Abstract
Mouse hepatitis virus (MHV) nucleocapsid (N) protein binds to the large, single-stranded, positive-sense viral genomic RNA to form a helical nucleocapsid structure in mature virions. In addition N protein binds the intracellular form of the genomic RNA, all of the MHV subgenomic mRNAs, and expressed non-MHV RNA transcripts to form ribonucleoprotein (RNP) complexes in infected cells. Among the intracellular viral RNP complexes, only the genomic RNP complex is packaged into virus particles. The present study demonstrated that N protein in the MHV virion nucleocapsid and in the intracellular genome-length RNP complex that bound to viral envelope M protein was tightly self-associated such that its association was retained even after extensive RNase A-treatment of the RNP complexes. The RNase A-resistant tight N protein association in the virion nucleocapsid was not mediated by an intermolecular disulfide bridge between N proteins. In contrast, N protein association in the majority of the intracellular RNP complexes was susceptible to RNase A-treatment. Because the RNP complexes that specifically interact with the M protein are selectively packaged into MHV particles, the present data suggested that there was a distinct difference between N protein association in viral genomic RNP complexes that undergo packaging into virus particles and the subgenomic RNP complexes that are not packaged into MHV particles.
Keywords: Coronavirus, Ribonucleoprotein (RNP) complexes, N protein
1. Introduction
Enveloped animal RNA viruses in the coronavirus, orthomyxovirus, paramyxovirus, filovirus, rhabdovirus, bunyavirus, and arenavirus families contain helical viral nucleocapsid. The biochemical properties and the virus-specific components of these helical nucleocapsids vary among the different viral families. For example, paramyxovirus viral RNA in the paramyxovirus nucleocapsid resists RNase treatment of the nucleocapsid (Duesberg, 1969, Kingsbury and Webster, 1969, Pons et al., 1969), yet orthomyxovirus and coronavirus nucleocapsids (Macneughton and Davies, 1978) fall apart under the same treatment. Furthermore, the protein composition of the coronavirus nucleocapsid, unlike those of the above-mentioned other six viral families, does not include the protein(s) that is needed for RNA polymerase function. The common feature in these viruses is that N protein (nucleocapsid protein or core protein) and viral RNA are the major components of the helical nucleocapsid.
Coronaviruses are important pathogens of man and animals (Wege et al., 1982). The etiological agent of severe acute respiratory syndrome, a disease that rapidly spread from its likely origin in Southern China to several other countries of the world, is identified as a new coronavirus (Drosten et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003). Coronavirus contains a single-stranded, positive-sense RNA genome of about 28–31 kb (Lai and Cavanagh, 1997) and is the only known positive-stranded animal RNA virus with a helical nucleocapsid genome. All coronaviruses contain three envelope proteins, M, S, and E; the first two are major envelope proteins, while the amount of E protein in virion is low (Godet et al., 1992, Smith et al., 1990, Yu et al., 1994). The S protein is not necessary for viral assembly (Holmes et al., 1981, Kim et al., 1997, Rottier et al., 1981), which occurs at the smooth membranes of the intermediate compartment, between the endoplasmic reticulum and the Golgi complex (Klumperman et al., 1994, Tooze et al., 1984). The M and E proteins are essential for viral envelope formation and release (Bos et al., 1996, Vennema et al., 1996). In infected cells, the virus produces an intracellular form of genomic RNA, mRNA 1, and six to eight species of subgenomic mRNAs (Lai et al., 1981, Leibowitz et al., 1981; Stern and Kennedy, 1980a, Stern and Kennedy, 1980b). These virus-specific mRNAs comprise a nested set with a common 3′-terminus (Lai et al., 1981, Leibowitz et al., 1981; Stern and Kennedy, 1980a, Stern and Kennedy, 1980b) and a common leader sequence of approximately 60–100 nucleotides at the 5′-end (Lai and Cavanagh, 1997).
In cells infected with the coronavirus, mouse hepatitis virus (MHV), the N protein not only binds to mRNA 1 to form a ribonucleoprotein (RNP) complex (intracellular genomic RNP complex), but also binds to all the subgenomic mRNAs to form subgenomic RNP complexes (Baric et al., 1988, Narayanan et al., 2000). Additionally in infected cells, MHV N protein can bind expressed non-MHV RNA transcripts to form RNP complexes (Cologna and Hogue, 2000, Cologna et al., 2000, Narayanan and Makino, 2001b). MHV N protein also binds to non-MHV RNA transcripts in vitro (Masters, 1992). While the binding of the N protein to MHV mRNAs has implications for viral RNA synthesis and viral mRNA translation (Baric et al., 1988, Compton et al., 1987, Kim and Makino, 1995, Tahara et al., 1994), the exact roles that N protein associations play in these RNP complexes are under characterized. Of the MHV RNP complexes, only the intracellular genomic RNP complex is efficiently packaged into MHV particles. In MHV dependency upon specific and selective packaging of intracellular genomic RNP complex is mediated by the selective interaction of one viral protein and one RNA element. The protein involved in packaging is the M protein (Narayanan et al., 2000), and the RNA element is a 190 nt-long RNA packaging signal (Narayanan and Makino, 2001b), which is present only in mRNA 1 but lacking in subgenomic mRNAs (Fosmire et al., 1992, van der Most et al., 1991). The interaction between M protein and the packaging signal leads to the subsequent packaging of only the genomic RNP complex from a pool of intracellular MHV RNP complexes (Narayanan and Makino, 2001b). While identification of the packaging signal of bovine coronavirus (Cologna and Hogue, 2000) and that of transmissible gastroenteritis virus (Escors et al., 2003) are reported, the RNA packaging mechanisms of other coronaviruses are rather poorly characterized.
