Abstract
Interferons represent a protein family with pleiotropic functions including immunomodulatory, cytostatic, and cytotoxic activities. Based on these effects, interferons are involved in innate as well as adaptive immunity, thereby shaping the tumor host immune responses. These cytokines, alone or in combination, have been successfully implemented for the treatment of some malignancies. However, it has been recently demonstrated that tumor cells could be resistant to interferon treatment, which may be associated with an escape of tumor cells from immune surveillance. Therefore, the aim of this chapter is to summarize the frequency of impaired interferon signal transduction, their underlying molecular mechanisms, and their clinical relevance.
Abbreviations: Ag, antigen; APC, antigen presenting cells; APM, antigen‐processing machinery; BH, bleomycin hydrolase; bp, base pairs; CIITA, class II transactivator protein; CLIP, class II invariant chain peptide; CTL, cytotoxic T lymphocyte; DC, dendritic cell; ER, endoplasmic reticulum; GAS, gamma‐interferon‐activated site; IFN, interferon; IFN‐γR1, interferon‐γ receptor‐1; IL, interleukin; IRF, interferon regulatory factor; ISG, interferon‐stimulated genes; ISGF3, IFN‐stimulated gene factor 3; ISRE, interferon‐stimulated response element; JAK, janus kinase; LPS, lipopolysaccharide; MAPK, mitogen‐activated protein kinase; MCA, methylcholanthrene; MHC, major histocompatibility complex; NF, nuclear factor; NK, natural killer; PKC, protein kinase C; RCC, renal cell carcinoma; SCLC, small‐cell lung carcinoma; SOCS, suppressor of cytokine signaling; STAT, signal transducer and activator of transcription; TA, tumor antigen; TAP, transporter associated with antigen processing; TCR, T cell receptor; TFBS, transcription factor‐binding sites; TNF, tumor necrosis factor; tpn, tapasin; TPPII, tripeptidyl peptidase II; TSA, trichostatin A; TYK, tyrosine kinase; UIRR, upstream interferon response region; USF1, upstream stimulatory factor 1; wt, wild type
Interferons (IFNs) represent proteins that are secreted from cells in response to various stimuli and provide the basis for the understanding of the evolution, structure, and function as well as the pathways of other cytokines and their receptors (Pestka, 2000, Pestka et al., 2004). They exert pleiotropic effects and are involved in host responses to bacterial and viral infection, in tumor surveillance mechanisms as well as in innate and adaptive immune responses (Decker et al., 2005, Pestka et al., 2004, Stetson and Medzhitov, 2006, Takaoka and Yanai, 2006). In addition, IFNs were the first cytokines used for the treatment of tumor patients. However, it has been suggested that tumor cells might develop either a transient or a permanent IFN insensitivity. This phenotype is linked to cytotoxicity resistance and might lead to escape of tumor cells from immune surveillance. We here summarize the current knowledge about (i) pleiotropic functions of IFNs that mediate various biological responses, (ii) mechanisms of action and transduction pathways, (iii) the effect of type I and type II IFNs on the expression levels of molecules involved in proper major histocompatibility complex (MHC) class I and class II antigen processing and presentation of tumor cells, (iv) the frequencies and the underlying molecular mechanisms of IFN resistance in tumors in association with alterations of the MHC class I and II antigen‐processing machinery, and (v) the clinical relevance of aberrant IFN signaling. The elucidation of the mechanisms leading to dysregulation of IFN signal transduction cascades triggering immune dysfunction and to tumor immune escape will benefit the design of strategies reversing these deficiencies, which could be of clinical relevance.
I. The Family of Interferons and Their Function
Interferons (IFNs) are a family of multifunctional cytokines, which were originally described as antiviral cytokines, thereby protecting cells from viral infection (Isaacs and Lindenmann, 1957). However, based on the current knowledge they exhibit a broad spectrum of activities including anti‐proliferative, immunomodulatory, anti‐inflammatory, apoptosis‐inducing, stress‐mediated effects as well as regulation of cell differentiation steps and angiogenesis (Amadori, 2007, Baccala et al., 2005, Theofilopoulos et al., 2005). The IFN family is divided into type I, type II, and type III IFNs. Type I IFNs consist of 13 IFN‐α members and single members of IFN‐β, IFN‐κ, IFN‐Ω, and IFN‐ε, respectively, which are all clustered on chromosome 9. In contrast, type II IFN is represented only by a single gene, IFN‐γ, encoded by chromosome 12 (Decker et al., 2005). Recently, type III IFNs have been discovered as a novel class of antiviral cytokines which are classified into IFN‐λ1, ‐λ2, and ‐λ3 (Oesterlund et al., 2007, Sheppard et al., 2003, Uze and Monneron, 2007). IFNs bind to two distinct cell surface receptors. Type I and II IFN signal through a common γ‐chain, thereby activating discrete, but related pathways leading to the transcriptional activation of the so‐called interferon‐stimulated genes (ISGs) (Table I ; Fig. 1 ). ISGs represent a functionally diverse group of genes involved in many cellular activities such as transcription, translation, regulation of cell cycle and apoptosis, intracellular communication as well as the processing and presentation of antigens. The transcriptional activity of ISGs is necessary to mediate the effect of IFNs.
Table I.
Features of the Major IFN Subtypes and Their Characteristics
| IFN‐α | IFN‐β | IFN‐γ | IFN‐λ | |
|---|---|---|---|---|
| Chromosomal localization | 9p21 | 9p21 | 12q14 | 19q/3 |
| Receptor | IFN‐αRI | IFN‐γ RI | IFN‐λR1 | |
| IFN‐αRII | IFN‐γ RII | IFN‐10R2 | ||
| Function | Antiviral | Antiviral | Antitumoral/Antiviral | Antiviral |
| Signal transduction pathways | JAK1, TYK2, STAT1‐5, PI3K, AKT, MAPK, NF‐κB, p53 | JAK1, 2; STAT1, 3, 5, PI3K, AKT, MAPK, NF‐κB | JAK 1, STAT1‐5 | |
The major characteristics of type I, II, and III IFN members; the localization of their genes; the components of the receptor complex; and the signal transduction pathways involved are summarized.
Fig. 1.
