. In the case of electrophoresis the transport time, τ, is given by [8]:
| (2) |
where μ is the electrophoretic mobility, E the electric field strength and τ is the time it takes to electrophorese over distance x. Using typical values for D (9.943 × 10−7 cm2/s, see above) and μ (15,000 μm2(V s)) [6], and under an electric field of 0.004 V/μm it appears that electrophoretic transport may be 150 times faster over a 500 μm distance. Even though the electrophoretic transport method presents a clear hybridization speed advantage, one critical disadvantage is that the sample solution containing the target DNA must be desalted before hybridization in order to establish an appropriate electric field. In a high salt solution, including most biological buffers, the electric field depth exists only in close proximity to the electrodes because the high concentration of ions nullifies the electric field in the area away from the electrodes. To facilitate rapid movement of DNA by an electric field, a low conductive buffer solution has to be used. In contrast, in molecular biology, to achieve efficient hybridization, one always works in high conductivity solutions. Therefore, desalting is not only counter intuitive, but also cumbersome. Another approach to enhance hybridization efficiency by Fan et al. consisted of using pumping of probe molecules dynamically past target molecules immobilized on magnetically suspended beads [9]. Others have accelerated the hybridization rate through convective mixing of hybridization fluid by using air bladders [10], or by cavitation-induced streaming between the DNA array slide and its cover slip [11]. Also, Pappaert et al. have developed a shear-driven micro-channel system to improve rapid DNA micro-array analysis [12] while Erickson et al. have enhanced the hybridization rate electrokinetically in the micro-channel [13]. Finally, Wang et al. combined a DNA micro-array with a microfluidic platform to enhance the mass transport of target DNA so as to make detection faster and to obtain a lower detection limit (LDL) for DNA hybridization. There are several advantages associated with microfluidic DNA arrays: they enable detection at the lowest possible DNA concentrations, allow for shorter times to achieve such sensitive detection because of enhanced mass transport, offer the ability to monitor several samples in parallel by using a multi-channel approach, reduce the potential for contamination by relying on an enclosed apparatus [4], and they hold the overall promise for integration of several functions in one apparatus (μ-TAS). In this paper, we investigate several factors that are important to DNA hybridization kinetics in PDMS microfluidic flow channels and demonstrate how to enhance the amount of hybridization in a fixed and short amount of time, typically 2 min. We also describe the effect of various volumetric flow rates of Cy3-labeled target DNA on hybridization with a fixed volume of the target DNA, the influence of the concentration of target DNA with fixed channel height and fixed volumetric flow rate, the contribution of the channel height in a fixed volumetric flow rate regime, and lastly the influence of the volumetric flow rate in a fixed amount of time.
2. Theoretical section
2.1. Target DNA transport in the bulk of the solution
In the microfluidic channel, the fluid flow is laminar with a Reynolds number, Re = U m H/ν, well below 2000. In the expression for Re, U m is the fluid mean velocity, H the channel height and ν is the fluid kinematic viscosity. The transport of target DNA molecules in a moving fluid is modeled as convective diffusion of species governed by two mechanisms: molecular diffusion of the target DNA molecules and the convection of target DNA in the direction of the fluid flow. For this type of transport, the Peclet number, Pe, a measure of the relative rate of convective to diffusive transport, is given as:
| (3) |
For hybridization in a flow channel as described here, with a very small DNA coefficient of diffusion, the Pe number is often greater than 1, as such, the transport of DNA molecules is dominated by convection. The immobilized DNA capture probes are located at the bottom of the flow channel. Upon reaction with those probes a diffusion-limited reaction is established when the rate of mass transport to the reactive surface is significantly slower than the rate of reaction with the DNA probes. In such case, as soon as the complementary target DNA molecules reach the reactive surface, they undergo a heterogeneous hybridization reaction resulting in a zero concentration of that target DNA in the sample directly adjacent to that surface. As a consequence of this local depletion of the target DNA, a thin diffusion layer, δ c, with a concentration gradient forms near the reactive surface. For a laminar fluid flow between two parallel plates of infinite width (applicable for a wide microfluidic channel with , where is the channel width), with a fully developed velocity profile and with the reactive surface located on the lower channel wall, the thickness of the diffusion layer can be estimated from the following Eq. [14]:
| (4) |
where x is the distance from the reactive surface edge. Hence, for the convective diffusion case, the diffusion layer thickness is proportional to the cubic root of the channel height (H) and inversely proportional to cubic root of the mean velocity (U m). This equation can be used to model the DNA hybridization in a microfluidic channel, the thinner the diffusion layer, the higher the depletion and the faster the hybridization.