Using two immunologically distinguishable types of N proteins, the present study explored N protein self-associations in the virion genomic RNP complex and in intracellular RNP complexes, including the subgenomic RNP complexes. Our data suggested a distinct difference between N protein associations in viral genomic RNP complexes that undergo packaging into virus particles and subgenomic RNP complexes that are not packaged into MHV particles.
2. Materials and methods
2.1. Viruses and cells
The plaque-cloned A59 strain of MHV (MHV-A59) (Lai et al., 1981) and its temperature-sensitive mutant Alb4 (Koetzner et al., 1992) were used for this study. MHV-A59 and Alb4 stock viruses were propagated in mouse DBT cells (Hirano et al., 1974) at 37 °C and 33 °C, respectively.
2.2. Preparation of anti-N-spacer antibody
A synthetic peptide, NH2-CPKPQRKGRRQAQEKKDEVD–COOH, was conjugated with keyhole lymphocyte hemocyanin and injected into two rabbits. Sera that were collected prior to immunization were used as preimmune sera. After three subsequent immunizations, sera were collected and used as anti-N-spacer antibody.
2.3. Labeling of viral proteins and purification of viruses
For labeling of MHV virion proteins, 100 μCi of tran 35S-label (ICN) was added to MHV-infected cells at 7.5 h p.i., and culture fluids were collected at 12 h p.i. (Kim and Makino, 1995). After brief centrifugation of the supernatant, released viruses were partially purified using ultracentrifugation on a discontinuous sucrose gradient consisting of 60, 50, 30 and 20% sucrose, as described previously (Fosmire et al., 1992). After centrifugation, virus particles at the interface of 30 and 50% sucrose were collected, and further purified on a 10–60% continuous sucrose gradient at 26,000 rpm for 18 h at 4 °C in a Beckman SW28 rotor. Purified viruses were pelleted through a 20% sucrose cushion in a Beckman SW28 rotor rotating at 38,000 rpm for 3 h at 4 °C.
2.4. Separation of viral nucleocapsid from envelope proteins
Pelleted purified viruses were suspended in high-salt buffer (0.1 M NaCl, 0.01 M Tris–HCl [pH 7.5], 0.001 M EDTA, 0.25 M KCl, 0.25% NP-40) and incubated on ice for 30 min. The radiolabeled viral proteins in the detergent-treated viruses were immunoprecipitated using MHV-specific monoclonal antibodies.
2.5. RNase A-treatment of viral nucleocapsid
Purified MHV was treated with the high-salt buffer. The sample was divided into two groups. In one group, the sample was incubated with 100 μg/ml of RNase A for 15 min at room temperature. In another group, the sample was kept on ice for 15 min. One-half of each group was incubated in 2X proteinase K buffer (0.02 M Tris–HCl [pH 7.8], 0.01 M EDTA, 1% SDS) and proteinase K at a final concentration for 10 mg/ml at 37 °C for 30 min. Subsequently, RNA was extracted with phenol–chloroform and precipitated with ethanol. The remaining half of each group was mixed with lysis buffer (1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS in PBS) (Narayanan et al., 2000) and used for immunoprecipitation analysis.
2.6. Immunoprecipitation of viral proteins and SDS–polyacrylamide gel electrophoresis (SDS–PAGE)
MHV-specific proteins from detergent-treated viruses were immunoprecipitated using an anti-M protein monoclonal antibody, J1.3 (Fleming et al., 1989), an anti-N protein monoclonal antibody, J3.3 (Fleming et al., 1989), anti-MHV polyclonal antiserum, anti-N-spacer antibody, or preimmune sera as described (Narayanan et al., 2000). Immunoprecipitated proteins were separated by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) as described previously (Narayanan et al., 2000).
2.7. Preparation of virus-specific RNA
The intracellular MHV-specific RNAs were labeled with 750 μCi of 32Pi and extracted from virus-infected cells as described previously (Makino et al., 1984).
2.8. Agarose gel electrophoresis of RNA and Northern (RNA) blotting
Radiolabeled RNAs were denatured and separated on a 1% agarose-formaldehyde gel as described previously (Makino et al., 1991). For Northern blot analysis, the non-radiolabeled RNAs were separated on a 1% denaturing agarose-formaldehyde gel and transferred onto nylon filters (Makino et al., 1991). Northern blot analyses were performed using digoxigenin-labeled, random-primed probes (Boehringer), one corresponded to the 5′-end of MHV genomic RNA and the other to the 3′-end of MHV genomic RNA; each probe was used in a separate Northern blot analysis. The separated RNAs were visualized using a DIG luminescent detection kit (Boehringer), according to the manufacturer’s protocol.
2.9. Isolation of intracellular genomic RNP complex from infected cells
Cell extracts from MHV-infected cells were prepared in lysis buffer. The anti-M protein monoclonal antibody J1.3 was added to the cell lysates, and the sample was incubated on ice for 3 h. Subsequently, protein A (Pansorbin cells, Calbiochem) was added to the sample and incubated on ice for 2 h. The immunoprecipitate was washed three times with lysis buffer. After the final wash, the immunoprecipitate was suspended in the high-salt buffer to release the coimmunoprecipitated intracellular genomic RNP complex from the antigen-antibody complex. After incubation on ice for 30 min, the samples were centrifuged at 10,000×g for 10 min at 4 °C. The supernatant that contained the intracellular genomic RNP complex was collected and used for further analysis.