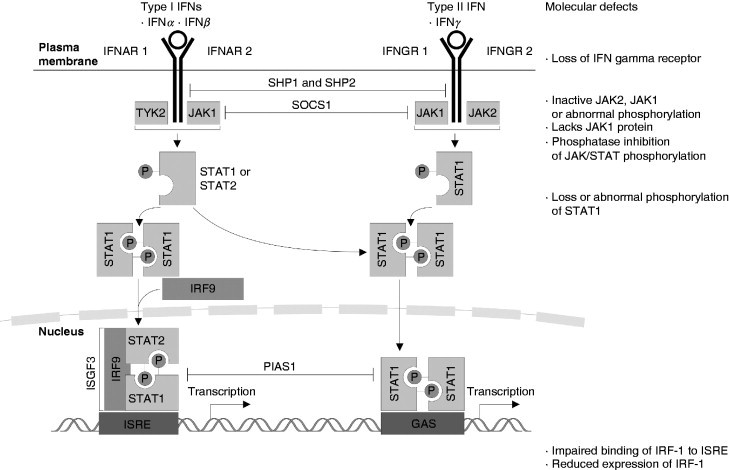
IFN signal transduction cascade and defects in this pathway. The type I and type II receptors are transmembrane glycoproteins whose extracellular domains serve as IFN‐binding sites, whereas their cytoplasmic domains associate with members of the JAK kinase family and initiate signal transmission (Dunn et al., 2006). Upon binding to their specific receptors both type I and type II IFNs induce a number of signal transduction cascades, which involve the phosphorylation of various components such as TYK2, JAKs, and STATs. After recruitment to the receptor, STATs become phosphorylated, form homo‐ or heterodimers, and migrate to the nucleus to bind to specific sequences in the promoter of target genes. Type I IFN‐induced signaling then induces homodimerization of STAT1 and heterodimerization of STAT1 and STAT2. STAT1 and STAT2 associate with the cytosolic transcription factor IFN‐regulatory factor 9 (IRF9), forming a trimeric complex known as IFN‐stimulated gene factor 3 (ISGF3) and activates transcription by binding to the ISREs. Type II IFN associates kinases, JAK1 and JAK 2 phosphorylate STAT1, which then forms homodimers, translocates to the nucleus, and activates transcription by binding to the GAS sequences. IFN‐mediated signaling is controlled by several mechanisms including dephosphorylation of IFN‐γR1, JAK1, and STAT1 (mediated by SH2‐domain‐containing protein tyrosine phosphatase 2, SHP2), inhibition of the JAKs (mediated by suppressor of cytokine signaling 1, SOCS1), proteasomal degradation of the JAKs, and inhibition of STAT1 (mediated by protein inhibitor of activated STAT1, PIAS1).
Because of their diverse activities, IFNs have been used for the treatment of various diseases such as chronic viral infections, like hepatitis C, multiple sclerosis, hematopoietic malignancies as well as solid tumors including renal cell carcinoma (RCC) and melanoma. The IFN therapy has been shown to reduce the rates of relapses and mortality by between 12 and 30% in tumor patients (Kirkwood et al., 2004). However, during the last decade no further progress concerning the adjuvant therapy of tumor patients has been achieved. Therefore, a better knowledge of the underlying molecular mechanisms of IFN action may lead to improved and more effective applications and the design of innovative, intelligent treatment strategies using IFNs alone or in combination with other therapeutics.
II. IFN Signal Transduction Pathways and Their Components
A wealth of information is available on the molecular processes underlying some of the IFN‐induced signaling cascades. Binding of IFNs to their specific receptors lacking intrinsic kinase activity induces oligomerization of receptor subunits triggering diverse signaling pathways (Fig. 1), thereby leading to the transcriptional regulation of a plethora of target genes (Kaur et al., 2005, Li et al., 2004, Schindler et al., 2007). The physiologic relevance of IFN‐dependent signal transduction cascades including the STAT/JAK pathway was established by generating and characterizing mice with targeted disruption of genes encoding STAT1/STAT2 or JAK1, respectively (Platanias, 2005, Ramana et al., 2002). Both type I and type II IFN receptors (IFN‐R) initiate the activation of the JAK/STAT cascade, which consists of four janus kinases (JAK1, JAK2, JAK3, and JAK4) and seven signal transducers and activators of transcription (STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, STAT5c; Shin‐Ya et al., 2005, Wright et al., 1995). STATs, SH2‐containing transcription factors, represent cytosolic proteins of 750–800 amino acids and are composed of (i) an extracellular domain that plays an important role in the association of STAT with receptor molecules, (ii) a ligand‐binding domain, and (iii) an intracellular domain that is responsible for the STAT dimer formation. STAT1 induces the expression of IFN‐responsive genes through the activation of IFN‐stimulated response element (ISRE)‐containing promoters (Yu and Jove, 2004). However, it has now become apparent that the activation of JAK–STAT pathways alone is not sufficient for the generation of all biological activities of IFNs. There exists accumulating evidence that several other IFN‐regulated signaling elements and cascades are required for the generation of many IFN responses. Some of them operate independently of the JAK–STAT pathway, whereas others cooperate with STATs to optimize the transcriptional regulation of target genes. These include in particular pathways linked to cellular stress and cell death like the mitogen‐activated protein kinase (MAPK), the stress‐induced kinase p38, and protein kinase C (PKC) signaling cascades. PKCs are known to be involved in both IFN‐α and ‐γ signaling pathways (Kwon et al., 2007). In this context, it is noteworthy that the IFN‐α, IFN‐β, and IFN‐γ cascades exhibit overlapping activities, but also clearly different features (Fig. 1; Levy et al., 1990).
A. IFN‐α‐Induced Signal Transduction Pathways
After the engagement with the type I IFN receptors (IFN‐αR), IFN‐α binding stimulates the cross‐linking between the IFN‐αR chain 1 (IFN‐αR1) and 2 (IFN‐αR2), thereby bringing the receptor‐associated kinases TYK2 and JAK1 into close proximity. This triggers the activation of JAK1 and TYK2 leading to the phosphorylation of Tyr‐466 of the IFN‐αR1, which serves as a docking site for STAT2. The activated kinase subsequently phoshorylates STAT2 and STAT1 on Tyr‐690 and Tyr‐701, respectively. Both phosphorylated STATs form a heterodimer and associate with the interferon regulatory factor (IRF)9, which does not undergo tyrosine phosphorylation to form the IFN‐stimulated gene factor 3 (ISGF3), which, in turn, translocates to the nucleus and binds specific elements known as ISREs that are present in the promoters of certain ISGs initiating the transcription of a broad variety of genes. In addition, phosphorylated STAT1, other STAT complexes, and combinations of different STAT‐containing complexes can be formed which translocate to the nucleus and bind to the IFN‐γ‐activated site (GAS) leading to the transcription of further genes (Caraglia et al., 2005). It is noteworthy that IFN‐α can also activate STAT3 and STAT5, but the role of STAT5 in the IFN‐α‐mediated activity has still to be elucidated (Uddin et al., 2003). In contrast, IFN‐γ mainly activates STAT5b. However, one can speculate that a fine balance between different STAT complexes might account for specific responses and represent a key mechanism for IFN‐α‐induced activities.