The mean velocity is determined by either the fluid flow rate (Q), the channel width and the channel height (H), or the pressure drop over the flow channel length, Δp, the channel length (L) and the dynamic viscosity (μ). The mean velocity can be expressed as:
| (5) |
2.2. Reaction kinetics of the DNA hybridization reaction
Target DNA from the sample immediately above the capture probe elements may hybridize with the immobilized DNA directly or may first be adsorbed onto the solid surface followed by 2D diffusion over the surface and hybridization. Taking into account only the direct DNA hybridization, the DNA heterogeneous hybridization reaction can be elucidated by considering the following chemical reaction [15]:
| (6) |
The symbol ssDNAImmob represents single stranded DNA molecules (DNA capture probes) immobilized on the solid surface that are available for hybridization. We model immobilized DNA molecules that are grouped/patterned and form capture probe elements set in a linear array. The target DNA molecules in the sample above the capture probes (represented by ssDNABulk) bind specifically to the DNA capture probes and form hybridized double stranded DNA molecules (represented by dsDNAHybridized) on the surface of the capture elements. The hybridization reaction rate is governed by the forward reaction rate constant, k F. While the disassociation reaction rate is governed by the reverse reaction rate constant, k R. A schematic illustration of the hybridization channel with the immobilized DNA capture probe elements is presented in Fig. 1 .
Fig. 1.
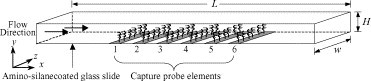
Schematic of the microfluidic flow channel system. The Corona Virus DNAs are immobilized on amino-silane coated glass slide forming capture probe elements 1 and 2, while the West Nile Virus DNAs are immobilized on the same slide forming capture probe elements 3–6. The microfluidic channel dimensions are: 15,000 μm long, 500 μm wide and either 50, 18 or 8 μm high.
The rate of the hybridization reaction is a function of all the concentrations of all species present in the overall chemical reaction (Eq. (6)) at any given time [16]. This rate can be represented by the rate law as [17]:
| (7) |
In Eq. (7), the square brackets represent the concentrations of the DNA molecules. The term [ssDNAImmob] is somewhat different yet, it represents the difference between the initial concentration of the capture probes [ssDNAImmob, initial], before hybridization, and the concentration of hybridized DNA at any given time t [dsDNAHybridized], which corresponds to the surface concentration of available probes.
3. Experimental
3.1. Microfluidic channel preparation
Polydimethylsiloxane (PDMS) microfluidic flow channels were fabricated using soft lithography as described by Duffy et al. [18]. SU-8/25 photoresist was used to fabricate the master mold. The resulting flow channels have a width of 500 μm, a length of 15 mm and are either 8, 18 or 50 μm high (see Fig. 2 ). After fabrication of the SU-8/25 master mold, the PDMS precursor and curing agent (Dow Corning, USA) were mixed thoroughly in a weight ratio of 10:1 and the mixture was degassed in a vacuum for 20 min. The mixture was then poured out on the master mold and cured in a 65 °C oven for 50 min. Finally, the PDMS microfluidic channel was peeled off from the SU-8/25 master mold. The PDMS structure was then put on top of a glass slide spotted with DNA reaction sites (see further below) to form a flow channel. To avoid non-selective binding a blocking scheme was carried out on the surface of the glass slides (see below), before they were covered with the PDMS microfluidic channels. As shown in Fig. 3 , our experimental set-up consists of three main parts: a syringe pump, an injector valve and a microfluidic flow channel. The syringe pump (Harvard Apparatus, USA) is the driving force that moves fluid into the micro-channel. We choose the injector valve (C22 Valco, USA) based on its ability to hold and inject small amounts of sample volume without generating air bubbles. The injector valve is equipped with either 6, 24, 42 or 60 μL of liquid injection loops (see the black arrow in Fig. 3) that can hold a mixture of target DNA and hybridization buffer solution. The inlet of the microfluidic channel is connected to the injector valve, which is in turn connected to the syringe pump by a fluorinated ethylene propylene tube. The outlet of the micro-channel is open to air. After hybridization, the PDMS microfluidic channel is detached from the glass slide. The glass slide is then washed first in 50 mL of wash buffer, followed by a wash with ultra-pure deionized water, injected into a centrifugation holding tube, and is finally dried by centrifugation.