3. Results
3.1. Experimental approach for analyzing N protein self-association in viral nucleocapsid
MHV helical nucleocapsid consists of N protein and viral genomic RNA. RNase A-treatment of virion nucleocapsid results in the degradation of genomic RNA (Macneughton and Davies, 1978). We assumed that N proteins in MHV virion nucleocapsid were tightly associated with each other, and asked whether this putative tight N protein association actually was maintained after RNase A-treatment of the virion nucleocapsid had extensively degraded the RNA. MHV-A59 and its temperature-sensitive mutant Alb4 (Koetzner et al., 1992) were used to address this question. MHV N protein consists of three domains, each of which is connected by a short spacer sequence (Koetzner et al., 1992). Alb4 replicates well at 33 °C, and its N protein has a deletion of 29-amino-acids within one of the spacer regions (Koetzner et al., 1992). We raised an anti-peptide antibody, anti-N-spacer antibody, against part of the specific 29-amino-acid sequence, with the expectation that this antibody would recognize only the MHV-A59 N protein and not the Alb4 N protein. We also expected that the MHV particles, released from cells coinfected with MHV-A59 and Alb4 at 33 °C, would contain a helical nucleocapsid composed of both MHV-A59 N protein and Alb4 N protein. We wanted to test whether the putative tight association of N proteins in the viral nucleocapsid was real enough to be maintained even after extensive degradation of the genomic RNA; hypothetically, RNase A-treatment of the virion nucleocapsid from coinfected cells followed by immunoprecipitation using anti-N-spacer antibody would likely result in the coimmunoprecipitation of Alb4 N protein with wt MHV-A59 N protein only if the two protein species had previously been associated with each other. Due to its deletion the Alb4 N protein migrates faster than MHV-A59 N protein in SDS–PAGE (Koetzner et al., 1992), which makes the identification of each N protein feasible. If the N protein monomers in the viral nucleocapsid are not associated with each other, then anti-N-spacer antibody most likely should immunoprecipitate only the MHV-A59 N protein from the same sample preparation.
Coronavirus M protein interacts with the virion nucleocapsid in virus particles (Escors et al., 2001). M protein not only binds to viral RNA (Narayanan et al., 2003, Sturman et al., 1980), but most probably also binds to N protein in the nucleocapsid (Escors et al., 2001, Kuo and Masters, 2002, Narayanan et al., 2000). A lateral M protein–M protein interaction exists in coronavirus particles (de Haan et al., 1998, de Haan et al., 2000). Accordingly, preparation of virion nucleocapsid samples devoid of M protein was essential for examining N protein self-association; any M protein–nucleocapsid interaction maintained in the sample might allow anti-N-spacer antibody to indirectly coimmunoprecipitate Alb4 N protein through an M protein–N protein interaction, masking the N protein association in the virion nucleocapsid. To establish experimental conditions, in which virion nucleocapsid no longer interacted with the M protein, purified MHV-A59 was incubated in high-salt buffer, which disrupts the interaction between the virion nucleocapsid and envelope M protein (Escors et al., 2001, Narayanan and Makino, 2001a), as described in Section 2. Subsequent sucrose gradient analysis of disrupted virus particles showed that viral genomic RNA and N protein were still associated in virion nucleocapsid, but virion nucleocapsid and envelope proteins M and S (data not shown) were not; this virus disruption procedure, therefore, was chosen as the basis for examining putative association among N proteins in virion nucleocapsid.
3.2. N protein self-association in the virion nucleocapsid
To study how and whether N protein self-associates in virion nucleocapsid, we first prepared purified MHV particles containing both MHV-A59 N protein and Alb4 N protein. These particles resulted from coinfecting DBT cells with MHV-A59 and Alb4 virus, each at an m.o.i. of 5. As controls, DBT cells were independently infected with either MHV-A59 or Alb4 at an m.o.i. of 5. Infected cells were incubated at 33 °C, and virus-specific proteins were radiolabeled with tran35S label from 7.5 to 12 h p.i. Culture fluid was collected at 12 h p.i., and radiolabeled MHV was purified using sucrose gradient centrifugation. Purified MHV was treated with high-salt buffer to dissociate nucleocapsid from the M protein. One-half of the sample was incubated with RNase A to digest MHV genomic RNA, while the other half was kept on ice. After RNase treatment, RNA was extracted from a portion of each sample. Northern blot analysis of extracted RNA samples showed intact virion RNA in the untreated sample, while no RNA signal was detected in the RNase-treated sample (data not shown), demonstrating that virion RNA was extensively degraded by RNase A-treatment. The remaining samples were used for immunoprecipitation analysis. Fig. 1A demonstrates the results of the RNase-treated samples. Anti-N-spacer antibody immunoprecipitated MHV-A59 N protein from purified MHV-A59 particles, but not Alb4 N protein from purified Alb4 virus particles, demonstrating that anti-N-spacer antibody recognized only MHV-A59 N protein. Anti-N protein monoclonal antibody J3.3 precipitated both MHV-A59 N protein and Alb4 N protein and, as expected, preimmune sera did not precipitate either of these proteins. Essentially the same results were obtained when untreated samples were used (data not shown). Analysis of samples from coinfected cells showed that J3.3, but not preimmune sera, precipitated MHV-A59 N protein and Alb4 N protein from both RNase-treated samples and untreated samples (Fig. 1B). These results were consistent with our expectations. Anti-N-spacer antibody immunoprecipitated both MHV-A59 N protein and Alb4 N protein from the untreated sample, as well as from the RNase-treated sample from coinfected cells (Fig. 1B), suggesting that the N proteins in the virion nucleocapsid were associated tightly enough that RNase A-treatment of the virion nucleocapsid did not disrupt N protein self-association. We obtained consistent results in three independent experiments. The intensity of coimmunoprecipitated Alb4 N protein band in the RNase-treated sample was about two to threefold lower than that in the untreated sample in these experimental triplicates (Fig. 1B, lanes 3 and 6), implying that N protein association in MHV virion nucleocapsid was at least partially sensitive to RNase A-treatment. Alternatively, some Alb4 N proteins involved in the RNase A-resistant N protein association were partially dissociated under our experimental conditions.
