Abstract
Overlapping genes represent an intriguing puzzle, as they encode two proteins whose ability to evolve is constrained by each other. Overlapping genes can undergo “symmetric evolution” (similar selection pressures on the two proteins) or “asymmetric evolution” (significantly different selection pressures on the two proteins). By sequence analysis of 75 pairs of homologous viral overlapping genes, I evaluated their accordance with one or the other model. Analysis of nucleotide and amino acid sequences revealed that half of overlaps undergo asymmetric evolution, as the protein from one frame shows a number of substitutions significantly higher than that of the protein from the other frame. Interestingly, the most variable protein (often known to interact with the host proteins) appeared to be encoded by the de novo frame in all cases examined. These findings suggest that overlapping genes, besides to increase the coding ability of viruses, are also a source of selective protein adaptation.
Keywords: Ancestral frame, De novo frame, Homologs, Non-synonymous nucleotide substitution, Selection pressure, Synonymous nucleotide substitution, Virus adaptation
Highlights
-
•
A dataset of 80 pairs of homologous overlapping genes from viruses is examined.
-
•
Its analysis reveals that half of overlapping genes undergo asymmetric evolution.
-
•
The most variable gene product is that encoded by the de novo overlapping gene.
-
•
Overlapping genes evolving asymmetrically are a source of selective protein adaptation.
1. Introduction
Many viruses produce novel genes inside pre-existing genes by overprinting of a de novo frame onto an ancestral frame (Atkins et al., 1979; Keese and Gibbs, 1992; Rancurel et al., 2009; Sabath et al., 2012). The high prevalence of overlapping genes in viruses has been attributed to the advantage of maximizing the gene information content of small viral genomes (Miyata and Yasunaga, 1978; Lamb and Orvath, 1991; Pavesi et al., 1997).
In detail, the gene-compression hypothesis states that the size of the viral capsid imposes a biophysical limit on the size of the viral genome, thus making overprinting the most adequate strategy to gain new function (Chirico et al., 2010). In alternative, the gene novelty hypothesis argues that the birth of overlapping genes is driven by selection pressures favoring evolutionary innovation (Brandes and Linial, 2016). This hypothesis is supported by the finding that overlaps, thought for a long time to be restricted to viruses, also occur in the large genomes of prokaryotic (Delaye et al., 2008; Fellner et al., 2015) and eukaryotic organisms (Szklarczyk et al., 2007; Bergeron et al., 2013; Vanderperre et al., 2013).
A particularly interesting feature of overlapping genes is that they represent an intriguing example of adaptive conflict. Indeed, they simultaneously encode two proteins whose freedom to change is constrained by each other (Sander and Schulz, 1979; Krakauer, 2000; Peleg et al., 2004; Allison et al., 2016), which would be expected to reduce the adaptive ability of the virus (Simon-Loriere et al., 2013).
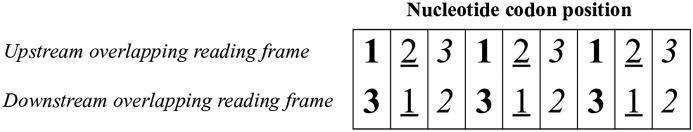
We would expect, in principle, that overlapping genes are subjected to strong evolutionary constraints, as a single nucleotide substitution can impair two proteins (see the codon position “21” in Fig. 1 ). A typical example of “constrained evolution” is that occurring in Hepatitis B virus (HBV), whose short genome (3.2 kb) contains a high percentage (50%) of overlapping coding regions (Mizokami et al., 1997; Zhang et al., 2010).
Fig. 1.
Orientation of overlapping genes, with the downstream frame having a shift of one nucleotide 3′ with respect to the upstream frame. There are 3 types of codon position (cp): cp13 (bold character), in which the first position of the upstream frame overlaps the third position of the downstream frame; cp21 (underlined character), in which the second position of the upstream frame overlaps the first position of the downstream frame; cp32 (italic character), in which the third position of the upstream frame overlaps the second position of the downstream frame. Based on the genetic code, a nucleotide substitution at first codon position causes an amino acid change in 95.4% of cases, at second codon position in 100% of cases, and at third codon position in 28.4% of cases. Thus, nucleotide substitutions at the codon positions “13” and “32” are usually non-synonymous in one frame and synonymous in the other. Nucleotide substitutions at the codon position “21” are almost all non-synonymous in both frames.
However, overlapping genes can also show a less conservative pattern of change, because of a high rate of non-synonymous substitutions in one frame (positive adaptive selection) with concurrent dominance of synonymous substitutions in the other (negative purifying selection). Examples of positive selection concern the overlapping genes that encode the tat and vpr proteins of simian immunodeficiency virus (Hughes et al., 2001), the p19 and p22 proteins of the tombusvirus family of plant viruses (Allison et al., 2016), and the ORF2 and ORF5 proteins of trichodysplasia spinulosa-associated polyomavirus (Kazem et al., 2016).
We can hypothesize for overlapping genes a first evolutionary model in which the two proteins they encode are subjected to similar selection pressures. When selection is strong both proteins (or protein regions) are highly conserved (e.g. the RNase domain of polymerase and the amino-terminal half of the X protein in HBV; see Fig. 4 in Mizokami et al., 1997). When selection is not too strong both proteins can vary considerably (e.g. the spacer domain of polymerase and the preS1/S2 domain of the surface protein in HBV; see Fig. 4 in Mizokami et al., 1997). This model is named “symmetric evolution”, because the number of amino acid substitutions of one protein is expected to be not significantly different from that of the other. It corresponds to the “shared model” by Fernandes et al. (2016).
In alternative, we can hypothesize for overlapping genes an evolutionary model in which the two proteins they encode are subjected to significantly different selection pressures. Support for this model, which implies adaptive selection on one frame and purifying selection on the other, was provided both by viral (Hughes et al., 2001; Fujii et al., 2001; Guyader and Ducray, 2002; Stamenković et al., 2016) and mammalian overlapping genes (Szklarczyk et al., 2007). This model is named “asymmetric evolution”, because the number of amino acid substitutions of one protein is expected to be significantly different from that of the other. It corresponds to the “segregated model” by Fernandes et al. (2016).
We recently assembled a dataset of 80 viral overlapping genes whose expression is experimentally proven (Pavesi et al., 2018), with the aim to provide a useful benchmark for systematic studies. A first analysis of the dataset revealed that overlapping genes differ significantly from non-overlapping genes in their nucleotide and amino acid composition (Pavesi et al., 2018). We also found that the vast majority of the 80 overlaps of the dataset have one or more homologs, suggesting further comparative studies.
In the present study, I investigated the evolution of viral overlapping genes by sequence analysis of 75 pairs of homologs. The first aim of the study was to determine which of the two evolutionary models described above is the prevailing one. The second aim was to identify the type of nucleotide substitution that significantly affects the pattern of symmetric/asymmetric evolution. Finally, the third aim was to assess whether the most variable protein (in the case of asymmetric evolution) is that encoded by the ancestral or the de novo frame.
2. Materials and methods
2.1. Selection criteria for homologous overlapping genes
I first extracted from the dataset of 80 overlapping genes experimentally proven (S1 Dataset from Pavesi et al., 2018) the amino acid sequence of the two proteins encoded by each overlap. For each protein, I searched for homologs against the non-redundant protein sequences NCBI database using BLASTP (Altschul et al., 1997). When BLASTP did not detect any homolog I used TBLASTN, which compared the protein query sequence against the nucleotide collection NCBI database translated in all reading frames. I used TBLASTN because the amino acid sequence of the protein encoded by one of the two overlapping frames (usually that discovered more recently) may not be reported in many viral genomes present in the NCBI database (Pavesi et al., 2018).