B. IFN‐γ Signal Transduction Cascade
IFN‐γ acts through a heterodimer consisting of the IFN‐γ receptor‐1 (IFN‐γR1) and IFN‐γR2 expressed on most cells, thereby upregulating specific genes. Binding of IFN‐γ initially leads to the formation of an IFN‐γR1 homodimer, which consecutively attracts the IFN‐γR2 chains. The IFN‐γR1 and ‐R2 homodimer is constitutively associated with JAK1 and JAK2, which phosphorylate the tyrosine 440 at the intracellular domain of the IFN‐γR1 serving as a docking site for the latent cytosolic transcription factor STAT1. STAT1 is subsequently phosphorylated on tyrosine 701 and serine 727 leading to the homodimerization of phospho‐STAT1 molecules. These form a complex named the γ‐activating factor (GAF) that translocates into the nucleus and upregulates the transcription of IFN‐γ‐regulated genes including in particular the interferon‐regulated factors (IRF)1 and IRF7 which represent transcriptional activators, whereas the constitutively expressed IRF2 generally acts as a transcriptional repressor (Harada et al., 1989). IRF1 subsequently activates the transcription of caspase genes involved in apoptosis next to genes encoded in the major histocompatibility complex (MHC) in particular components of the MHC class I and class II antigen‐processing machinery (APM) as well as β2‐microglobulin (β2‐m) located on chromosome 15. The molecules of the antigen‐processing pathway are required for the initiation and triggering of proper CD4+or CD8+T‐cell responses, respectively. In addition, STAT1 and IRF1 cooperate with the ubiquitously expressed transactivating factor upstream stimulatory factor (USF)1 to activate the transcription of the class II transactivator protein promoter IV (CIITA–PIV) that controls the expression of MHC class II molecules (Chen et al., 2007).
III. The MHC Class I and Class II Antigen‐Processing Pathways
The expression of MHC class I and class II molecules is critical for the presentation of antigens and essential for the generation of an adaptive immune response (Cresswell et al., 2005, Jensen, 2007). In the last decades, CD8+ cytotoxic T lymphocytes (CTL) have been implicated as main effector cells in antitumor responses. They recognize and attack tumor cells presenting intracellular antigens derived from different nonself peptides on their surface through the interaction of the T‐cell receptor (TCR) with MHC class I peptide complexes.
A. The MHC Class I Antigen‐Processing Pathway
The generation and presentation of these antigens (Ag) requires a coordinated expression of several genes (Fig. 2A ). Briefly, endogenously synthesized proteins are cleaved by the multicatalytic proteasome complex, in particular the IFN‐γ‐regulated proteasome subunits, such as the low molecular weight proteins (LMP)2, LMP7, and LMP10. These peptides are further trimmed by cytosolic enzymes such as, for example, the tripeptidyl peptidase (TPP)II and the bleomycin hydrolase (BH) generating the correct N‐terminus (Kloetzel, 2004, Rock et al., 2004). Then the peptides are transported from the cytosol into the endoplasmic reticulum (ER) via the transporter associated with antigen processing (TAP), a heterodimer consisting of the TAP1 and TAP2 subunits. In the lumen of the ER the MHC class I assembly occurs, which is assisted by various chaperones such as calnexin, calreticulin, the oxido thiol reductase ERp57, and tapasin (tpn). Tpn facilitates the peptide loading onto MHC class I molecules. After successful peptide loading, MHC molecules are released from the peptide loading complex and the trimer consisting of MHC class I heavy chain (HC)/β2‐m/peptide is then transported through the trans‐Golgi apparatus to the cell surface and presented to CD8+ CTL. Thus, proper expression of the major components of the complex MHC class I APM components is obligatory for effective T‐cell recognition of tumors (Groettrup et al., 1996, Jensen, 2007, Seliger et al., 2006).
Fig. 2.
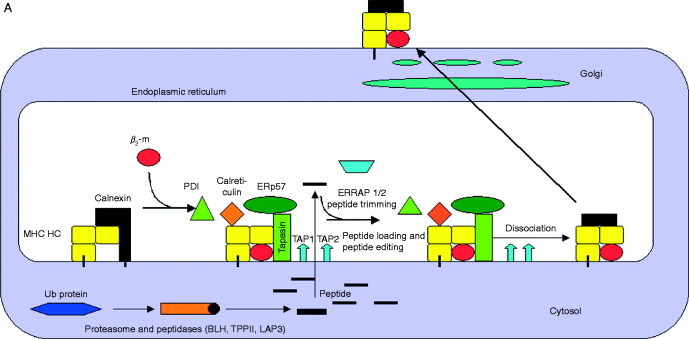
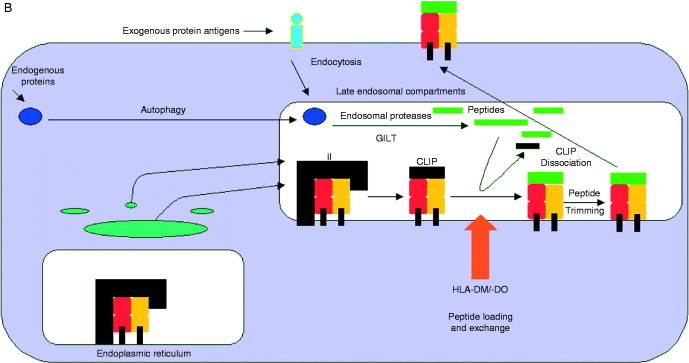
Schematic diagram of the MHC class I and class II APM. (A) MHC class I pathway. MHC class I heavy‐chain assembly with β2‐m, which is assisted by various chaperones such as calnexin and calreticulin. The MHC/β2‐m dimer is incorporated into the peptide loading complex (PLC) in the ER. In the cytosol, endogenous peptides are generated by the proteasome, which were further trimmed by other peptidases and then transported into the ER via the heterodimeric TAP. ERAP is involved in the final aminoterminal trimming of peptides. The loading of MHC class I molecules with peptides is further assisted by the chaperone tapasin which is also a component of the PLC. Upon peptide loading, the PLC dissociates and then transported via the trans Golgi to the cell surface and there exposed to CD8+ cytotoxic T lymphocytes. (B) MHC class II pathway. MHC class II molecules assemble in the ER with the invariant chain (li), which contains an endosomal targeting signal. This complex is then transported to the endosomal compartment and there the Ii is cleaved by a number of proteases leaving only the CLIP fragment, which occupies the peptide‐binding groove. HLA‐DM and ‐DO catalyze the release of CLIP, which is exchanged by antigenic peptides. HLA‐DM edit the repertoire of the MHC class II‐peptide complexes, which are then transported to the cell surface for recognition by CD4+ T lymphocytes. Exogenous proteins are internalized into the endosomal pathway by different mechanisms then unfolded and cleaved which is catalyzed by different proteases. In addition, the yielded peptides are further trimmed after binding to MHC class II molecules.