Fig. 1.
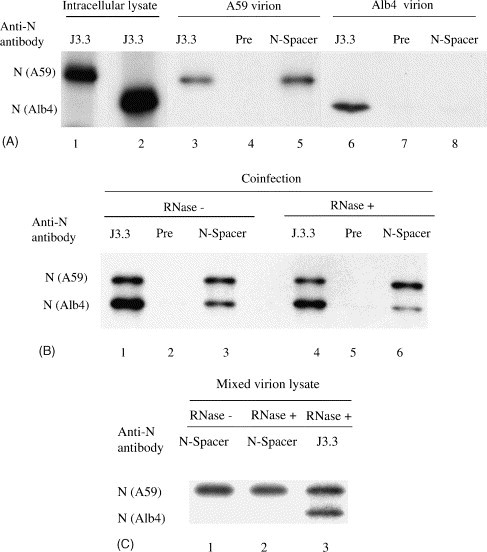
Characterization of a homotypic N protein interaction in virion nucleocapsid. (A) Purified MHV-A59 or Alb4 were disrupted by high-salt buffer, treated with RNase A and then immunoprecipitated with anti-N protein monoclonal antibody J.3.3 (lanes 3, 6), preimmune serum (lanes 4, 7), or anti-N-spacer antibody (lanes 5, 8). Radiolabeled intracellular extracts from MHV-A59-infected cells (lane 1) and Alb4-infected cells (lane 2) were also immunoprecipitated with anti-N protein monoclonal antibody J3.3. The immunoprecipitated samples were separated by SDS–PAGE. (B) Purified MHV from the cells coinfected with MHV-A59 and Alb4 were disrupted by high-salt buffer, and then treated with RNase A (lanes 4–6) or mock-treated (lanes 1–3). The samples were separated by SDS–PAGE after immunoprecipitation with anti-N protein monoclonal antibody J3.3. (lanes 1, 4), preimmune serum (lanes 2, 5), or anti-N-spacer antibody (lanes 3, 6). (C) Purified MHV-A59 and Alb4 were independently lysed with high-salt buffer, and then each virion lysate containing a similar level of N protein was mixed and treated with RNase A (lane 2, 3) or mock-treated (lane 1). The samples were then immunoprecipitated with anti-N-spacer antibody (lanes 1, 2) or J3.3 (lane 3).
The above data suggested the presence of an RNase A-resistant N protein association in the virion nucleocapsid. To eliminate the possibility that Alb4 and MHV-A59 N proteins had associated after lysis and RNAse treatment, we asked if anti-N-spacer antibody could coimmunoprecipitate Alb4 N protein from an in vitro mixture of MHV-A59 nucleocapsid and Alb4 nucleocapsid that had been treated with RNase. Cells were independently infected with MHV-A59 and Alb4, and progeny virions were purified and lysed with high-salt buffer. Each virion lysate containing a similar level of N protein was mixed and treated with RNase A. Subsequent immunoprecipitation analysis showed that anti-N-spacer antibody precipitated only MHV-A59 N protein (Fig. 1C), demonstrating that de novo N protein association did not occur in vitro after RNase treatment of nucleocapsids. Thus, coimmunoprecipitation of Alb4 N protein, along with MHV-A59 N protein after RNase treatment of the sample from the coinfected cells, demonstrated that an RNase A-resistant N protein association existed in the virion nucleocapsid.
A small population of MHV virion N protein forms trimers that are stabilized by intermolecular disulfide bonds (Robbins et al., 1986). We wondered whether the RNase A-resistant N protein association in the virion nucleocapsid represented the disulfide-linked N protein trimers reported previously (Robbins et al., 1986). Purified MHV from coinfected cells was disrupted by high-salt buffer treatment, and then incubated in the presence of RNase A as described above. The sample was then subjected to immunoprecipitation analysis using anti-N-spacer antibody, anti-N protein monoclonal antibody J3.3, or preimmune serum. The immunoprecipitates were analyzed on non-reducing SDS–PAGE (Fig. 2 ). J3.3, but not preimmune serum, immunoprecipitated both monomeric MHV-A59 N protein and Alb4 N protein. The anti-N-spacer antibody coimmunoprecipitated monomeric Alb4 N protein, along with MHV-A59 N protein, in non-reducing SDS–PAGE. In addition to the monomeric form of N protein, J3.3 and anti-N-spacer antibody immunoprecipitated N protein trimers of about 140 kDa. J3.3 precipitated three N protein trimers, while anti-N-spacer antibody precipitated two N protein trimers (Fig. 2, lanes 1 and 3, arrowheads). Although we did not further characterize these trimers, the slowest migrating signal and the fast migrating signal (Fig. 2, lane 1, asterisk) probably represented N protein trimer that was formed by MHV-A59 N protein and those formed by Alb4 N protein, respectively; the fast migrating N protein trimer was not detected in the sample immunoprecipitated by anti-N-spacer antibody, because anti-N-spacer antibody could not immunoprecipitate Alb4 N protein trimer. The band migrating between those two N protein trimers probably represented mosaic trimers formed by both MHV-A59 N protein and Alb4 N protein. If the RNase A-resistant N protein association was mediated solely by intermolecular disulfide linkage of N proteins, as reported previously (Robbins et al., 1986), then coimmunoprecipitated Alb4 N protein should only appear in the trimeric form of about 140 kDa in non-reducing SDS–PAGE. Yet a monomeric, roughly 50 kD, form of Alb-4 is clearly apparent; evidently the majority of N protein association that was resistant to RNase A-treatment was not held together by disulfide linkages of N protein trimers.
Fig. 2.