The selection of homologous overlapping genes was based on three criteria. The first was an equal length of the homolog. It was met in the great majority of cases (72 out of 80). In the remaining cases, the homolog was only slightly shorter than the query sequence. The exception was the overlap capsid protein/Assembly Activating Protein (AAP) of adeno-associated virus-2, whose homolog encodes an AAP 9 amino acids shorter in the amino-terminal region and 26 amino acids shorter in the carboxy-terminal region.
The second criterion was a homolog yielding, for both the encoded proteins, an alignment with no insertion/deletion (indel) or with a minimal number of indels. In the latter case, I imposed the rule that indel(s) must be located at the same amino acid position in the alignments of the two pairs of proteins (see for example the overlap polymerase/2b protein of Spinach latent virus, which is the first overlap in Supplementary File S1). By imposing this rule, I could align the two homologous nucleotide sequences in full accordance with the corresponding protein sequences. The alignment of protein sequences was carried out with Clustal Omega (Sievers and Higgins, 2014).
The third criterion concerned the cases in which I found multiple homologs meeting the two criteria described above. In these cases, I selected the most distantly related homolog, with the aim to cover the largest evolutionary space. The choice to select only one homolog for each overlapping gene was due to the fact that collection of a larger sample of homologs is limited to a few overlaps, mainly those occurring in virus species that are human pathogens (e.g. influenza and hepatitis viruses or SARS and Ebola viruses).
3. Results
3.1. Creation of a dataset of 80 homologous overlapping genes
The search for homologs yielded a dataset of 80 pairs of homologous overlapping genes (Supplementary File S1). Thirty-seven homologs came from a different virus species, in accordance with the ICTV taxonomy (King et al., 2018) (https://talk.ictvonline.org/taxonomy/). The mean nucleotide identity between overlaps and homologs was 70.7%, with a standard deviation (sd) of 9.4%. The remaining 43 homologs came from isolates belonging to the same virus species. In this case, the mean nucleotide identity between overlaps and homologs was 89.6% (sd = 7.1%).
For each pair of homologous overlapping genes, the Supplementary File S1 contains the following information: i) the nucleotide sequence of the upstream frame and that of the homolog; ii) the amino acid sequence of the protein encoded by the upstream frame (Up1) and that of the protein encoded by the homolog (Up2); iii) the nucleotide sequence of the downstream frame (shifted of one nucleotide 3’ with respect to the upstream frame) and that of the homolog; iv) the amino acid sequence of the protein encoded by the downstream frame (Down1) and that of the protein encoded by the homolog (Down2); v) the alignment of Up1 with Up2 and the percent amino acid identity; vi) the alignment of Down1 with Down2 and the percent amino acid identity; vii) the chi-square analysis, which compared by a 2 x 2 contingency-table the number of the amino acid identities and differences in the Up1-Up2 alignment with that in the Down1-Down2 alignment (cut-off of significance = 3.84; 1 degree of freedom; P < 0.05).
3.2. Half of overlapping genes evolve in accordance with the asymmetric model
I carried out a preliminary analysis using the t-Student test for paired data. For each pairs of homologous overlaps, I counted the number of amino acid identities between Up1 and Up2 and that between Down1 and Down2. I then calculated the absolute value of the difference between them. The null hypothesis was a mean difference between paired observations close to zero, indicating that overlapping genes evolve in accordance with the symmetric model. The null hypothesis was rejected (t-Student = 5.91; 79 degrees of freedom; P = 10−5), indicating that overlapping genes can also evolve in accordance with the alternative asymmetric model.
In order to identify which and how many overlapping genes undergo symmetric or asymmetric evolution, I then compared the amino acid diversity between Up1 and Up2 to that between Down1 and Down2. I used the contingency-table chi-square test (Snedecor and Cochran, 1967) with a cut-off value of 3.84 for 1 degree of freedom (P < 0.05).
I classified a pair of homologous overlaps as a case of symmetric evolution, if the number of amino acid substitutions in the Up1-Up2 alignment did not significantly differ from that in the Down1-Down2 alignment (chi-square <3.84). An example is given by the overlap NS1 protein/NS2 protein from Dendrolimus punctatus densovirus. For the NS1 protein, I found 73 identities and 86 differences when compared to the homolog from Hordeum marinum Itera-like densovirus. For the NS2 protein, I found 71 identities and 88 differences, yielding a chi-square value (0.01) largely below the cut-off of significance.
In alternative, I classified a pair of homologous overlaps as a case of asymmetric evolution, if the number of amino acid substitutions in the Up1-Up2 alignment was significantly different from that in the Down1-Down2 alignment (chi-square >3.84). An example is given by the overlap movement protein/replicase from Turnip yellow mosaic virus. For the movement protein, I found 302 identities and 323 differences when compared to the homolog from Watercress white vein virus. For replicase, I found 454 identities and 171 differences, yielding a chi-square value (76.3) largely above the cut-off of significance.
The chi-square test was highly sensitive. For example, I found that the overlap capsid protein/p31 protein from Maize chlorotic mottle virus undergoes asymmetric evolution, in spite of a nucleotide identity with the homolog extremely high (96.7%). Indeed, the number of amino acid differences between p31 and homolog (12 out of 149 sites) was significantly higher than that between capsid and homolog (2 out of 149 sites) (chi-square = 6.07; P = 0.01). Based on this finding, I set the upper limit of sensitivity of the chi-square test to a nucleotide identity between overlap and homolog of 97%. This filter limited the analysis to 75 (out of 80) pairs of homologous overlaps.
Overall, I found that 38 overlapping genes evolve in accordance with the asymmetric model (significantly different selection pressures on the two proteins). The highest chi-square value (113.8) concerned the overlap from Apple stem grooving virus, which encodes the 36kD movement protein and the polyprotein linker-domain. Indeed, the amino acid diversity between linker-domain and homolog (39%; 125 differences and 195 identities) was ten-fold higher than that between movement protein and homolog (4%; 13 differences and 307 identities).
I found that the remaining 37 overlapping genes evolve in accordance with the symmetric model (similar selection pressures on the two proteins). The occurrence of similar selection pressures can yield two highly conserved proteins. For example, analysis of the overlap 3a protein/3b protein from human SARS coronavirus revealed that the amino acid diversity between 3a and homolog is remarkably low (5.3%; 6 differences and 108 identities), as well as that between 3b and homolog (8.8%; 10 differences and 104 identities).
However, the occurrence of similar selection pressure can also yield two proteins with a remarkably less conserved pattern of change. This is the case of the overlap from Spinach latent virus, which encodes the zinc-finger domain of polymerase and the 2b protein. Sequence analysis revealed that the amino acid diversity between zinc-finger domain and homolog is considerably high (47%; 47 differences and 54 identities), as well as that between 2b and homolog (44%; 44 differences and 57 identities).