B. The MHC Class II Antigen‐Processing Pathway
Recently, it has been demonstrated that CD4+ T cells are also important for proper antitumor immune responses (Drozina et al., 2005, Jensen, 2007). These T cells recognize via their TCR antigens presented on MHC class II molecules. In contrast to MHC class I antigens which are expressed on all nucleated adult cells, the expression of the heterodimeric MHC class II molecules also representing transmembrane glycoproteins is highly restricted and preferentially found on the cell surface of professional antigen presenting cells (APCs). However, MHC class II antigen expression can be induced in other cell types by various cytokines, in particular IFN‐γ. MHC class II expression is mainly controlled by the class II transactivator protein (CIITA), which acts as a master regulator for its coordinated constitutive and IFN‐γ‐induced expression which also involves PKC delta (Kwon et al., 2007, Giroux et al., 2003). CIITA interacts with the transcription factors RFX, NFY, and CREB (van den Elsen et al., 2004), thereby forming an enhanceosome governing the MHC class II transcription. In addition, a coordinated expression of various MHC class II APM components exists. Mainly exogenous antigens are phagocytosed by APCs, directed then to lysosomes where they are cleaved into small peptide fragments (Fig. 2B). MHC class II antigens are assembled in the ER. The peptide‐binding groove of these molecules is initially occupied by the invariant chain which is degraded into the class II invariant chain peptide (CLIP) fragment by a series of key cleavage events, thereby protecting the MHC class II‐binding groove. The loading of MHC class II molecules with exogenously derived peptides is assisted by the chaperone‐like components HLA‐DM and ‐DO, which results in an exchange of the CLIP fragment by these antigens. HLA‐DM is editing the peptides presented to CD4+T cells by catalyzing multiple rounds of peptide exchanges possibly favoring the most stable complexes. The peptide‐loaded MHC class II molecules are then transported to the cell surface and presented to CD4+T lymphocytes. In professional APC, exogenous antigens can gain access to the MHC class I pathway through distinct cross‐presentation mechanisms. Furthermore, the endosomal MHC class II loading pathway could also receive peptides derived from endogenous antigens through autophagy and other mechanisms (Dengjel et al., 2005, Schmid et al., 2007).
C. Regulatory Elements of the MHC Class I and Class II APM Promoters and their IFN Inducibility
The promoters of the MHC class I and class II APM components have been intensely characterized and exert some similarities, but also unique properties. Concerning the promoter of MHC class I APM components, some of them contain TATA and CAAT boxes, whereas others completely lack these regulatory domains in the promoters). In addition, it is noteworthy that both TAP1 and LMP2 are transcribed from a shared bidirectional promoter of only 596 base pairs (bp) separating their ATG translation initiation codon (Wright et al., 1995). The promoter of the major MHC class I APM components contain a combination of distinct transcription factor‐binding sites (TFBS), like Sp1, CREB, the nuclear factor (NF)‐κB, E2F, and p300, but all exhibit IFN‐response elements, which hint toward their regulation by IRFs (Brucet et al., 2004, Chatterjee‐Kishore et al., 1998, Chatterjee‐Kishore et al., 2000; Fig. 3). In terms of the MHC class II pathway, the promoters of the invariant chain, HLA‐DM/‐DO and the MHC class II HC, respectively, contain similar, but also distinct transcription factor‐binding sites, whereas all of them contain an IFN‐response element in their promoter. An exception is represented by CIITA, which is regulated by multiple promoters differing in their TFBS composition. There exist three tissue‐specific promoters for CIITA, pI, pII, and pIII. One promoter controls the constitutive CIITA expression in dendritic cells (DC), whereas another is specific for the constitutive expression in B cells. The CIITA–PIV regulates the induction of CIITA expression in different cell types. It contains several cis elements including a putative NF‐κB site overlapping with an AP2 site, the IFN‐γ activating sequence (GAS), the E box, and an IRF element (Dong et al., 1999, Muhlethaler‐Mottet et al., 1998). Thus, the activity of the different MHC class I and II APM component promoters can be induced, but to a different extent, by type I and type II IFNs, respectively. IFN‐γ is a stronger inducer when compared to type I IFNs, whereas a combination of both substances exerts additive or even synergistic effects on MHC class I and II APM components. Activation of adaptive immune responses by IFN, in particular IFN‐γ, is partially due to transcriptional activation of genes encoding the MHC class I and class II antigens and respective APM components such as the invariant chain, HLA‐DM/‐DO, CIITA, TAP, tpn, the LMPs, and ERAP1/2.
Fig. 3.
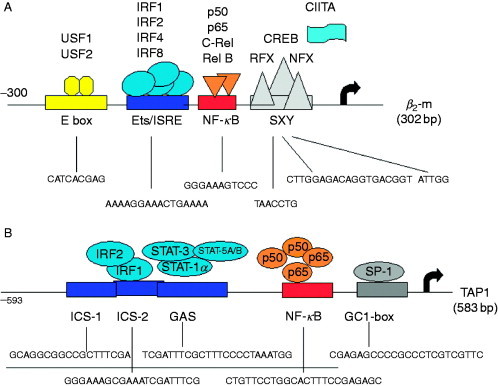
Promoter structure of major APM components. The structure of representative promoters of the major APM components is schematically illustrated, demonstrating a number of transcription factor‐binding sites such as NF‐κB, AP1, SP1, and CREB as well as interferon regulatory response elements (ISRE), which are involved in the inducibility by this cytokines.