Fig. 2.
Picture of the PDMS master molding whose microfluidic flow channels were fabricated using SU-8/25. Each channel was 15,000 μm in length, 500 μm in width and either 50, 18 or 8 μm in height.
Fig. 3.
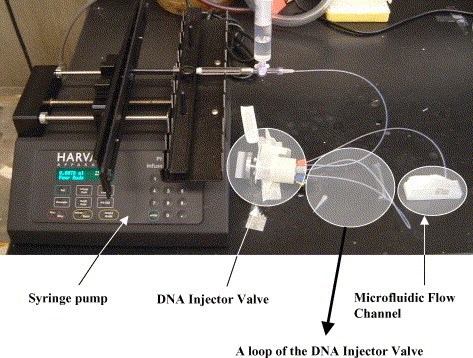
Experimental set-up.
3.2. DNA probe immobilization
The amino-silane coated glass slides, 25 mm × 75 mm, were purchased from VWR Inc. (USA). All oligoneucliotides, including West Nile Virus DNA, Corona Virus DNA and Cy3-labeled target DNA, complementary to West Nile Virus DNA, were purchased from Biosource, Inc. The sequences for the West Nile Virus DNA and Corona Virus DNA capture probes and the Cy3-labeled DNA target probe, are detailed in Table 1 . The composition of the spotting buffer solution is 50% (v/v) dimethyl sulfoxide and 50% (v/v) TE buffer (10 mM Tris–HCl, pH 7.4, 1 mM EDTA). The concentration of DNA capture probe in this mixture is 5 μM. As shown in Fig. 1, West Nile Virus DNA was immobilized in a linear array, along the axis of the flow channel on the amino-silane coated glass slide (probe elements 3–6). The Corona Virus DNA was immobilized next to the West Nile Virus DNA spots as two non-specific sites (probe elements 1 and 2). The DNA array spots are 1 mm apart and each spot size is 500 μm × 500 μm. After the slide was blotted dry, it was exposed to 100 mJ of UV light (UV Stratalinker 1800, USA) to enhance the binding between the phosphate groups of the capture probe DNA backbone and the amino-silane coated slide with its free amine groups [19], [20], [21].
Table 1.
Capture probe and target probe sequences used for hybridization experiments
| Name | Sequences |
|---|---|
| West Nile Virus DNA | 5′-ACTCAGGAGG AGGTGTCGAG GGCTT |
| GGGCC TCCAAAAACT GGGTTACATC CTGCGTGAAG TTGGCACCCG-3′ | |
| Cy3-labeled target DNA | 3′-CCCGG AGGTTTTTGA CCCAATGTAG-5′-Cy3 |
| Corona Virus DNA | 5′-GATAATATGTT AAAGAACCTG ATGGCCGATG TTGATGATCC TAAATTGATGG-3′ |
3.3. Blocking scheme
A blocking scheme is implemented to deactivate the unused surface area of the glass slides in order to prevent undesirable non-specific binding of target probe molecules on the slide. In our procedure, we deposited 500 μL of blocking buffer solution (1% bovine serum albumin (BSA), 5× saline sodium citrate (SSC) and 0.1% sodium dodecyl sulfate (SDS)), and then covered the entire surface with cover slips (1 cm2 surface area each) to help the blocking buffer solution to spread evenly on the surface as well as to prevent the evaporation of the blocking buffer solution. The slide was then incubated in an oven at 37 °C for 15 min, washed with ultra-pure deionized water, and dried by centrifugation (Sorvall RT7, USA).