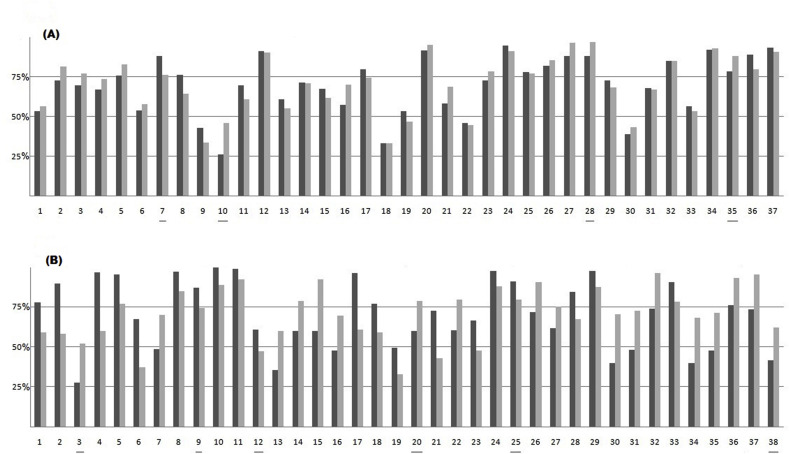
The analysis of amino acid diversity in the 75 pairs of homologous overlapping genes is summarized in Fig. 2 . It shows, for each overlap, the percent amino acid (aa) identity of the two encoded proteins with those encoded by the homolog. The subset of the 37 overlapping genes under symmetric evolution (Fig. 2A) contains 31 overlaps in which both proteins have high conservation (aa identity >50%), 5 overlaps in which both proteins have poor conservation (aa identity <50%) and 1 overlap with a protein having an aa identity above 50% and the other below 50%. The subset of the 38 overlapping genes under asymmetric evolution (Fig. 2B) contains 24 overlaps in which both proteins have high conservation (aa identity >50%), 1 overlap in which both proteins have poor conservation (aa identity <50%) and 13 overlaps with a protein having an aa identity above 50% and the other below 50%.
Fig. 2.
Analysis of the amino acid diversity in the 75 pairs of homologous overlapping genes. Each pair of columns shows: i) the percent amino acid identity between the protein encoded by the upstream frame of the overlap and that encoded by the homolog (dark column); ii) the percent amino acid identity between the protein encoded by the downstream frame of the overlap (shifted of one nucleotide 3′ with respect to the upstream frame) and that encoded by the homolog (gray column). The horizontal line separates well-conserved homologous pairs (aa identity >50%) from not well-conserved homologous pairs (aa identity <50%). (A) Subset of the 37 overlapping genes under symmetric evolution. (B) Subset of the 38 overlapping genes under asymmetric evolution. The numbering of overlapping genes is in accordance with that given in Supplementary Table S1. The underlined numbers indicate the overlaps in which the pattern of symmetric evolution (4 cases out of 37) or that of asymmetric evolution (6 cases out of 38) was not confirmed by chi-square analysis of the nucleotide diversity.
Finally, a list of the 75 overlapping genes, classified in accordance with the symmetric or asymmetric model (37 and 38 cases, respectively), is given in Supplementary Table S1.
3.3. Validation of the model of symmetric/asymmetric evolution by analysis of the pattern of nucleotide substitutions in homologous overlapping genes
In accordance with Wei and Zhang (2014), I first classified the nucleotide sites of each overlapping gene into four categories depending on the impact of potential mutations on the two encoded proteins. The four categories are referred as NN, SN, NS, and SS sites, respectively, where N stands for non-synonymous change and S stands for synonymous change. That is, if all potential mutations at a site cause non-synonymous change in both proteins, it is a NN site, and so on. I then classified the nucleotide substitutions occurring in the homolog into four categories: NN, SN, NS, and SS. Using the contingency-table chi-square test, I compared the number of SN and NS sites in each overlapping gene with the number of SN and NS substitutions in the homolog.
Under symmetric evolution, I would expect a chi-square value below the cut-off of significance (3.84; 1 degree of freedom), that is a full concordance between the number of SN and NS sites and that of SN and NS substitutions. For example, in the overlap ORF4/ORF5 from Barley yellow striate mosaic virus I counted 49 SN sites and 51 NS sites. In the homolog from Maize yellow striate virus, I classified 23 nucleotide substitutions into the SN category and 28 substitutions into the NS category. The chi-square test yielded a value (0.08) largely below the cut-off of significance.
Under asymmetric evolution, I would expect a chi-square above the cut-off of significance, that is a significant discordance between the number of SN and NS sites and that of SN and NS substitutions. For example, the overlap capsid protein/NS4 protein from Bluetongue virus (serotype 10) has 56 SN sites and 46 NS sites. The homolog from Bluetongue virus (serotype 16) has 5 nucleotide substitutions belonging to the SN category and 29 substitutions to the NS category. The chi-square test yielded a value (15.07) largely above the cut-off of significance.
The analysis of the pattern of nucleotide substitutions in the 75 pairs of homologous overlaps revealed 39 and 36 cases of symmetric and asymmetric evolution, respectively (Supplementary Table S2). This result was in accordance with that obtained previously (from analysis of the amino acid diversity, see Supplementary Table S1) in the 87% of cases (65 out of 75). Overall, I found a total of 33 overlaps under symmetric evolution (they are marked with a single asterisk in Supplementary Tables S2a) and a total of 32 overlaps under asymmetric evolution (they are marked with a double asterisk in Supplementary Table S2b). A list of the 32 overlapping genes under asymmetric evolution is given in Table 1 .
Table 1.
List of the 32 overlapping genes evolving in accordance with the asymmetric model.