D. Mechanisms of Impaired MHC Class I and APM Component Expression in Tumors
Decrease in or absence of MHC class I molecules has been observed in a diversity of human tumor types (Garrido, 2001, Garrido et al., 1993, Garrido et al., 1997). An increasing proportion of tumors were found with total or selective HLA allelic losses supporting the theory that altered HLA expression phenotypes represent a major mechanism of tumor escape from T‐cell recognition due to downmodulation of presentation of immunodominant tumor antigens. Distinct HLA class I abnormalities, including total loss or downregulation of HLA class I antigens (Paschen et al., 2003), HLA haplotype loss (Ramal et al., 2000), HLA locus or allele loss (Jimenez et al., 2001) has been described in tumors originating from different tissues and multiple molecular mechanisms have been identified as responsible for these changes (Garrido and Algarra 2001). The mechanisms that underlie total or partial loss of HLA class I antigens (Table II ) include mutations of the β2‐microglobulin (β2‐m) gene (Perez et al., 1999) and loss of heterozygosity (LOH) of MHC genes (Maleno et al., 2004). Other causes of total HLA class I downregulation comprise defects in the regulation of different components of the MHC class I antigen processing. Structural defects of APM components cannot be corrected by cytokine treatment; therefore, it does not restore HLA class I surface antigen expression. T‐cell‐based therapy may not be effective due to the irreversible loss of HLA class I molecules. This is important when selecting the appropriate immunotherapy for a given cancer patient.
Table II.
Mechanisms of Impaired MHC Class I Expression
|
Abnormalities in the expression of various MHC class I APM components occur at a high frequency in human tumors of distinct origin like small‐cell lung carcinoma (SCLC), melanoma, colon carcinoma, breast carcinoma, renal cell carcinoma, and hematological malignancies and are frequently associated with malignant transformation (Table II). This phenotype allows the tumor cells to evade recognition by MHC class I‐restricted, tumor antigen (TA)‐specific CTL. Mutations in different APM components appear to be a rare event postulating that dysregulation rather than structural alterations is the major cause for aberrant APM component expression (Fernandez et al., 2000, Ramal et al., 2000, Seliger et al., 2006; Table II). This hypothesis is supported by experiments (i) identifying only few mutations in these molecules, (ii) characterizing the APM promoter activity in tumors, (iii) determining posttranscriptional regulatory mechanisms, and (iv) treating tumor cells with IFNs to analyze whether deficiencies of APM component expression could be overcome by cytokines. Indeed, impaired APM component expression of tumor cells could be often restored by IFN‐α/β and/or IFN‐γ treatment. The IFN‐mediated upregulation of APM components often results in enhanced MHC class I surface expression, which is required for the generation of an effective antitumor‐specific immune response. Indeed, the IFN‐induced upregulation of APM components improves antitumor‐specific CTL responses (Seliger et al., 1997, Tajima et al., 2004) and therefore represent a valuable strategy for the treatment of patients with APM component deficiencies. However, in some cases, tumors remain insensitive to IFN treatment despite the lack of structural alterations in APM components, rather suggesting an impaired IFN signal transduction.
IV. Defective IFN Inducibility of APM Components in Tumors
A. Frequency of IFN Resistance
The unresponsiveness to IFN treatment was analyzed in a number of different tumor types and according to Kaplan et al. (1998) can be frequently found in human cancers. Approximately 33% of 33 melanoma and 17 non‐adenocarcinoma lung tumor cell lines analyzed exhibit a quantitative reduction in IFN‐γ sensitivity, while 2 out of 17 lung adenocarcinoma cell lines were totally unresponsive to IFN‐γ. These data were extended in a recent study in which 57 melanoma cell lines were analyzed for the ability to upregulate MHC class I surface antigens in response to stimulation with IFN‐γ. A total unresponsiveness to IFN‐γ was found in 2 out of 57 melanoma cell lines (Rodriguez et al., 2007b). However, the number of tumor types and tumor samples analyzed for IFN resistance is still limited and requires further studies in order to determine the frequency, relevance, and molecular mechanisms of these deficiencies. It is noteworthy that an impaired IFN‐γ response despite a functional IFN‐α induction may exist. On the other hand, a lack of IFN‐α responsiveness can also be found in the presence of IFN‐γ sensitivity, suggesting that the IFN signal transduction cascades are not coordinately regulated in tumor cells.
B. Mechanisms of IFN Insensitivity
The importance and involvement of IFN signal transduction pathways in the transcriptional regulation of APM promoters have been established, but there exists only limited information about the underlying molecular mechanisms of defective IFN‐inducible APM component expression. The impairment could occur at different steps along the IFN signal transduction pathways and might involve sequence abnormalities and/or different regulatory processes such as transcriptional, posttranscriptional, and epigenetic control (Fig. 1; Table III ). The physiological relevance of the STAT/JAK and PI3K pathway has been established in mice with a targeted disruption of these genes. The lack of JAK1 activity was associated with a loss of IFN‐γ to induce growth arrest and apoptosis as well as an increased tumorgenicity (Sexl et al., 2003). However, the observed IFN‐γ response with respect to growth inhibition might also be attributable to the IFN‐γ inducibility of LMP2 (Hayashi et al., 2006). So far, there exists only limited information regarding the molecular mechanisms of IFN resistance in tumors (Huang et al., 2002, Lesinski et al., 2007, Wellbrock et al., 2005, Wong et al., 1997). Based on the current knowledge that STAT1 and IRF1 are involved in the transcriptional regulation of the dual TAP1 and LMP2 promoter, the loss of TAP1 and LMP2 expression may be attributable to deficiencies of these regulatory factors. Regarding the IFN‐α resistance of RCC cell lines, it is associated with a defective induction of STAT1 that could be restored by the addition of a supernatant from PMA‐stimulated peripheral mononuclear cells (Brinckmann et al., 2002). This effect appears to be mediated by IFN‐γ although other cytokines might also be involved in this process.
Table III.
Defects Involved in IFN Resistance
|
In addition, the loss of the IFN‐γ‐mediated upregulation of MHC class I APM components in some RCC cell lines appears to be due to the lack of IRF1‐ and STAT1‐binding activities upon IFN‐γ stimulation. The STAT1, JAK1, and JAK2 proteins were expressed but not phosphorylated in the presence of IFN‐γ. The IFN‐γ‐mediated inducibility was not restored by gene transfer of JAK1 and/or JAK2 into RCC cells, whereas JAK1 overexpression increased both TAP1 and LMP2 expression independent of IFN‐γ. Therefore, the loss of TAP1 and LMP2 induction was associated with a defect of an early step in the IFN‐γ signal transduction pathway (Dovhey et al., 2000).