3.4. DNA hybridization
3.4.1. DNA hybridization in a microfluidic flow channel
What follows is a description of a typical assay sequence. A 2.5 mL glass luer head syringe (Hamilton, USA) is filled with a wash buffer solution—0.2× SSC and 0.1% SDS and is pumped into the microfluidic channel. Meanwhile, Cy3-labeled target DNA in 1× hybridization buffer solution (50% formamide, 10% SSC and 0.2% SDS) is injected into a loop of the injector valve. When the injector valve switch is turned on, the wash buffer solution pushes the target-DNA in the hybridization buffer solution through the chosen loop and into the microfluidic channel without generating bubbles.
3.4.2. Passive DNA hybridization
For the passive hybridization reference experiments a solution that consists of 0.5 μL of Cy3-labeled target DNA and 6 μL of 1× hybridization buffer—50% formamide, 10% SSC and 0.2% SDS was dropped onto the DNA capture probe elements. The DNA array was then covered with glass cover slips and incubated for 6 min at 37 °C in an oven and washed, first with wash buffer—0.2× SSC and 0.1% SDS and then with ultra-pure deionized water before detection. The temperature for passive hybridization was maintained at 37 °C as this gives the best signal to noise ratio for the short oligonucleotides we used.
3.5. Detection
After peeling the PDMS microfluidic chip from the glass slide and washing, the slide was scanned using the ScanArray Express (Perkin-Elmer, USA) incorporating a choice of different lasers, filters, a barcode reader and an autoloader. QuantArray Imaging Software was used for image capture and analysis. We selected a green HeNe 543.5 nm excitation laser for Cy3 measurement. The value for the laser power and photomultiplier tube (PMT) gain of the ScanArray Express was set to 90 and 80, respectively. The pixel resolution was 10 μm and the scan rate 50% of full speed for better signal to noise ratio. For the measurement, the average intensity of each spot was analyzed.
4. Results and discussion
4.1. Influence on DNA hybridization of volumetric flow rate with fixed target DNA volume
We injected buffered Cy3-labeled target DNA in the microfluidic flow channel, using the following volumetric flow rates: 1, 4, 7 and 10 μL/min. The total volume of Cy3-labeled target DNA was kept constant at 6 μL. The height of the channel was 50 μm and the concentration of the target DNA 50 pM. As shown in Fig. 4 , a slow flow rate of 1 μL/min leads to the highest fluorescence intensity, while the intensities for flow rates of 7 μL/min and up level off at a lower value. At first this result might seem surprising, since based on Eq. (4), the concentration gradient (Δc/Δy) at the surface increases as the flow rate increases resulting in a reduction in diffusion layer thickness (δ) from 16 μm for a flow rate of 1 μL/min to 7.4 μm for a flow rate of 10 μL/min. Thus, the diffusion flux of target DNA to capturing DNA should increase. However, the increase of volumetric flow rate results in an increase in the Peclet number. For example, the Peclet number with a flow rate of 1 μL/min and a channel height of 50 μm is 337, while the Peclet number for 10 μL/min with a 50 μm height is 3370. Thus, convective transport becomes relatively more dominant than molecular diffusion transport when the flow rate increases; therefore, performing the experiment with a fixed volume, the sample is pumped through the hybridization channel in a shorter amount of time. As a result, a large number of target DNA molecules will pass over the capture probes instead of diffusing into them. From Fig. 4a, we observe that there is no binding of the Cy3-labeled target DNA on the non-specific Corona Virus DNA sites. This result is expected, as bases of Cy3-labeled target DNA are supposed to pair only with bases of West Nile Virus, since they are the complementary base pairs. For Corona Virus DNA, the bases are not complementary to the bases of Cy3-labeled target DNA. Based on this result, we can assume there is no non-specific binding in the microfluidic flow channel.
Fig. 4.