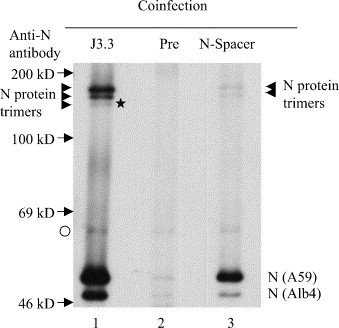
Characterization of a homotypic N protein interaction in virion under non-reducing conditions. Purified MHV from the cells coinfected with MHV-A59 and Alb4 was disrupted by high-salt buffer and then treated with RNase A. The samples were separated by non-reducing SDS–PAGE after immunoprecipitation with anti-N protein monoclonal antibody J3.3. (lane 1), preimmune serum (lane 2), or anti-N-spacer antibody (lane 3). The arrowheads indicate the N protein trimers. The asterisk represents the putative Alb4 N protein trimer. The arrows represent the positions of the 14C-labeled marker protein bands. The origin of the band, marked by open circle, is unknown.
3.3. Characterization of intracellular RNP complexes
In infected cells MHV N protein binds to all the MHV mRNAs, as well as to expressed non-MHV RNA transcripts (Cologna and Hogue, 2000, Cologna et al., 2000, Narayanan and Makino, 2001b), to form intracellular RNP complexes (Baric et al., 1988, Narayanan et al., 2000). To determine whether N proteins in the intracellular RNP complexes were also involved in the RNase A-resistant association, DBT cells were infected independently with MHV-A59 or Alb4 at an m.o.i. of 5, or coinfected with MHV-A59 and Alb4 at an m.o.i. of 5 for each virus. Infected cells were incubated at 33 °C, and virus-specific proteins were radiolabeled with tran 35S label from 8 to 8.5 h p.i. At 8.5 h p.i., cell extracts were prepared using high-salt buffer. Half of each sample was incubated with RNase A, while the remaining half was kept on ice. Subsequently, the RNaseA-treated and untreated samples were halved again to check for RNA degradation and to use in immunoprecipitation analysis. As expected, the RNase-untreated samples showed intact MHV mRNAs, and the RNase-treated samples showed complete degradation of all of the MHV mRNAs (data not shown). Radioimmunoprecipitation analysis of the remaining halves of the RNase-treated and untreated samples was used to examine RNase A-resistant N-protein interaction (Fig. 3 ). J3.3 immunoprecipitated MHV-A59 N protein from the MHV-A59-infected cell extracts, Alb4 N protein from Alb4-infected cell extracts, and both N proteins from the extracts of the coinfected cells. Anti-N-spacer antibody immunoprecipitated MHV-A59 N protein from the MHV-A59-infected cells, but did not pull down Alb4 N protein from Alb4-infected cell extracts. Intracellular N protein of both MHV strains can be detected as two closely migrating species (Koetzner et al., 1992, Stohlman and Lai, 1979). In all cases RNase A-treatment did not alter the immunoprecipitation results. These results were consistent with our expectations. Anti-N-spacer antibody coimmunoprecipitated both MHV-A59 N protein and Alb4 N protein from the mock-treated cell extracts of coinfected cells (Fig. 3A, lane 9). In marked contrast, anti-N-spacer antibody immunoprecipitated only MHV-A59 N protein, but not Alb4 N protein from the RNase-treated cell extracts from the coinfected cells (Fig. 3B, lane 9). These data demonstrated that the N protein association in the majority of intracellular RNP complexes was susceptible to RNase A-treatment.
Fig. 3.
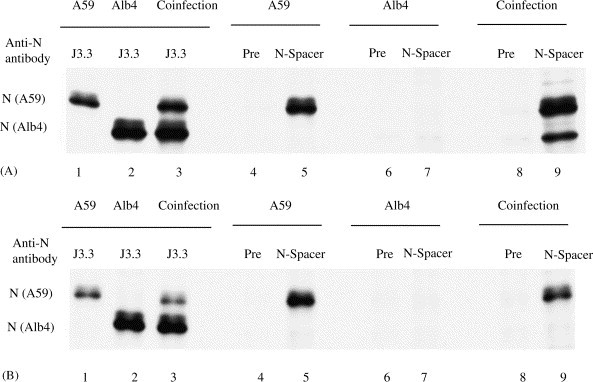
Characterization of a homotypic N protein interaction in intracellular RNP complexes. (A) Cell extracts were prepared using high-salt buffer from MHV-A59-infected cells (lanes 1, 4, 5), Alb4-infected cells (lanes 2, 6, 7) and coinfected cells (lanes 3, 8, 9). The samples were separated by SDS–PAGE after immunoprecipitation with anti-N protein monoclonal antibody J3.3. (lanes 1–3), preimmune serum (lanes 4, 6, 8), or anti-N-spacer antibody (lanes 5, 7, 9). (B) Essentially the same experimental methods were used as in (A), except that cell extracts were treated with RNase A prior to immunoprecipitation.