| Genome ac. number (homolog) | Virus species | Overlapping gene | Chi-square analysis of amino acid substitutions | Chi-square analysis of nucleotide substitutions | Most variable protein |
|---|---|---|---|---|---|
| NC_001366 (EU542581) | Theiler's murine encephalomyelitis virus | polyprotein/L* | 11.60 | 6.79 | L* |
| NC_004102 (JQ061474) | Hepatitis C virus | polyprotein/F (ARFP) | 36.27 | 25.11 | F |
| NC_002021 (CY109232) | Influenza A virus | RdRp (subunit PB1)/PB1-F2 | 32.36 | 20.61 | PB1-F2 |
| NC_002022 (KY614903) | Influenza A virus | RdRp (subunit PA)/PA-X | 6.83 | 8.06 | PA-X |
| NC_001498 (KM089831) | Measles virus | V/phosphoprotein (P) | 10.80 | 8.60 | phosphoprotein |
| NC_001552 (KF687311) | Sendai virus | phosphoprotein (P)/C′ | 18.52 | 9.01 | phosphoprotein |
| NC_024473 (JX121105) | Vesicular stomatitis New Jersey virus | phosphoprotein (P)/C′ | 4.50 | 4.35 | C′ |
| NC_008311 (JQ658375) | Murine norovirus | capsid protein (VP1)/VF1 | 20.57 | 17.50 | VF1 |
| NC_003627 (JX286709) | Maize chlorotic mottle virus | capsid protein/p31 | 6.07 | 4.85 | p31 |
| NC_002568 (MSBMVCCG) | Sesbania mosaic virus | Px/polyprotein P2ab (protease domain) | 7.33 | 4.03 | Px |
| NC_001749 (KC310737) | Encephalomyocarditis virus | 2B*/polyprotein | 8.85 | 11.72 | 2B* |
| NC_006008 (KP821839) | Bluetongue virus | capsid protein (VP6)/NS4 | 20.49 | 15.07 | capsid protein |
| NC_001409 (NC_006946) | Apple chlorotic leaf spot virus | movement protein/capsid protein | 9.50 | 3.91 | movement protein |
| NC_001749 (EU553489) | Apple stem grooving virus | movement protein (36 kd)/polyprotein (linker domain) | 113.83 | 69.34 | polyprotein (linker domain) |
| NC_005224 (NC_005227) | Puumala virus | nucleocapsid protein/non-structural protein NSs | 5.72 | 3.85 | non-structural protein NSs |
| NC_001427 (NC_015396) | Chicken anemia virus | capsid protein (VP2)/apoptin (VP3) | 6.26 | 4.96 | apoptin |
| NC_004674 (KC795968) | East African cassava mosaic virus | replication associated protein (Rep, AC1)/AC4 | 12.88 | 11.82 | AC4 |
| NC_001412 (NC_015051) | Beet curly top virus | movement protein (V3)/V2 | 4.56 | 4.57 | movement protein |
| NC_001401 (KP733795) | Adeno-associated virus-2 | capsid protein (VP1)/AAP (Assembly Activating Protein) | 10.87 | 5.80 | AAP |
| NC_001401 (AY530620) | Adeno-associated virus-2 | capsid protein (VP1)/X protein | 9.20 | 13.09 | X protein |
| NC_014126 (KU885997) | Providence virus | p130/replicase (p104) | 102.76 | 104.30 | p130 |
| NC_001554 (NC_007729) | Tomato bushy stunt virus | p19/p22 | 6.60 | 6.75 | p19 |
| NC_003608 (DQ392986) | Hibiscus chlorotic ringspot virus | p28/p23 | 15.09 | 11.90 | p23 |
| NC_003608 (DQ392986) | Hibiscus chlorotic ringspot virus | capsid protein/p25 | 15.00 | 14.62 | p25 |
| NC_004366 (NC_027710) | Tobacco bushy top virus | movement protein (ORF3)/movement protein (ORF4) | 40.04 | 33.30 | ORF3 |
| NC_004063 (JQ001816) | Turnip yellow mosaic virus | movement protein (p69)/replicase | 76.32 | 60.61 | movement protein |
| NC_001915 (NC_030242) | Infectious pancreatic necrosis virus | VP5/polyprotein | 23.62 | 19.81 | VP5 |
| NC_011505 (JX416217) | Rotavirus A | phosphoprotein (NSP5)/NSP6 | 4.09 | 4.38 | NSP6 |
| NC_001841 (KU877879) | Sweet potato feathery mottle virus | P1N-PISPO/polyprotein | 35.86 | 17.07 | P1N-PISPO |
| NC_001549 (JN662633) | Simian immunodeficiency virus | vif protein/vpx protein | 6.46 | 5.78 | vif protein |
| NC_001607 (AF136236) | Borna disease virus | X protein/phosphoprotein (P) | 6.51 | 6.69 | X protein |
| NC_006497 (GU830910) | Infectious salmon anemia virus | P6 (ORF2)/P7 (ORF1) | 31.16 | 27.78 | P6 |
These findings were not affected by the fact that some homologs came from a different virus species, while others from an isolate within the same virus species. Under symmetric evolution, I found 14 and 19 overlaps with the homolog within and between species, respectively. Under asymmetric evolution, I found 18 and 14 overlaps with the homolog within and between species, respectively.
Finally, a further validation of the model of symmetric/asymmetric evolution was provided by a correlation test between the chi-square value from analysis of amino acid substitutions and the distribution of nucleotide substitutions at the codon positions “32” and “13” (Fig. 1). Given the orientation of overlapping genes in our dataset (Fig. 1), a substitution at the codon position “32” (cp32) is usually synonymous in the upstream frame and always non-synonymous in the downstream frame, while a substitution at the codon position “13” is almost always non-synonymous in the upstream frame and usually synonymous in the downstream frame.
Under symmetric evolution, the number of substitutions at the codon position “32” is expected to be close to that at the codon position “13”, yielding a similar distribution of the amino acid substitutions in the two pairs of homologous proteins. Under asymmetric evolution, the number of substitutions at the codon position “32” is expected to be significantly higher (or lower) than that at the codon position “13”, yielding a different distribution of the amino acid substitutions in the two pairs of homologous proteins.
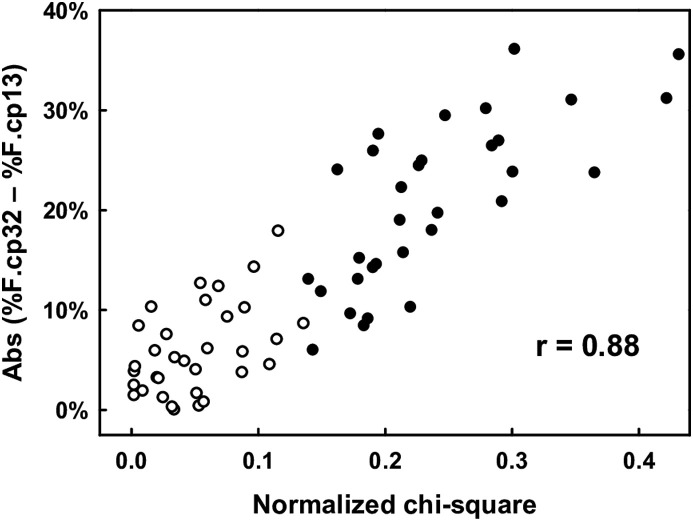
By comparing the upstream frame of each overlap with that of the homolog, I calculated the absolute value (Abs) of the difference between the percent frequency (%F) of substitutions at the codon position “32” (%F.cp32) and that at the codon position “13” (%F.cp13). I then carried out a correlation test between Abs (%F.cp32 – %F.cp13) and the chi-square value from analysis of amino acid substitutions. As the chi-square test depends on the extent of the sample (here the length of the protein encoded by the overlap), I normalized the chi-square value in accordance with the Cohen's rule (Cohen, 1988). Normalization was the square root of the ratio between the chi-square value and the overall length of the two proteins encoded by the overlap (e.g. the highest chi-square value, 113.83, was converted into the highest normalized chi-square value, 0.42).
I found a significantly positive correlation between Abs (%F.cp32 – %F.cp13) and the normalized chi-square value (r = 0.88; t-Student = 14.36; one tailed P < 0.00001; 63 degrees of freedom) (Fig. 3 ). As expected, this result indicates that asymmetric evolution is significantly affected by an unbalanced distribution of the nucleotide substitutions at the codon positions “32” and “13”.
Fig. 3.
Correlation between the normalized chi-square value (from analysis of amino acid substitutions) and the absolute value (Abs) of the difference between the percent frequency (%F) of nucleotide substitutions at the codon position “32” (%F.cp32) and that at the codon position “13” (%F.cp13). Empty circles indicate the 33 overlapping genes under symmetric evolution. Black circles indicate the 32 overlapping genes under asymmetric evolution.
3.4. In overlapping genes under asymmetric evolution the most variable protein is that encoded by the ancestral or the de novo frame?
To answer the question, I investigated the genealogy of the 32 overlapping genes under asymmetric evolution. Identifying which gene is ancestral and which one is de novo (the genealogy of the overlap) can be done by examining their phylogenetic distribution, under the assumption that the gene with the most restricted distribution is the de novo one (Rancurel et al., 2009). This approach yielded a set of 34 overlapping genes with a reliably predicted genealogy (see Table 1 in Sabath et al., 2012 and Table 1 in Pavesi et al., 2013). This set included 16 out of the 32 overlaps under asymmetric evolution.