(a) Different flow rates at 1, 4, 7 and 10 μL/min at room temperature while keeping Cy3-labeled target DNA volume constant at 6 μL. The concentration of target DNA is 50 pM and the height of microfluidic flow channel is 50 μm. Fluorescence signals in the first row correspond to 1 μL/min, 4 μL/min in the second row, 7 μL/min in the third row and 10 μL/min in last row. Note that there is no indication of binding at the Corona Virus DNA sites, the control or inside the ellipses. (b) Fluorescence intensity vs. a flow rate of 50 pM of target DNA in the hybridization solution. The number of experiments done (n) is equal to 6.
4.2. Influence of concentration of target DNA on DNA hybridization
4.2.1. Microfluidic flow channel hybridization
In this experiment, we fixed the volumetric flow rate at 1 μL/min, the channel height at 50 μm and the total volume at 6 μL. The Cy3-labeled target DNA concentration varied from 50 pM to 50 nM. From Fig. 5 , it is clear that the intensity of the fluorescence increases with increasing concentration of target DNA up to 50 nM and reaches close to plateau. This result is in agreement with the rate law, presented earlier in Eq. (7), in which the binding rate increases with increasing target concentration in the sample until the system reaches equilibrium where the hybridization and dissociation reactions reach a steady state. Also as the target concentration increases, there are a larger number of target molecules near the capture probe element available for hybridisation; thus, a larger number will participate in the heterogeneous hybridization reaction. No fluorescence signal could be picked up on the Corona Virus DNA sites (figure is not shown).
Fig. 5.
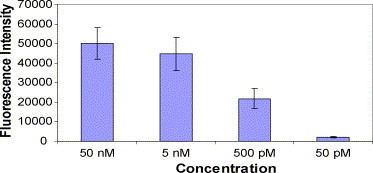
Fluorescence intensity vs. concentration of Cy3 target DNA varying from 50 nM to 50 pM in the microfluidic flow channel hybridization. The flow rate was 1 μL/min, the channel height was 50 μm, the sample volume was 6 μL and the temperature was room temperature. The number of experiments done (n) is equal to 6.
4.2.2. Passive hybridization
The passive hybridization control experiments were carried out under the same conditions as the microfluidic flow channel hybridization tests. The concentration was varied from 50 nM to 50 pM, and the incubation time was 6 min at 37 °C. As shown in Fig. 6 , the fluorescence intensity for passive hybridization follows the same trend as that of the microfluidic flow hybridization channel, which increases with increasing concentration of the Cy3-labeled target DNA. No fluorescence signal could be picked up on the Corona Virus DNA sites (figure is not shown).
Fig. 6.
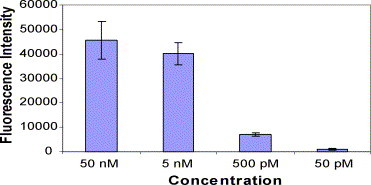
Fluorescence intensity vs. concentration of Cy3 target DNA varying from 50 nM to 50 pM in the passive hybridization. The sample volume was 6 μL, temperature was 37 °C and the incubation time was 6 min. The number of experiments done (n) is equal to 6.
4.2.3. Comparison between hybridization in a flow channel and passive hybridization
In Fig. 7 , we compare the fluorescence intensities in passive hybridization with those of hybridization in a flow channel at various concentrations of Cy3-labeled DNA target probe. One clearly observes from Fig. 7 that the microfluidic flow channel tests produce higher hybridization intensity signals throughout the whole concentration range compared to those in passive hybridization. However, the intensity differences at lower concentrations are significantly higher than those at higher concentrations. The microfluidic flow channel generates fluorescence intensities three times as high as those for passive hybridization at lower concentrations, while at higher concentrations, there is not much difference between the two cases. This is explained by the fact that at higher concentrations the hybridization becomes more reaction rate limited and is less controlled by either diffusive or convective transport of target DNA. While at lower concentrations, the transport of the lower number of molecules to the capture probe is diffusion controlled and convection enhances the hybridization rate compared to the passive case where transport occurs solely by diffusion. Therefore, the higher the target concentration the lower the difference between the passive and flow through hybridization signals intensities.
Fig. 7.
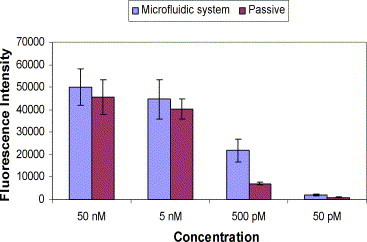
Comparison between passive hybridization and microfluidic hybridization. The number of experiments done (n) is equal to 6.