3.4. Characterization of the intracellular genomic RNP complex
Among all intracellular RNP complexes formed by MHV-specific mRNAs and N protein, only the intracellular genomic RNP complex binds to the M protein (Narayanan et al., 2000). Binding of the intracellular genomic RNP complex to the M protein drives the specific and selective packaging of mRNA 1, which has the same structure as genomic RNA, into MHV particles (Narayanan et al., 2000, Narayanan and Makino, 2001b). Because N protein association in virion nucleocapsid was resistant to RNase A-treatment, we wondered whether N proteins might exhibit RNase A-resistant association in the intracellular genomic RNP complex too. The intracellular genomic RNP complex represents only a fraction of intracellular RNP complexes, hence analysis of the total intracellular RNP complexes shown above might not be sensitive enough to detect the putative RNase A-resistant N protein association in the intracellular genomic RNP complex. Samples containing an enriched amount of intracellular genomic RNP complex were prepared by taking advantage of the selective binding property of the M protein to the intracellular genomic RNP complex. 32P-radiolabeled cell extracts from MHV-A59-infected cells were prepared at 8.5 h p.i. using the lysis buffer, as described previously (Narayanan et al., 2000). The anti-M protein monoclonal antibody J1.3 was used to coimmunoprecipitate the intracellular genomic RNP complex from the infected cell extracts. The non-MHV specific monoclonal antibody, anti-H2K, was used as a negative control (Narayanan et al., 2000). Subsequently, the interaction between the M protein and the intracellular genomic RNP complex was disrupted by treating the immunoprecipitates with high-salt buffer. To confirm that intracellular genomic RNP complex was isolated using this procedure, RNA was extracted from the sample that was obtained after high-salt treatment of the immunoprecipitates. Agarose gel electrophoresis of the sample showed the presence of mRNA 1, but not the subgenomic mRNA species (Fig. 4 ). The subgenomic mRNA species were not detected even after a 5 times longer exposure of the gel (data not shown), demonstrating that intracellular genomic RNP complex was released from the M protein after high-salt treatment of the J1.3-immunoprecipitate. As expected, no RNA signal was detected when the anti-H2K-immunoprecipitate was used. These data demonstrated that this experimental method could be used for the selective enrichment of intracellular genomic RNP complex.
Fig. 4.
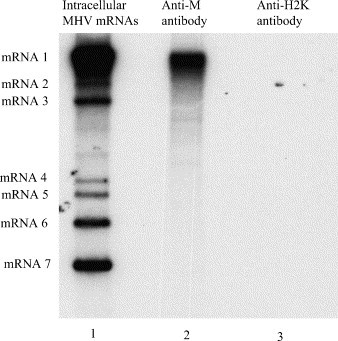
Separation of the intracellular genomic RNP complex from other MHV intracellular subgenomic RNP complexes. 32P-radiolabeled cell extracts from MHV-A59-infected cells were prepared using the lysis buffer. The samples were incubated with either anti-M protein monoclonal antibody, J1.3 (lane 2) or anti-H2K antibody (lane 3). After immunoprecipitation, the sample was suspended in the high-salt buffer to release the coimmunoprecipitated intracellular genomic RNP complex from the antigen-antibody complex. The antigen-antibody complex was removed by centrifugation. RNA was extracted from the supernatant and examined by agarose-formaldehyde gel electrophoresis. Lane 1 represents 32P-labeled MHV-A59 mRNAs.
To examine whether the N protein association in the intracellular genomic RNP complex was RNase A-resistant, the cell extracts from coinfected cells were immunoprecipitated with either anti-M protein monoclonal antibody or anti-H2K antibody. The immunoprecipitates were treated with high-salt buffer to release the coimmunoprecipitated intracellular genomic RNP complex from M protein. Then the sample containing intracellular genomic RNP complex was divided into two halves. One-half of the sample was treated with RNase A and the other half was untreated. After RNase treatment, the samples were immunoprecipitated with anti-N protein monoclonal antibody, preimmune serum or anti-N-spacer antibody. Anti-N protein monoclonal antibody J3.3 immunoprecipitated both the MHV-A59 N protein and Alb4 N protein from the untreated sample, while the anti-N-spacer antibody also immunoprecipitated both the MHV-A59 N protein and Alb4 N protein, with a higher amount of the former than the latter. These results were similar to those from the analyses of the untreated virion helical nucleocapsid (see Fig. 1). As expected, J3.3 immunoprecipitated both MHV-A59 and Alb4 N proteins, while the preimmune sera failed to immunoprecipitate either of these proteins from the RNase-treated sample. More importantly, anti-N-spacer antibody coimmunoprecipitated Alb4 N protein along with the MHV-A59 N protein from the RNase-treated sample (Fig. 5 ). The intensity of coprecipitated Alb4 N protein band in the RNase-treated sample was slightly lower than that in the untreated sample (Fig. 5), which was similar to the data obtained from analysis of the virion helical nucleocapsid using anti-N-spacer antibody (Fig. 1). RNase treatment of the virion lysates in vitro did not lead to N protein association (see Fig. 1C), suggesting that likewise Alb4 N protein association with the MHV-A59 N protein in vitro after RNase treatment of the intracellular genomic RNP complex would be unlikely. We concluded that the RNase A-resistant N protein association existed in the intracellular genomic RNP complex that bound to the M protein.
Fig. 5.
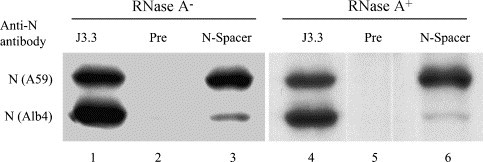
Characterization of a homotypic N protein interaction in an intracellular genomic RNP complex. Coinfected cells were radiolabeled and cell extracts were prepared using lysis buffer. Intracellular genomic RNP complex was immunoprecipitated with anti-M monoclonal antibody, and then released from the antigen-antibody complexes by high-salt buffer treatment of the immunoprecipitates. One-half of the intracellular genomic RNP complex was incubated with RNase A (lanes 4–6), while the other half was mock-treated (lanes 1–3). After RNase treatment, the samples were immunoprecipitated with anti-N protein monoclonal antibody J3.3. (lanes 1, 4), preimmune serum (lanes 2, 5) or anti-N-spacer antibody (lanes 3, 6), and analyzed by SDS–PAGE.