Another approach to infer the genealogy of overlapping genes is the codon-usage method. It is based on the assumption that the ancestral gene, which has co-evolved over a long period of time with the other viral genes, has a distribution of synonymous codons significantly closer to that of the viral genome than the de novo gene (Keese and Gibbs, 1992; Sabath et al., 2012; Pavesi et al., 2013; Willis and Masel, 2018). Due to the shortness of most overlapping genes, the method has been improved, with the aim to evaluate the correlation between the codon-usage patterns of overlapping and non-overlapping genes with a minimal loss of information (Pavesi, 2015).
Using the improved version of the codon-usage method (Pavesi, 2015), I could predict the genealogy of 18 out of the 32 overlapping genes under asymmetric evolution. In 11 cases, the prediction by codon-usage was concordant with that established by the phylogenetic method. In the remaining 7 cases, the prediction was provided only by the codon-usage method (Supplementary Table S3).
The overlap p130/p104 of Providence virus is notable, as the ancestral frame p104 was acquired from another viral genome by distant horizontal gene transfer (Pavesi et al., 2013), which makes the codon usage an unreliable predictor of the genealogy. The prediction yielded by phylogenetics is supported by the finding that p104, unlike p130, has a wide phylogenetic distribution (Pavesi et al., 2013).
Overall, I collected a set of 23 overlapping genes, all under asymmetric evolution and with known genealogy (15 overlaps with a shift of the de novo frame of one nucleotide 3′ with respect to the ancestral frame and 8 overlaps with a shift of two nucleotides 3’). Interestingly, I found that in all cases the most variable protein is that encoded by the de novo gene (Table 2 ).
Table 2.
List of the 23 overlapping genes with known genealogy and evolving in accordance with the asymmetric model.
| Virus species and genome ac. number | Overlapping gene (protein products) | Most variable protein | De novo protein | Length of overlapping and non-overlapping part of the de novo protein | De novo protein predicted by: |
|---|---|---|---|---|---|
| Theiler's murine encephalomyelitis virus (NC_001366) | polyprotein (leader protein, 72 aa; capsid protein VP4; 71 aa; C-end of capsid protein VP2; 13 aa)/L* | L* | L* (suppressor of interferon response) | 156 aa; 0 aa | Phylogeny and codon usage |
| Hepatitis C virus (NC_004102) | polyprotein (core protein, 151 aa)/F (ARFP) | F (ARFP) | F (ARFP) (suppressor of interferon response) | 151 aa; 0 aa | Codon usage |
| Influenza A virus (NC_002021) | RNA-dependent RNA polymerase (subunit PB1)/PB1-F2 | PB1-F2 | PB1-F2 (suppressor of interferon response; apoptosis facor)) | 87 aa; 0 aa | Phylogeny and codon usage |
| Influenza A virus (NC_002022) | RNA-dependent RNA polymerase (subunit PA)/PA-X | PA-X | PA-X (degradation of host mRNA) | 61 aa; 0 aa | Codon usage |
| Puumala virus (NC_005224) | nucleocapsid protein/non-structural protein NSs | non-structural protein NSs | non-structural protein NSs (suppressor of interferon response) | 90 aa; 0 aa | Codon usage |
| Infectious pancreatic necrosis virus (NC_001915) | VP5/polyprotein (N-end half of capsid protein VP2, 131 aa) | VP5 | VP5 (suppressor of interferon response) | 131 aa; 0 aa | Phylogeny and codon usage |
| Borna disease virus (NC_001607) | X protein/phosphoprotein (P) | X protein | X protein (antagonist of interferon response) | 71 aa; 16 aa | Codon usage |
| Infectious salmon anemia virus (NC_006497) | P6 (ORF2)/P7 (ORF1) | P6 (ORF2) | P6 (ORF2) (antagonist of interferon response) | 183 aa; 51 aa | Codon usage |
| Murine norovirus (NC_008311) | capsid protein (VP1)/VF1 (virulence factor 1) | VF1 (virulence factor 1) | VF1 (antagonist of interferon response; apoptosis factor) | 213 aa; 0 aa | Phylogeny and codon usage |
| Apple chlorotic leaf spot virus (NC_001409) | movement protein/capsid protein | movement protein | movement protein (suppressor of RNA silencing) | 105 aa; 355 aa | Phylogeny |
| Tomato bushy stunt virus (NC_001554) | p19/p22 | p19 | p19 (suppressor of RNA silencing) | 172 aa; 0 aa | Phylogeny |
| Turnip yellow mosaic virus (NC_004063) | movement protein (p69)/replicase (C-end region, 63 aa; methyltransferase domain; 156 aa; downstream region, 407 aa) | p69 | p69 (suppressor of RNA silencing) | 626 aa; 0 aa | Phylogeny and codon usage |
| East African cassava mosaic virus (NC_004674) | replication associated protein AC1 (two-thirds C-end of DNA binding domain; 77 aa)/AC4 | AC4 | AC4 (suppressor of RNA silencing) | 77 aa; 0 aa | Phylogeny |
| Chicken anemia virus (NC_001427) | capsid protein (VP2)/apoptin (VP3) | apoptin | apoptin (apoptosis factor) | 119 aa; 0 aa | Phylogeny |
| Maize chlorotic mottle virus (NC_003627) | capsid protein/p31 | p31 | p31 | 149 aa; 69 aa | Phylogeny and codon usage |
| Tobacco bushy top virus (NC_004366) | movement protein (ORF3)/movement protein (ORF4) | ORF3 | ORF3 | 220 aa; 17 aa | Phylogeny and codon usage |
| Hibiscus chlorotic ringspot virus (NC_003608) | capsid protein/p25 | p25 | p25 | 224 aa; 0 aa | Phylogeny and codon usage |
| Adeno-associated virus- 2 (NC_001401) | capsid protein (VP1)/AAP (Assembly Activating Protein) | AAP | AAP | 169 aa; 35 aa | Phylogeny and codon usage |
| Adeno-associated virus- 2 (NC_001401) | capsid protein (VP1)/X protein | X protein | X protein | 155 aa; 0 aa | Codon usage |
| Rotavirus A (NC_011505) | phosphoprotein (NSP5)/NSP6 | NSP6 | NSP6 | 92 aa; 0 aa | Codon usage |
| Hibiscus chlorotic ringspot virus (NC_003608) | p28/p23 | p23 | p23 | 209 aa; 0 aa | Phylogeny and codon usage |
| Apple stem grooving virus (NC_001749) | movement protein (36 kd)/polyprotein (linker domain) | linker domain | linker domain | 320 aa; 0 aa | Phylogeny and codon usage |
| Providence virus (NC_014126) | p130/replicase (p104) | p130 | p130 | 893 aa; 327 aa | Phylogeny |
3.5. Symmetric and asymmetric evolution in the same overlap: the case of the overlap polymerase/large envelope protein of hepatitis B virus (HBV)
Chi-square analysis indicated that the overlap polymerase/large envelope protein of HBV evolves in accordance with the symmetric model (Supplementary Tables S1 and S2). On the other hand, theoretical and experimental studies (Pavesi, 2015; Lauber et al., 2017) demonstrated that this long overlap (1167 nt) is subjected to modular evolution, as the spacer domain of polymerase and the S domain of the large envelope protein originated de novo by overprinting. Thus, the overlap can be subdivided into two regions: a 5′ region (480 nt), in which the spacer domain of polymerase (de novo gene product) overlaps the Pre-S domain of envelope (ancestral gene product), and a 3’ region (687 nt), in which the reverse transcriptase domain of polymerase (ancestral gene product) overlaps the S domain of envelope (de novo gene product).