4.3. Influence on DNA hybridization of channel height in the case of fixed volumetric flow rate (Q) and fixed velocity (Um)
The heights of the channel we used in these experiments were 8, 18 and 50 μm. The concentration of Cy3-labeled target DNA was held constant at 50 pM. Cy3-labeled target DNA was delivered to each microfluidic flow channel in a constant volume of 6 μL. In Fig. 8 , we demonstrate that at a fixed volumetric flow rate of 1 μL/min, the fluorescence intensity increases as the channel height becomes smaller. This might seem to contradict our previous finding that increase in flow rates results in a decrease in hybridization signal. However, even though the flow velocity increases as we decrease the height of the channel, the Peclet number still holds at around 335 in all three channels. In other words, the molecular diffusive transport of DNA molecules in smaller microfluidic flow channels plays a more significant role in contrast to our previous finding regarding the influence of volumetric flow rate in a fixed target volume. Therefore, a decrease of total diffusion distance, by reducing channel height for target DNA molecules, enhances the hybridization signal. When using a fixed velocity of 0.67 mm/s, the fluorescence intensity is much higher than in the case of fixed volumetric flow rate as the channel height decreases (see Fig. 8). This is expected because the Peclet numbers with a constant flow velocity in 18 and 8 μm height are 121 and 54, respectively, which is much lower than 350 seen with a fixed volumetric flow rate. Thus, molecular diffusion plays the dominant role in this flow regime. However, to reach a fixed velocity for a lower height channel, 18 and 8 μm, we used 0.36 and 0.16 μL/min, respectively, so that the hybridization detection time for each channel increased from 6 to 16.7 min and 37.5 min.
Fig. 8.
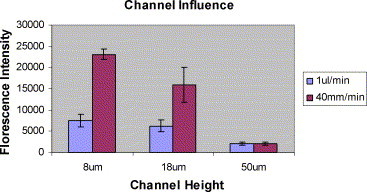
Influence of channel height on DNA hybridization in constant volumetric flow rate, 1 μL/min, and a constant velocity 40 mm/min. The number of experiments done (n) is equal to 6.
4.4. Influence of various volumetric flow rates in a fixed amount of time
Assuming there is enough target DNA, we may use a continuous flow regime to optimize the DNA hybridization speed and lower detection limit. We also investigated the hybridization speed for a fixed amount of time (hopefully as short as possible) at various flow rates. To study the influence of various flow rates at a fixed time of 2 min, hybridization of 10, 7, 4 and 1 μL/min were compared (see Fig. 9 a). The experiments were carried out at a fixed concentration of 50 pM of Cy3-labeled target DNA. The results show that the West Nile Virus DNA capture probes have a greater chance to hybridize with Cy3-labeled target DNA at a higher flow rate (10 μL/min versus 1 μL/min), and at a lower height (18 μm versus 50 μm) at a duration of 2 min (see Fig. 9b). The reason can be attributed to a larger number of target DNA molecules being delivered into the microfluidic flow channel at larger flow rates when time is held constant. Secondly, the total diffusion distance of target DNA molecules to capture probes is decreased when the channel height is reduced. We summarize all the results above in Table 2 .
Fig. 9.
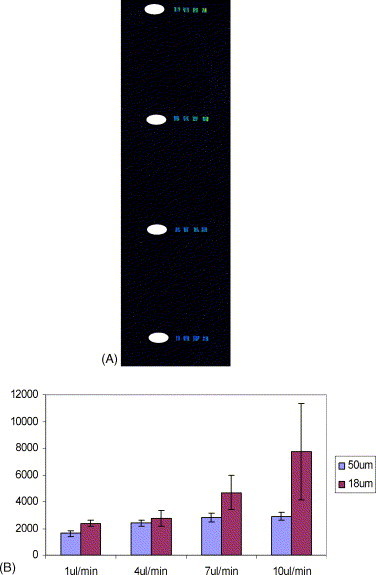
(a) Continuous flow rates at 1, 4, 7 and 10 μL/min at room temperature while keeping time constant at 2 min period. The concentration of target DNA is 50 pM and the height of microfluidic flow channel is 50 μm. Fluorescence signals in the first row correspond to 10 μL/min, 7 μL/min in the second row, 4 μL/min in the third row and 1 μL/min in last row. Note that there is no indication of binding at the Corona Virus DNA sites, the control or inside the ellipses. (b) Fluorescence intensity vs. continuous flow at different heights for 2 min. Flow rate of continuous flow were 1, 4, 7 and 10 μL/min. The concentration of target DNA was 50 pM. The number of experiments done (n) is equal to 6.