4. Discussion
We compared MHV N protein self-association in intracellular subgenomic RNP complexes, in the intracellular genomic RNP complex that associates with the M protein, and in the virion helical nucleocapsid. Purified MHV released from cells coinfected with MHV-A59 and Alb4 was reacted with anti-N-spacer antibody and resulted in a coimmunoprecipitation of the N proteins, which strongly suggests that in the virus, these two N proteins had been associated tightly enough such that they were resistant to RNase A-treatment of the nucleocapsid. At least the majority of RNase-resistant N protein association in the virion nucleocapsid was not due to intermolecular disulfide linkage of N protein trimers (Fig. 2). The N protein association in the majority of the intracellular RNP complexes was RNase-sensitive, whereas the RNase-resistant N protein association was observed exclusively with intracellular genomic RNP complex that bound to the M protein. These data suggested that N protein self-association differed between specifically packaged intracellular genomic RNP complexes and those subgenomic RNP complexes that are not packaged into MHV particles.
A small population of the N protein in MHV particles exists as trimers that are linked by intermolecular disulfide bonds (Robbins et al., 1986). Peng et al. characterized a series of MHV recombinant viruses, each of which contained chimeric MHV and bovine coronavirus N protein, and suggested the possibility of an N protein–N protein interaction in coronavirus (Peng et al., 1995). These published studies and our study described here established that the N protein in the MHV nucleocapsid existed as multimers through N protein association. Currently, the type of bonds stabilizing the RNase A-resistant N protein association in the viral nucleocapsid is unclear. If RNase A-treatment of the viral RNP complex completely digests viral RNAs, then direct N protein–N protein binding mediates the tight N protein association. If some very small RNA segments are not digested even after extensive RNase A-treatment of the samples, then these undigested RNA fragments may have some role in the tight N protein association.
Association of N proteins in the majority of intracellular RNP complexes was susceptible to RNase A-treatment, whereas that in the intracellular genomic RNP complex that bound to the M protein and virion nucleocapsid was resistant to RNase A-treatment. These data imply that RNase A-resistant N protein association is established in infected cells prior to the release of MHV from the cells. What is the mechanism that selectively induces the RNase A-resistant N protein association in the genomic RNP complex? One possibility is that mRNA 1, but not subgenomic mRNAs, contains a specific nucleation site for N protein, and the binding of the N protein to this nucleation site may induce a specific conformational change in the N protein that may serve as a nucleation event for the cooperative binding of the N protein to other regions of mRNA 1. This putative conformational change in the N protein may lead to self-association of N proteins and formation of the helical nucleocapsid structure. Because MHV mRNAs have a 3′-coterminal nested-set structure, the most likely position of this putative nucleation site is between the 5′-end leader sequence and the 3′ end of gene 1. Another possibility for the RNase A-resistant N protein association may be that M protein binds to N protein in the intracellular genomic RNP complex initiating the tight N protein association. A current model of MHV RNA packaging into particles proposes that the intracellular genomic RNP complex, consisting of mRNA 1 and N protein, binds to M protein through selective and specific binding of the 190 nt-long MHV packaging signal to M protein that already has accumulated and probably oligomerized in the intermediate compartment. After the binding of M protein to the packaging signal, N protein in the intracellular genomic RNP complex interacts with the oligomerized M protein. Subsequently, the M protein-mRNA 1 RNP complex undergoes virion morphogenesis in concert with E protein (Narayanan et al., 2003). Binding of the M protein to the N protein in the intracellular genomic RNP complex may trigger a conformational change in the N protein that induces the RNase A-resistant tight N protein association.
Another question that remains to be addressed is the biological significance of the RNase A-resistant N protein association in the intracellular genomic RNP complex and virion helical nucleocapsid. Coexpression of MHV E protein and the M protein, which contains a specific mutation at its C-terminal region, does not result in the formation of VLPs (de Haan et al., 1998), while a recombinant MHV that contains the M protein with the same mutation is infectious (Kuo and Masters, 2002). A possible interpretation of these data is that the intracellular genomic RNP complex binds to the mutated M protein and compensates for the loss of the M protein function(s) in assembly; the N protein multimer of the intracellular genomic RNP complex, which has the RNase A-resistant N protein association, might interact with mutated M protein, thereby compensating for the loss of function by promoting MHV assembly. Biologically, tightly self-associated N protein in virion nucleocapsid may have an advantage or function during uncoating. Incoming viral nucleocapsid likely dissociates from the envelope M protein, and perhaps multimeric N protein in the virion nucleocapsid may promote efficient release of nucleocapsid from the M protein. Supporting this idea, Alb4 is more heat labile than MHV-A59 (Koetzner et al., 1992), suggesting that the incubation of Alb4 at high temperature disrupts a function of Alb4 N protein that is required early in infection. High temperature may induce a conformational change in the structure of the N protein in Alb4 nucleocapsid such that the release of the altered nucleocapsid from the M protein during the viral uncoating process is rendered inefficient.
Acknowledgements
We thank Paul Masters for Alb4. This work was supported by Public Health Service grant AI29984 from the National Institutes of Health. K.N. was supported by a McLaughlin fellowship.