I carried out a chi-square analysis of the 2 regions of the overlap independently, under the hypothesis that they may have been subject to different evolutionary pressures. This analysis revealed that the 5’ region of the overlap undergoes asymmetric evolution, because the amino acid diversity of the spacer domain (33.7%; 54 differences and 106 identities) is significantly higher than that of the Pre-S domain (19.4%; 31 differences and 129 identities) (chi-square = 7.75; P = 0.005). Asymmetric evolution was confirmed by analysis of the pattern of nucleotide substitutions (chi-square = 10.13; P = 0.001).
In addition, chi-square analysis revealed that the 3’ region of the overlap undergoes symmetric evolution, as the amino acid diversity of the reverse transcriptase domain (7.4%; 17 differences and 212 identities) does not significantly differ from that of the S domain (11.8%; 27 differences and 202 identities) (chi-square = 2.04; P = 0.15). Symmetric evolution was confirmed by analysis of the pattern of nucleotide substitutions (chi-square = 1.61; P = 0.20).
With the aim to further validate these findings, I carried out a further analysis using, as homolog, the most distantly related overlap of woolly monkey HBV (79.9% of nucleotide identity). Again, chi-square analysis of the amino acid and nucleotide diversity revealed asymmetric evolution in the 5′ region and symmetric evolution in the 3’ region. Details of both analyses are reported in the Supplementary File S2.
Finally, the finding that the spacer domain of polymerase (de novo gene product) is significantly more variable than the Pre-S domain (ancestral gene product) confirms that the most variable protein, under asymmetric evolution, is usually that encoded by the de novo gene.
4. Discussion
Several researchers have developed methods for estimating the strength of selection pressure on overlapping genes (Pedersen and Jensen, 2001; Sabath et al., 2008; de Groot et al., 2008; Mir and Schober, 2014; Wei and Zhang, 2014). All methods evaluate, in both overlapping frames, the ratio of non-synonymous nucleotide substitutions to synonymous nucleotide substitutions (dn/ds) by correctly taking into account the problem of the interdependence between sequences imposed by the overlap. The aim is to assess if there is neutral evolution or positive selection in one frame (dn/ds higher than 1) and purifying selection (strong constraints) in the other frame (dn/ds lower than 1).
However, the only method having an accessible implementation is that by Sabath et al. (2008). Yet, the method has some limitations, as it restricts the analysis to the homologous overlaps in which the two encoded proteins have both an amino acid diversity smaller than 50% or greater than 5%. In the dataset examined here (see the first 75 pairs of homologous overlaps in Supplementary File S1), these limitations would have considerably reduced the size of the sample from 75 to 43 pairs of homologous overlaps.
I thus chose an approach focused, at first instance, on the evaluation of the amino acid diversity of homologous overlapping proteins, which is the final result of the complex pattern of the interdependent nucleotide substitutions that occur in dual-coding regions. Unlike previous studies, limited to a few virus species (Sabath et al., 2012; Zaaijer et al., 2007; Liang et al., 2010; Shukla and Hilgenfeld, 2015; Brayne et al., 2017), I examined a large dataset of 75 overlaps from 59 virus species.
A possible limitation of the study concerns the selection criteria for homologous overlapping genes. In particular, the first two stringent criteria (an equal length of the homolog and an alignment with a minimal number of indels) led to exclusion, for some overlaps, of highly divergent homologs. An example is given by the overlap P3N-PIPO/polyprotein of Turnip mosaic virus, in which the length of the P3N-PIPO protein is quite variable among the different potyvirus species, ranging from 60 to 115 amino acids (Hillung et al., 2013). Thus, the dataset used in this study likely underestimates the sequence diversity of overlapping genes, as it was created mainly to ensure a high quality in the homologous relationship.
The finding that 32 out of 65 overlapping genes (Table 1) undergo asymmetric evolution is striking, as well as that the most variable protein is encoded by the de novo gene in all cases examined (Table 3). In particular, I would point out the overlap ORF3/ORF4 from Tobacco bushy top virus, which encodes two proteins entirely nested within each other. This peculiar arrangement is similar to that of the overlap p19/p22 from Tomato bushy stunt virus, in which the de novo p19 protein shows a previously unknown structural fold an a previously unknown mechanism of binding to small interfering RNAs (Vargason et al., 2003; Baulcombe and Molnár, 2004; Scholthof, 2006). I believe that structural or functional studies on the de novo ORF3 protein from Tobacco bushy top virus could reveal new interesting features.
In addition, I would point out the overlap polymerase (PB1 subunit)/PB1-F2 protein of human influenza A virus. It shows, when compared to the homolog from duck, a sixteen-fold increase of substitutions at the codon position “32” (89.2%) with respect to the codon position “13” (5.4%). This yields only 3 amino acid differences between the two PB1 subunits and as many as 35 differences between the two PB1-F2 proteins. Interestingly, the de novo PB1-F2 protein has been shown to largely contribute to viral pathogenicity by a pleiotropic effect (Chen et al., 2001; Varga et al., 2011; Yoshizumi et al., 2014).
Several other de novo proteins under asymmetric evolution are known to play a role in viral pathogenicity. Eight de novo proteins (ARFP, VP5, L*, X, VF1, PB1-F2, P6, and NSs) act as suppressor or antagonist of the interferon response by the host (Park et al., 2016; Lauksund et al., 2015; Sorgeelos et al., 2013; Wensman et al., 2013; McFadden et al., 2011; Varga et al., 2011; García-Rosado et al., 2008; Jääskeläinen et al., 2007). Four de novo proteins (p19, p69, AC4, and movement protein) act as suppressor of RNA silencing (Silhavy et al., 2002; Chen et al., 2004; Chellappan et al., 2005; Yaegashi et al., 2008). Two de novo proteins (apoptin and PB1-F2) act as apoptosis factor (Noteborn et al., 1994; Chen et al., 2001). Finally, the de novo protein PA-X has the ability to selectively degrade the host RNA-polymerase II transcripts (Khaperskyy et al., 2016).
However, another possible limitation of the study depends on the fact that the subset of overlapping genes evolving asymmetrically and with known genealogy (23 overlaps) is too small to conclude that the de novo protein is always the preferred target of selection. Furthermore, overlapping genes are subjected to a variety of selection pressures that are independent of the orientation of the overlapping frames relative to one another. Thus, it is hypothetically possible that an ancestral protein may be significantly more variable than a de novo protein under peculiar selective constraints.
Despite this limitation, our findings suggest that the birth of new overlapping genes, besides to increase the coding ability of small viral genomes (Chirico et al., 2010), is also a valuable source of selective protein adaptation.
Declaration of interest
None.