Table 2.
Overall results from the experiments
| Flow rate (Q) | Target concentration | Channel height | Target volume | Run-time | Hybridization intensity |
|---|---|---|---|---|---|
| ↑ | Constant | Constant | Constant | ↓ | ↓ |
| Constant | ↑ | Constant | Constant | Constant | ↑ |
| Constant | Constant | ↓ | Constant | Constant | ↑ |
| ↑ | Constant | ↓ | ↑ | Constant | ↑ |
5. Conclusion
Our experimental focus was the exploration of DNA hybridization kinetics by manipulating essential parameters such as volumetric flow rate (Q), target DNA concentration, height of flow channel, flow mean velocity (U m) and the amount of hybridization time in the microfluidic flow channel. While it is possible to improve the intensity of the hybridization signal in a microfluidic flow channel by hybridizing for a long time, this is not always practical or convenient. What is desirable is to obtain a high intensity signal in the shortest amount of time for any given target concentration. To increase the intensity for a low fixed target DNA concentration, it is possible to reduce the height of channel with slower volumetric flow rates or with fixed pressure so that the target DNA has more time to diffuse toward the capture probes. However, this results in a slower assay. For faster and more sensitive hybridization detection, continuous flow of target DNA with a higher flow rate and with a lower channel height enhances the hybridization signal and significantly reduces the detection time.
Acknowledgement
We would like to thank Dr. J. Tilles, at University of California, Irvine Medical Center, for use of their facility and resources and to thank Mr. Tsung-Hsi Hsieh at University of California, Irvine, for his assist of making PDMS mold.
References
- 1.Madou M.J. second ed. CRC Press; Boca Raton, FL: 2002. Fundamentals of Microfabrication. [Google Scholar]
- 2.Sheehan A.D., Quinn J., Daly S., Dillon P., O’Kennedy R. The development of novel miniaturized immuno-sensing devices: a review of a small technology with a large future. Anal. Lett. 2003;36(3):511–537. [Google Scholar]
- 3.Heller M.J. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Vaidya B., Rarquar H.D., Stryjewske W., Hammer R., McCarley R., Cheng Y., Barany F., Soper S.A. Microarrays assembled in microfluidic chips fabricated from poly(methyl methacrylate) for the detection of low-abundant DNA mutations. Anal. Chem. 2003;75:1130–1140. doi: 10.1021/ac020683w. [DOI] [PubMed] [Google Scholar]
- 5.Nkodo A.E., Garnier J.M., Tinland B., Ren H., Desruisseaux C., McCormick L.C., Drouin G., Slater G.W. Diffusion coefficient of DNA molecules during free solution electrophoresis. Electrophoresis. 2001;22:2424–2432. doi: 10.1002/1522-2683(200107)22:12<2424::AID-ELPS2424>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 6.Kassegne S.K., Reese H., Hodko D., Yang J.M., Sarkar K., Smolko D., Swanson P., Raymond D.E., Heller M.J., Madou M.J. Numerical modeling of transport and accumulation of DNA on electronically active biochips. Sens. Actuators B. 2003;94:81–98. [Google Scholar]