Furthermore, an association of impaired STAT1 phosphorylation with the loss of IFN‐mediated HLA class I induction was also found in melanoma cell lines (Rodriguez et al., 2007b). The absence of STAT1 phosphorylation was at least partially due to the constitutive expression of the suppressor of cytokine signaling (SOCS)‐1 protein, which could be mediated by the JAK2 kinase inhibition via the SOCS phosphatase. SOCS‐1 modulates the IFN‐γ‐mediated signaling by binding to the autophosphorylation site of JAK2 and by targeting bound JAK2 to the proteasome for degradation (Waiboci et al., 2007). In addition, SOCS‐1 expression correlates with melanoma progression and confers growth advantage (Komyod et al., 2007, Li et al., 2004). In another study, the IFN‐γ resistance was associated with SOCS3 expression. The resistant cell lines differed from the sensitive cells by a constitutive expression of SOCS3, by the absence or a low degree of SOCS1–3 activation following IFN‐γ treatment, and by a short duration of the cytokine activatory signal (Fojtova et al., 2007). The expression of IFN‐γ‐responsive genes is also reduced in the choriocarcinoma cells JEG3 and JAR in comparison to the epithelial cell line Hela (Choi et al., 2007). This is mediated by a compromised tyrosine phosphorylation of JAK2 and STAT1 at tyrosine 701 and the reduced expression of IRF1. In addition, inhibition of the tyrosine phosphatases results in increased JAK1 and STAT1 phosphorylation and IFN‐γ‐induced gene expression in these cells (Choi et al., 2007). The impaired expression of IRF1 and deficient phosphorylation of STAT1 were also observed in primary trophoblast cell lines suggesting that these defects are of clinical relevance.
Besides the posttranslational regulation of components of the IFN signal cascades, the absence of the IFN‐γ‐mediated MHC class I expression can be controlled by epigenetic alterations in this pathway. Indeed, methylation affects the binding of IRF1 leading to an abrogation of the IRF1 transactivation (Rodriguez et al., 2007b). Treatment with the demethylating agent 2′5′‐deoxyazacytidine (DAC) restored the IRF1 expression and consecutively led to the reconstitution of the IFN‐γ‐mediated MHC class I inducibility. Other studies have identified that the IFN unresponsiveness is attributed to low expression of STAT1 rather than to an absence of its phosphorylation (Abril et al., 1998, Xi et al., 2006). The absence of STAT1 expression has been correlated with the methylation of its promoter (Xi et al., 2006). Finally, there exists evidence that genetic instability in tumor cells may lead to modulation of the expression of the IFN‐γR, which in some cases has been reported to be associated with cancer prognosis. For instance, the loss of IFN‐γR independently predicts poor prognosis in ovarian cancer and may be responsible for the limited success in the outcome of treatment of ovarian cancer with IFN‐γ (Duncan et al., 2007).
C. Involvement of the IFN Pathways in Tumor Surveillance In Vivo
The multiple activities of IFNs on tumor cells might coordinate the antitumor immune responses so that the early recognition and/or elimination of cancer cells by the innate immune system transitions to immune attack by the adaptive immune system (Dunn et al., 2006). The IFN‐γ on the tumor cell immunogenicity mediate the immune response directed against tumor cells through distinct mechanisms. IFN‐γ can downregulate the expression of the NKG2D ligands and at the same time increase the expression of MHC class I molecules (Bui et al., 2006). In vitro treatment with IFN‐γ decreased the death by NK cells independently from the expression of HLA class I molecules, whereas an increased MHC class I expression increased the sensibility CTL‐mediated lysis. Besides these in vitro results, there also exist information that abnormalities in the IFN signaling occurs in vivo. LMP2–/– mice exhibit an impaired proteasome function and 36% of female LMP2–/– mice develop uterine leiomyosarcomas by 12 months of age. Thus, the development of spontaneous human uterine leiomyosarcomas might be probably due to defects in early steps of the IFN signal cascade. Indeed, the defective TAP1 and LMP2 expression in these tumors is associated with a G871E mutation in the ATP‐binding region of the JAK1 kinase domain, thereby affecting JAK1 kinase activity, but neither JAK1 expression and production nor its degradation (Hayashi et al., 2006). This allows the tumor cells to evade antitumor‐specific immunity. In different tumor types, immunosuppression associated with STAT3 activation and STAT3‐mediated inhibition of DC function has been reported (Yu and Jove, 2004). The biological function of STAT1 and STAT3 differs in terms of cell growth and induction of an antitumor immune response. Whereas STAT1 abrogates growth and mediates antitumor effects, STAT3 promotes cell proliferation and tumorigenicity as it has been shown in melanoma and head neck squamous carcinoma. In both tumor entities, STAT3 expression is associated with tumor progression and mediates immune suppression. In addition, unphosphorylated or phosphorylated STAT1 and STAT3 are coordinately upregulated by both IFN‐α and IFN‐γ and may represent a marker for the dynamic mechanism of melanoma progression and host response. Using methylcholanthrene (MCA) and untreated IFN‐γR–/–, a significant tumor development was observed in the IFN‐γR control mice. The crossing of IFN‐γR1 and STAT1–/– mice with p53–/– mice resulted in a spontaneous and more rapid tumor development in particular teratomas, hemangiomas, and chondrocytomas, whereas lymphoid tumors generally develop in IFN‐γ‐sensitive p53–/– mice. Interestingly, the IFN‐γ‐sensitive tumor cells transfected with the dominant negative IFN‐γR mutant grew faster than untransfected tumors and were not rejected upon their treatment with lipopolysaccharide (LPS) effectively eliminating control tumors (Marques et al., 2004). Furthermore, downregulation of the IFN‐γR in association with loss of Fas function is linked to tumor progression (Yang et al., 2008). Thus, the IFN‐γ responsiveness is an important mechanism in the control of tumor growth. An increased responsiveness to metastases‐promoting agents might be induced by many mediators in the microenvironment of melanoma including type I and type II IFNs. Both cytokines cooperate with TNF‐α, which involves a positive interplay between JAK1 and PKC signal transduction (Bianchini et al., 2006). These data suggest that multiple signals were generated by the host inflammatory cells, which are accompanied by cooperate with the invasive properties of tumor cells. Therefore, strategies targeting this cross‐talk among tumor and host cells in the microenvironment are needed to prevent tumor growth.