Acknowledgements
The author is grateful to Alessio Peracchi (University of Parma) and Alberto Vianelli (University of Insubria) for helpful suggestions. Special thanks to Xinzhu Wei (University of Michigan) for valuable comments and suggestions and to Gianmarco Del Vecchio for preparing the figures. The author thanks the anonymous referees and the Editor Alexander E. Gorbalenya for their helpful feedback and suggestions. The study was financed by the MIUR (Ministero dell’Università e della Ricerca).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2019.03.017.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Allison J.R., Lechner M., Hoeppner M.P., Poole A.M. Positive selection or free to vary? Assessing the functional significance of sequence change using molecular dynamics. PLoS One. 2016;11:e0147619. doi: 10.1371/journal.pone.0147619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J.F., Steitz J.A., Anderson C.W., Model P. Binding of mammalian ribosomes to MS2 phage RNA reveals an overlapping gene encoding a lysis function. Cell. 1979;18:247–256. doi: 10.1016/0092-8674(79)90044-8. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C., Molnár A. Crystal structure of p19--a universal suppressor of RNA silencing. Trends Biochem. Sci. 2004;29:279–281. doi: 10.1016/j.tibs.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Bergeron D., Lapointe C., Bissonnette C., Tremblay G., Motard J., Roucou X. An out-of-frame overlapping reading frame in the ataxi-1 coding sequence encodes a novel ataxin-1 interacting protein. J. Biol. Chem. 2013;288:21824–21835. doi: 10.1074/jbc.M113.472654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N., Linial M. Gene overlapping and size constraints in the viral world. Biol. Direct. 2016;11:26. doi: 10.1186/s13062-016-0128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayne A.B., Dearlove B.L., Lester J.S., Kosakovsky Pond S.L., Frost S.D. Genotype specific evolution of hepatitis E virus. J. Virol. 2017;91 doi: 10.1128/JVI.02241-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P., Vanitharani R., Fauquet C.M. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Calvo P.A., Malide D., Gibbs J., Schubert U., Bacik I., Basta S., O'Neill R., Schickli J., Palese P., Henklein P., Bennink J.R., Yewdell J.W. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- Chen J., Li W.X., Xie D., Peng J.R., Ding S.W. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microma in host gene expression. Plant Cell. 2004;16:1302–1313. doi: 10.1105/tpc.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico N., Vianelli A., Belshaw R. Why genes overlap in viruses. Proc. Biol. Sci. 2010;277:3809–3817. doi: 10.1098/rspb.2010.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. second ed. Lawrence Eribaum Associates; New York, NY: 1988. Statistical Power and Analysis for the Behavioral Sciences; p. 223. [Google Scholar]
- de Groot S., Mailund T., Lunter G., Hein G. Investigating selection in viruses: a statistical alignment approach. BMC Bioinf. 2008;9:304. doi: 10.1186/1471-2105-9-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaye L., Deluna A., Lazcano A., Becerra A. The origin of a novel gene through overprinting in Escherichia coli. BMC Evol. Biol. 2008;8:31. doi: 10.1186/1471-2148-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellner L., Simon S., Scherling C., Witting M., Schober S., Polte C., Schmitt-Kopplin P., Keim D.A., Scherer S., Neuhaus K. Evidence for the recent origin of a bacterial protein-coding, overlapping orphan gene by evolutionary overprinting. BMC Evol. Biol. 2015;15:283. doi: 10.1186/s12862-015-0558-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J.D., Faust T.B., Strauli N.B., Smith C., Crosby D.C., Nakamura R.L., Hernandez R.D., Frankel A.D. Functional segregation of overlapping genes in HIV. Cell. 2016;167:1762–1773. doi: 10.1016/j.cell.2016.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y., Kiyotani K., Yoshida T., Sakaguchi T. Conserved and non-conserved regions in the Sendai virus genome: evolution of a gene possessing overlapping reading frames. Virus Gene. 2001;47:47–52. doi: 10.1023/a:1008130318633. [DOI] [PubMed] [Google Scholar]
- García-Rosado E., Markussen T., Kileng O., Baekkevold E.S., Robertsen B., Mjaaland S., Rimstad E. Molecular and functional characterization of two infectious salmon anaemia virus (ISAV) proteins with type I interferon antagonizing activity. Virus Res. 2008;133:228–238. doi: 10.1016/j.virusres.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Guyader S., Ducray D.G. Sequence analysis of Potato leafroll virus isolates reveals genetic stability, major evolutionary events and differential selection pressure between overlapping reading frame products. J. Gen. Virol. 2002;83:1799–1807. doi: 10.1099/0022-1317-83-7-1799. [DOI] [PubMed] [Google Scholar]
- Hillung J., Elena S.F., Cuevas J.M. Intra-specific variability and biological relevance of P3N-PIPO protein length in potyviruses. BMC Evol. Biol. 2013;13:249. doi: 10.1186/1471-2148-13-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L., Westover K., da Silva J., O'Connor D.H., Watkins D.I. Simultaneous positive and purifying selection on overlapping reading frames of the tat and vpr genes of simian immunodeficiency virus. J. Virol. 2001;75:7966–7972. doi: 10.1128/JVI.75.17.7966-7972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jääskeläinen K.M., Kaukinen P., Minskaya E.S., Plyusnina A., Vapalahti O., Elliott R.M., Weber F., Vaheri A., Plyusnin A. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 2007;79:1527–1536. doi: 10.1002/jmv.20948. [DOI] [PubMed] [Google Scholar]
- Kazem S., Lauber C., van der Meijden E., Kooijman S., Kravchenko A.A., the TrichSpin Network, Feltkamp M.C.W., Gorbalenya A.E. Limited variation during circulation of a polyomavirus in the human population involves the COCO-VA toggling site of Middle and Alternative T-antigen(s) J. Virol. 2016;487:129–140. doi: 10.1016/j.virol.2015.09.013. [DOI] [PubMed] [Google Scholar]
- Keese P.K., Gibbs A. Origins of genes: "big bang" or continuous creation? Proc. Natl. Acad. Sci. U.S.A. 1992;89:9489–9493. doi: 10.1073/pnas.89.20.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaperskyy D.A., Schmaling S., Larkins-Ford J., McCormick C., Gaglia M.M. Selective degradation of host RNA polymerase II transcripts by influenza A virus PA-X host shutoff protein. PLoS Pathog. 2016;12:e1005427. doi: 10.1371/journal.ppat.1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.M.Q., Lefkowitz E.J., Mushegian A.R., Adams M.J., Dutilh B.E., Gorbalenya A.E., Harrach B., Harrison R.L., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M.L., Rubino L., Sabanadzovic S., Sanfaçon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Davison A.J. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch. Virol. 2018;163:2601–2631. doi: 10.1007/s00705-018-3847-1. [DOI] [PubMed] [Google Scholar]
- Krakauer D.C. Stability and evolution of overlapping genes. Evolution. 2000;54:731–739. doi: 10.1111/j.0014-3820.2000.tb00075.x. [DOI] [PubMed] [Google Scholar]
- Lamb R.A., Orvath C.M. Diversity of coding strategies in influenza viruses. Trends Genet. 1991;7:261–266. doi: 10.1016/0168-9525(91)90326-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Seitz S., Mattei S., Suh A., Beck J., Herstein J., Börold J., Salzburger W., Kaderali L., Briggs J.A.G., Bartenschlager R. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe. 2017;22:387–399. doi: 10.1016/j.chom.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauksund S., Greiner-Tollersrud L., Chang C.J., Robertsen B. Infectious pancreatic necrosis virus proteins VP2, VP3, VP4 and VP5 antagonize IFNa1 promoter activation while VP1 induces IFNa1. Virus Res. 2015;196:113–121. doi: 10.1016/j.virusres.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.W., Tian F.L., Huang B., Zhuang W.Z. Selection characterization on overlapping reading frame of multiple-protein-encoding P gene in Newcastle disease virus. Vet. Microbiol. 2010;144:257–263. doi: 10.1016/j.vetmic.2009.12.029. [DOI] [PubMed] [Google Scholar]