- 7.Heller M.J., Forster A.H., Tu E. Active microelectronic chip devices which utilize controlled electrophoretic fields for multiplex DNA hybridization and other genomic applications. Electrophoresis. 2000;21(1):157–164. doi: 10.1002/(SICI)1522-2683(20000101)21:1<157::AID-ELPS157>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 8.Eisenberg D., Crothers D. The Benjamin/Cummings Publishing Company, Inc.; Menlo Park, CA: 1979. Physical Chemistry with Applications to the Life Sciences. [Google Scholar]
- 9.Fan Z.H., Mangru S., Granzow R., Heaney P., Ho W., Hong Z., Kumar R. Dynamic DNA hybridization on a chip using paramagnetic beads. Anal. Chem. 1999;71:4851–48598. doi: 10.1021/ac9902190. [DOI] [PubMed] [Google Scholar]
- 10.Adey N.B., Lei M., Howard M.T., Jensen J.D., Mayo D.A., Butel D.L., Coffin S.C., Moyer T.C., Slade D.E., Spute M.K., Hancock A.M., Eisenhoffer G.T., Dalley B.K., McNeely M.R. Gains in sensitivity with a device that mixes microarray hybridization solution in a 25-μm-thick chamber. Anal. Chem. 2002;74:6413–6417. doi: 10.1021/ac026082m. [DOI] [PubMed] [Google Scholar]
- 11.Liu R.H., Lenigk R., Druyor-Sanchez R.L., Yang J., Grodzinski P. Acoustic micromixer for enhancement of DNA biochip systems. Anal. Chem. 2003;75:1911–1917. doi: 10.1021/ac026267t. [DOI] [PubMed] [Google Scholar]
- 12.Pappaert K., Vanderhoeven J., Van Hummelen P., Dutta B., Clicq D., Baron G.V., Desmet G. Enhancement of DNA micro-array analysis using a shear-driven micro-channel flow system. J. Chromatogr. 2003;A 1014:1–9. doi: 10.1016/s0021-9673(03)00715-5. [DOI] [PubMed] [Google Scholar]
- 13.Erickson D., Liu X., Krull U., Li D. Electrokinetically controlled DNA hybridization microfluidic chip enabling rapid target analysis. Anal. Chem. 2004;76:7269–7277. doi: 10.1021/ac049396d. [DOI] [PubMed] [Google Scholar]
- 14.Lavich V.G. Prentice-Hall; Englewood Cliff, NJ: 1962. Physicochemical Hydrodynamics. [Google Scholar]
- 15.Okahata Y., Kawase M., Niikura K., Ohtake F., Furusawa H., Ebara Y. Kinetic measurements of DNA hybridisation an an oligonucleotide-immobilized 27 MHz quartz crystal microbalance. Anal. Chem. 1998;70(7):1288–1296. doi: 10.1021/ac970584w. [DOI] [PubMed] [Google Scholar]
- 16.Atkins P.W. sixth ed. Oxford University Press; Oxford/Melbourne/Tokyo: 1998. Physical Chemistry. [Google Scholar]
- 17.Erickson D., Li D.Q., Krull U.J. Modeling of DNA hybridization kinetics for spatially resolved biochips. Anal. Biochem. 2003;317(2):186–200. doi: 10.1016/s0003-2697(03)00090-3. [DOI] [PubMed] [Google Scholar]
- 18.Duffy D.C., Gills H.L., Lin J., Sheppard N.F., Kellogg G.J. Microfabricated centrifugal microfluidic systems: characterization and multiple enzymatic assays. Anal. Chem. 1999;71(20):4669–4678. [Google Scholar]
- 19.Franssen-van Hal N.L.W., Vorst S., Kramer E., Hall R.D., Keijer J. Factors influencing cDNA microarray hybridization on silylated glass slides. Anal. Biochem. 2002;308:5–17. doi: 10.1016/s0003-2697(02)00203-8. [DOI] [PubMed] [Google Scholar]
- 20.Hegde P., Qi R., Abernathy K., Gay C., Dharap S., Gaspard R., Hughes J.E., Snesrud E., Lee N., Quackenbush J. A concise guide to cDNA microarray analysis. BioTechniques. 2000;29:548–562. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- 21.Diehl F., Beckmann B., Kellner N., Hauser N.C., Diehl S., Hoheisel J.D. Manufacturing DNA microarrays from unpurified PCR products. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/gnf078. e38. [DOI] [PMC free article] [PubMed] [Google Scholar]