D. Effect of IFN Signaling on MHC Class II Components
The chimeric RET/PTC (rearranged in transformation/papillary thyroid carcinoma) oncoproteins were constitutively expressed in papillary thyroid cancer and are able to phosphorylate the Y107 of STAT1, which is accompanied by IRF1 expression (Hwang et al., 2004). This is associated with an enhanced transcription of CIITA and consequently with MHC class II expression of papillary thyroid carcinoma cells and explain the immune cell infiltration of RET/PTC‐positive cancers. Furthermore, a synergistic activity of TNF‐α and IFN‐γ on CIITA was found in thyroid carcinoma (Rahat et al., 2001) The CIITA‐independent MHC class I expression could be upregulated by histone deacetylases like Trichostatin A (TSA) (Chou et al., 2005, Gialitakis et al., 2006). CIITA was refractory to IFN induction in many tumors. In colorectal and gastric carcinoma cells, CIITA is silenced by epigenetic mechanisms resulting in the lack of IFN‐γ‐induced MHC class II expression (Satoh et al., 2004). In order to correlate the IFN unresponsiveness with the expression profile of ISGs, cDNA microarray analyses were employed using a customized microarray consisting of 850 ISG (Holko and Williams, 2006). Expression of genes associated with transcription precedes the expression of genes involved in signal transduction, whereas no differences in the STAT1 induction were observed. However, subtle alterations in the expression profile might be responsible for the insensitivity to this cytokine. The maintenance of transcriptional activation following IFN treatment appeared to enhance IFN sensitivity.
V. Clinical Relevance of Aberrant IFN Signaling
IFNs have been used in various clinical settings, since they are potent negative regulators of cell growth either by modulating the cell cycle or by inducing pro‐apoptotic genes. IFN‐α has been extensively studied in the treatment of various malignancies during the last two decades demonstrating improved clinical outcome of hematological malignancies (chronic myeloid leukemia, cutaneous T‐cell lymphoma, hairy‐cell leukemia, multiple myeloma), solid tumors including malignant melanoma, renal‐cell carcinoma (RCC), AIDS‐related Kaposi's sarcoma, and viral syndromes (hepatitis C, hepatitis B, severe acute respiratory syndrome). IFN‐γ has shown positive results in the treatment of chronic granulomatous disease, multiple sclerosis, and severe malignant osteopetrosis (Parmar and Platanias, 2003, for review). However, the resistance to IFNs has been described, which limits their anticancer activity. The impaired expression of IFN‐responsive genes might have important implications not only in immunotherapy but also in transplantation, pregnancy, and the development of tumors such as choriocarcinoma. Despite proven clinical efficacy in malignancies, viral infections, and multiple sclerosis, a substantial number of patients fail to develop positive clinical response to IFN therapy. Although IFN‐α2b is a clinically active therapeutic agent for malignant melanoma and RCC, only 15–20% patients with metastatic melanoma respond to IFN therapy (Marincola et al., 1995). Other reviews report even lower response rates of only 6% of treated melanoma patients (Quesada et al., 1985, Umeda and Niijima, 1986). In RCC, the best results of IFN treatment as determined by the response rate and the duration of the effect were obtained in patients with a previous nephrectomy without chemotherapy, in a good functional state, and with preferentially lung metastasis. In these patients the survival rate increased from 49 to 115 weeks upon IFN‐α administration (Logothetis, 1992). Despite these positive results, there exist many aspects of these response factors which are not well understood. Actually, none of these factors has been proved to be associated in an unambiguous way with the cytokine response and the patients' survival. The key aspect may be the right selection of patients, since currently all of them independent of previous nephrectomy and the presence of metastasis are enrolled into the treatment with poor clinical outcome. Unlike type I IFNs, IFN‐γ has not been approved for cancer treatment by the FDA. IFN‐γ produces numerous antitumor effects and plays a central role in promoting natural immune responses directed against developing tumors. However, its practical application in immunotherapeutic protocols has been very limited. In clinical trials, an improved survival was observed in patients with ovarian cancer of stage Ic–IIIc treated with IFN‐γ (Windbichler et al., 2000), when IFN was intravesically administered to patients with transitional‐cell bladder carcinoma (Giannopoulos et al., 2003) or when IFN was used in isolated‐limb perfusion of individuals with non‐melanoma cancers of the extremities (Lienard et al., 1998). However, no effect was detected upon IFN‐γ treatment of patients with metastatic RCC (Gleave et al., 1998), advanced colon cancer (Wiesenfeld et al., 1995), or small‐cell lung cancer (Jett et al., 1994). The limited success of the therapeutic use of IFN‐γ might reflect the inability to target IFNs in the right place with an efficient concentration (Dunn et al., 2006). Despite the proven pivotal role of endogenously produced antitumor immunity of IFN‐γ in animal models, the limited success of this cytokine in cancer immunotherapy trials in humans might be explained by the resistance of tumor cells to IFN‐γ (Kaplan et al., 1998, Rodriguez et al., 2007a, Wong et al., 1997). In this context, it is important to note that unlike type I IFNs, IFN‐γ has a direct effect on tumor cells during the antitumor immune response supporting the relevance of IFN‐γ in the cancer immunoediting process (Dunn et al., 2004). The targets of the immunologic unresponsiveness represent genes encoding components of the MHC APM components or the constituents of the IFN‐γR signaling pathway. In this context, in two recent studies from our laboratory, the physiological relevance of HLA class I surface expression during the tumor rejection process in patients receiving different protocols of immunotherapy was assessed (Cabrera et al., 2007; Carretero et al., submitted). In the first study, a significant difference in the immunotherapeutic response of patients exhibiting metastases with low levels of MHC class I surface antigens and those with high levels of MHC class I expression was detected. In a second trial, the impact of cytokine unresponsiveness was demonstrated by determination of HLA class I antigen expression levels on metastatic melanoma lesions during the course of the disease in one patient undergoing IFN‐α 2b and autologous vaccination plus BCG (M‐VAX). BCG triggers the IL‐12/IFN‐γ axis and induces upregulation of genes associated with antigen presentation (Feinberg et al., 2004, Saban et al., 2007). The level of the MHC class I antigen expression was dependent on the IFN response since neither of the progressor metastases increased the expression of HLA class I antigens after vaccination. However, a significant increase in the HLA class I surface expression was detected in the regressor metastases. Therefore, the HLA class I surface antigen on tumor cells significantly contributed to the therapeutic effect of BCG. In connection with these findings, downregulation of HLA class I surface antigens in cancer cells has been considered a significant risk factor for recurrence in patients with intravesical BCG immunotherapy for bladder cancer (Kitamura et al., 2006).