- McFadden N., Bailey D., Carrara G., Benson A., Chaudhry Y., Shortland A., Heeney J., Yarovinsky F., Simmonds P., Macdonald A., Goodfellow I. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 2011;7:e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir K., Schober S. Selection pressure in alternative reading frames. PLoS One. 2014;9:e108768. doi: 10.1371/journal.pone.0108768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T. Evolution of overlapping genes. Nature. 1978;272:532–535. doi: 10.1038/272532a0. [DOI] [PubMed] [Google Scholar]
- Mizokami M., Orito E., Ohba K., Ikeo K., Lau J.Y., Gojobori T. Constrained evolution with respect to gene overlap of hepatitis B virus. J. Mol. Evol. 1997;44:S83–S90. doi: 10.1007/pl00000061. [DOI] [PubMed] [Google Scholar]
- Noteborn M.H., Todd D., Verschueren C.A., de Gauw H.W., Curran W.L., Veldkamp S., Douglas A.J., McNulty M.S., van der EB A.J., Koch G. A single chicken anemia virus protein induces apoptosis. J. Virol. 1994;68:346–351. doi: 10.1128/jvi.68.1.346-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.B., Seronello S., Mayer W., Ojcius D.M. Hepatitis C virus frameshift/alternate reading frame protein suppresses interferon responses mediated by pattern recognition receptor retinoic-acid-inducible gene-I. PLoS One. 2016;11:e0158419. doi: 10.1371/journal.pone.0158419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A., De Iaco B., Granero M.I., Porati A. On the informational content of overlapping genes in prokaryotic and eukaryotic viruses. J. Mol. Evol. 1997;44:625–631. doi: 10.1007/PL00006185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A., Magiorkinis G., Karlin D.G. Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of deltaretroviruses. PLoS Comput. Biol. 2013;9:e1003162. doi: 10.1371/journal.pcbi.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi A. Different patterns of codon usage in the overlapping polymerase and surface genes of hepatitis B virus suggest a de novo origin by modular evolution. J. Gen. Virol. 2015;96:3577–3586. doi: 10.1099/jgv.0.000307. [DOI] [PubMed] [Google Scholar]
- Pavesi A., Vianelli A., Chirico N., Bao Y., Blinkova O., Belshaw R., Firth A., Karlin D. Overlapping genes and the proteins they encode differ significantly in their sequence composition from non-overlapping genes. PLoS One. 2018;13:e0202513. doi: 10.1371/journal.pone.0202513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen A.M., Jensen J.L. A dependent-rates model and an MCMC-based methodology for the maximum-likelihood analysis of sequences with overlapping reading frames. Mol. Biol. Evol. 2001;18:763–766. doi: 10.1093/oxfordjournals.molbev.a003859. [DOI] [PubMed] [Google Scholar]
- Peleg O., Kirzhner V., Trifonov E., Bolshoy A. Overlapping messages and survivability. J. Mol. Evol. 2004;59:520–527. doi: 10.1007/s00239-004-2644-5. [DOI] [PubMed] [Google Scholar]
- Rancurel C., Khosravi M., Dunker A.K., Romero P.R., Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N., Landan G., Graur D. A method for the simultaneous estimation of selection intensities in overlapping genes. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N., Wagner A., Karlin D. Evolution of viral proteins originated de novo by overprinting. Mol. Biol. Evol. 2012;29:3767–3780. doi: 10.1093/molbev/mss179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander C., Schulz G.E. Degeneracy of the information contained in amino acid sequences: evidence from overlaid genes. J. Mol. Evol. 1979;13:245–252. doi: 10.1007/BF01739483. [DOI] [PubMed] [Google Scholar]
- Scholthof H.B. The Tombusvirus-encoded P19: from irrelevance to elegance. Nat. Rev. Microbiol. 2006;5:405–411. doi: 10.1038/nrmicro1395. [DOI] [PubMed] [Google Scholar]
- Shukla A., Hilgenfeld R. Acquisition of new protein domains by coronaviruses: analysis of overlapping genes coding for proteins N and 9b in SARS coronavirus. Virus Gene. 2015;50:29–38. doi: 10.1007/s11262-014-1139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Silhavy D., Molnár A., Lucigli A., Szittya G., Hornyik C., Tavazza M., Burgyán J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Holmes E.C., Pagan I. The effect of gene overlapping on the rate of RNA virus evolution. Mol. Biol. Evol. 2013;30:1916–1928. doi: 10.1093/molbev/mst094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedecor G.W., Cochran W.G. Iowa State University Press; Ames, IA: 1967. Statistical Methods; p. 228. [Google Scholar]
- Sorgeloos F., Jha B.K., Silverman R.H., Michiels T. Evasion of antiviral innate immunity by Theiler's virus L* protein through direct inhibition of RNase L. PLoS Pathog. 2013;9:e1003474. doi: 10.1371/journal.ppat.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenković G.G., Ćirković V.S., Šiljić M.M., Blagojević J.V., Knežević A.M., Joksić I.D., Stanojević M.P. Substitution rate and natural selection in parvovirus B19. Sci. Rep. 2016;6:35759. doi: 10.1038/srep35759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R., Heringa J., Pond S.K., Nekrutenko A. Rapid asymmetric evolution of a dual-coding tumor suppressor INK4a/ARF locus contradicts its function. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12807–12812. doi: 10.1073/pnas.0703238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderperre B., Lucier J.F., Bissonnette C., Motard J., Tremblay G., Vanderperre S., Wisztorski M., Salzet M., Boisvert F.M., Roucou X. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z.T., Ramos I., Hai R., Schmolke M., García-Sastre A., Fernandez-Sesma A., Palese P. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 2011;7:e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason J.M., Szittya G., Burgyan J., Hall T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- Wei X., Zhang J. A simple method for estimating the strength of natural selection on overlapping genes. Genome Biol. Evol. 2014;7:381–390. doi: 10.1093/gbe/evu294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensman J.J., Munir M., Thaduri S., Hörnaeus K., Rizwan M., Blomström A.L., Briese T., Lipkin W.I., Berg M. The X proteins of bornaviruses interfere with type I interferon signalling. J. Gen. Virol. 2013;94:263–269. doi: 10.1099/vir.0.047175-0. [DOI] [PubMed] [Google Scholar]
- Willis S., Masel J. Gene birth contributes to structural disorder encoded by overlapping genes. Genetics. 2018;210:303–313. doi: 10.1534/genetics.118.301249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaegashi H., Tamura A., Isogai M., Yoshikawa N. Inhibition of long-distance movement of RNA silencing signals in Nicotiana benthamiana by Apple chlorotic leaf spot virus 50 kDa movement protein. Virology. 2008;382:199–206. doi: 10.1016/j.virol.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Yoshizumi T., Ichinohe T., Sasaki O., Otera H., Kawabata S., Mihara K., Koshiba T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat. Commun. 2014;5:4713. doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]
- Zaaijer H.L., van Hemert F.J., Koppelman M.H., Lukashov V.V. Independent evolution of overlapping polymerase and surface protein genes of hepatitis B virus. J. Gen. Virol. 2007;88:2137–2143. doi: 10.1099/vir.0.82906-0. [DOI] [PubMed] [Google Scholar]
- Zhang D., Chen J., Deng L., Mao Q., Zheng J., Wu J., Zeng C., Li Y. Evolutionary selection associated with the multi-function of overlapping genes in the hepatitis B virus. Infect. Genet. Evol. 2010;10:84–88. doi: 10.1016/j.meegid.2009.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.