References
- Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 1988;62(11):4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218(1):52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R., Hogue B.G. Identification of a bovine coronavirus packaging signal. J. Virol. 2000;74(1):580–583. doi: 10.1128/jvi.74.1.580-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R., Spagnolo J.F., Hogue B.G. Identification of nucleocapsid binding sites within coronavirus-defective genomes. Virology. 2000;277(2):235–249. doi: 10.1006/viro.2000.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton S.R., Rogers D.B., Holmes K.V., Fertsch D., Remenick J., McGowan J.J. In vitro replication of mouse hepatitis virus strain A59. J. Virol. 1987;61(6):1814–1820. doi: 10.1128/jvi.61.6.1814-1820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72(8):6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74(11):4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Duesberg P.H. Distinct subunits of the ribonucleoprotein of influenza virus. J. Mol. Biol. 1969;42(3):485–499. doi: 10.1016/0022-2836(69)90237-x. [DOI] [PubMed] [Google Scholar]
- Escors D., Izeta A., Capiscol C., Enjuanes L. Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J. Virol. 2003;77(14):7890–7902. doi: 10.1128/JVI.77.14.7890-7902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane m protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75(3):1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J.O., Shubin R.A., Sussman M.A., Casteel N., Stohlman S.A. Monoclonal antibodies to the matrix (E1) glycoprotein of mouse hepatitis virus protect mice from encephalitis. Virology. 1989;168(1):162–167. doi: 10.1016/0042-6822(89)90415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosmire J.A., Hwang K., Makino S. Identification and characterization of a coronavirus packaging signal. J. Virol. 1992;66(6):3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., L’Haridon R., Vautherot J.F., Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188(2):666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N., Fujiwara K., Hino S., Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch. Gesamte Virusforsch. 1974;44(3):298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- Holmes K.V., Doller E.W., Sturman L.S. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115(2):334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Makino S. Two murine coronavirus genes suffice for viral RNA synthesis. J. Virol. 1995;69(4):2313–2321. doi: 10.1128/jvi.69.4.2313-2321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Narayanan K., Makino S. Assembled coronavirus from complementation of two defective interfering RNAs. J. Virol. 1997;71(5):3922–3931. doi: 10.1128/jvi.71.5.3922-3931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D.Y., Webster R.G. Some properties of influenza virus nucleocapsid. J. Virol. 1969;4:219–225. doi: 10.1128/jvi.4.3.219-225.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J. Coronavirus M proteins accumulate in the golgi complex beyond the site of virion budding. J. Virol. 1994;68(10):6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetzner C.A., Parker M.M., Ricard C.S., Sturman L.S., Masters P.S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 1992;66(4):1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76(10):4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Brayton P.R., Armen R.C., Patton C.D., Pugh C., Stohlman S.A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J. Virol. 1981;39(3):823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J.L., Wilhelmsen K.C., Bond C.W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology. 1981;114(1):39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macneughton M.R., Davies H.A. Ribonucleoprotein-like structures from coronavirus particles. J. Gen. Virol. 1978;39(3):545–549. doi: 10.1099/0022-1317-39-3-545. [DOI] [PubMed] [Google Scholar]
- Makino S., Joo M., Makino J.K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 1991;65(11):6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Hirano N., Fujiwara K. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology. 1984;139(1):138–151. doi: 10.1016/0042-6822(84)90335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch. Virol. 1992;125(1-4):141–160. doi: 10.1007/BF01309634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Chen C.-J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77(5):2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74(17):8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Makino S. Characterization of nucleocapsid-M protein interaction in murine coronavirus. Adv. Exp. Med. Biol. 2001;494:577–582. doi: 10.1007/978-1-4615-1325-4_85. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Makino S. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 2001;75(19):9059–9067. doi: 10.1128/JVI.75.19.9059-9067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D., Koetzner C.A., McMahon T., Zhu Y., Masters P.S. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J. Virol. 1995;69(9):5475–5484. doi: 10.1128/jvi.69.9.5475-5484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons M.W., Schulze I.T., Hirst G.K., Hauser R. Isolation and characterization of the ribonucleoprotein of influenza virus. Virology. 1969;39(2):250–259. doi: 10.1016/0042-6822(69)90045-2. [DOI] [PubMed] [Google Scholar]
- Robbins S.G., Frana M.F., McGowan J.J., Boyle J.F., Holmes K.V. RNA-binding proteins of coronavirus MHV: detection of monomeric and multimeric N protein with an RNA overlay-protein blot assay. Virology. 1986;150(2):402–410. doi: 10.1016/0042-6822(86)90305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottier P.J., Horzinek M.C., van der Zeijst B.A. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J. Virol. 1981;40(2):350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.R., Boursnell M.E., Binns M.M., Brown T.D., Inglis S.C. Identification of a new membrane-associated polypeptide specified by the coronavirus infectious bronchitis virus. J. Gen. Virol. 1990;71(Pt 1):3–11. doi: 10.1099/0022-1317-71-1-3. [DOI] [PubMed] [Google Scholar]
- Stern D.F., Kennedy S.I. Coronavirus multiplication strategy. I. Identification and characterization of virus-specified RNA. J. Virol. 1980;34(3):665–674. doi: 10.1128/jvi.34.3.665-674.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.F., Kennedy S.I. Coronavirus multiplication strategy. II. Mapping the avian infectious bronchitis virus intracellular RNA species to the genome. J. Virol. 1980;36(2):440–449. doi: 10.1128/jvi.36.2.440-449.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohlman S.A., Lai M.M. Phosphoproteins of murine hepatitis viruses. J. Virol. 1979;32(2):672–675. doi: 10.1128/jvi.32.2.672-675.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L.S., Holmes K.V., Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 1980;33(1):449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara S.M., Dietlin T.A., Bergmann C.C., Nelson G.W., Kyuwa S., Anthony R.P., Stohlman S.A. Coronavirus translational regulation: leader affects mRNA efficiency. Virology. 1994;202(2):621–630. doi: 10.1006/viro.1994.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Tooze S., Warren G. Replication of coronavirus MHV-A59 in sac-cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 1984;33(2):281–293. [PubMed] [Google Scholar]
- van der Most R.G., Bredenbeek P.J., Spaan W.J. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J. Virol. 1991;65(6):3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15(8):2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege H., Siddell S., ter Meulen V. The biology and pathogenesis of coronaviruses. Curr. Top. Microbiol. Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- Yu X., Bi W., Weiss S.R., Leibowitz J.L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]