Based on these results, a better understanding of the molecular mechanisms by which tumors modulate the cytokine signaling may be essential for the development of immunotherapeutic strategies with the aim to enhance MHC class I surface antigen expression in tumor cells. The balance of STAT phosphorylation versus SOCS expression might be crucial in the activation of immunologic response through APM and MHC class I transactivation (Wang et al., 2007). For instance, the effects of high‐dose IFN are associated with immunologic processes such as an upregulation of TAP1, TAP2, tpn, and LMP2. The STAT1 and STAT2 pathways in melanoma cells are sensitized to IFN‐α by pretreatment of the cells with IFN‐γ. Thus, the biological response to IFN‐α might be mediated by a direct effect on melanoma cells and suggests also a potential role for IFN‐γ in the treatment of this disease (Carson, 1998). In addition, it has recently been demonstrated that IFN‐α treatment of patients with cutaneous melanoma significantly modulates the balance of STAT1/STAT3 in tumor cells and host lymphocytes. This results in an upregulation of TAP2 and an increased immune response (Wang et al., 2007).
VI. Conclusions
An increased knowledge of the factors responsible for the resistance to IFNs might lead to an improved use of these cytokines in malignant diseases. The application of the molecular analysis of tumor tissues has now advanced to the point where better classification schemes and prognostic variables are used leading to an optimization of specific treatment programs and patients' selection. The identification of tumor lesions with the capacity to upregulate MHC gene expression will determine the ability to present new antigenic peptides to T lymphocytes favoring regression of primary or metastatic tumor lesions. In contrast, the identification of tumors with MHC irreversible genetic lesions will maintain an unaltered MHC expression, thereby not exposing new antigenic peptides to T cells, which subsequently favors tumor and/or metastases progression. We propose that suppression of IFN signaling in tumors contributes to tolerance by inhibiting expression of genes encoding subunits of HLA class I/II antigens and/or components of the MHC class I/II APM that could be detrimental to successful antitumor responses.
Acknowledgments
This work was supported by grants from the Fondo de Investigaciones Sanitarias (FIS), Red Genomica del Cancer (RETIC RD 06/0020), Plan Andaluz de Investigacion (Group CTS 143), Consejeria Andaluz de Salud (SAS), Proyecto de Excelencia de Consejeria de Innovacion (CTS 695), Proyecto de investigacion I+D (SAF 2007–63262) in Spain; and from the Integrated European Cancer Immunotherapy project (OJ2004/C158, 518234) and by grants from the Deutsche Forschungsgemeinschaft DFG SE581 9‐1/2 and 11‐1 (B.S). In addition, we thank Tarish Abbas for providing Figure 3 and Anne Wasilewski for excellent secretarial help.
References
- Abril E., Real L.M., Serrano A., Jimenez P., Garcia A., Canton J., Trigo I., Garrido F., Ruiz‐Cabello F. Unresponsiveness to interferon associated with STAT1 protein deficiency in a gastric adenocarcinoma cell line. Cancer Immunol. Immunother. 1998;47:113–120. doi: 10.1007/s002620050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadori M. The role of IFN‐alpha as homeostatic agent in the inflammatory response: A balance between danger and response? J. Interferon Cytokine Res. 2007;27:181–189. doi: 10.1089/jir.2006.0110. [DOI] [PubMed] [Google Scholar]
- Baccala R., Kono D.H., Theofilopoulos A.N. Interferons as pathogenic effectors in autoimmunity. Immunol. Rev. 2005;204:9–26. doi: 10.1111/j.0105-2896.2005.00252.x. [DOI] [PubMed] [Google Scholar]
- Bianchini F., Mannini A., Mugnai G., Ruggieri S., Calorini L. Expression of a metastatic phenotype in IFNs‐primed/TNFalpha‐activated B16 murine melanoma cells: Role of JAK1/PKCdelta signal transduction factors. Clin. Exp. Metastasis. 2006;23:203–208. doi: 10.1007/s10585-006-9030-1. [DOI] [PubMed] [Google Scholar]
- Brinckmann A., Axer S., Jakschies D., Dallmann I., Grosse J., Patzelt T., Bernier T., Emmendoerffer A., Atzpodien J. Interferon‐alpha resistance in renal carcinoma cells is associated with defective induction of signal transducer and activator of transcription 1 which can be restored by a supernatant of phorbol 12‐myristate 13‐acetate stimulated peripheral blood mononuclear cells. Br. J. Cancer. 2002;86:449–455. doi: 10.1038/sj.bjc.6600066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucet M., Marques L., Sebastian C., Lloberas J., Celada A. Regulation of murine Tap1 and Lmp2 genes in macrophages by interferon gamma is mediated by STAT1 and IRF‐1. Genes Immun. 2004;5:26–35. doi: 10.1038/sj.gene.6364035. [DOI] [PubMed] [Google Scholar]
- Bui J.D., Carayannopoulos L.N., Lanier L.L., Yokoyama W.M., Schreiber R.D. IFN‐dependent down‐regulation of the NKG2D ligand H60 on tumors. J. Immunol. 2006;176:905–913. doi: 10.4049/jimmunol.176.2.905. [DOI] [PubMed] [Google Scholar]
- Cabrera T., Lara E., Romero J.M., Maleno I., Real L.M., Ruiz‐Cabello F., Valero P., Camacho F.M., Garrido F. HLA class I expression in metastatic melanoma correlates with tumor development during autologous vaccination. Cancer Immunol. Immunother. 2007;56:709–717. doi: 10.1007/s00262-006-0226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraglia M., Marra M., Pelaia G., Maselli R., Caputi M., Marsico S.A., Abbruzzese A. Alpha‐interferon and its effects on signal transduction pathways. J. Cell. Physiol. 2005;202:323–335. doi: 10.1002/jcp.20137. [DOI] [PubMed] [Google Scholar]
- Carson W.E. Interferon‐alpha‐induced activation of signal transducer and activator of transcription proteins in malignant melanoma. Clin. Cancer Res. 1998;4:2219–2228. [PubMed] [Google Scholar]
- Chatterjee‐Kishore M., Kishore R., Hicklin D.J., Marincola F.M., Ferrone S. Different requirements for signal transducer and activator of transcription 1alpha and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J. Biol. Chem. 1998;273:16177–16183. doi: 10.1074/jbc.273.26.16177. [DOI] [PubMed] [Google Scholar]
- Chatterjee‐Kishore M., Wright K.L., Ting J.P., Stark G.R. How Stat1 mediates constitutive gene expression: A complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Gilbert C.A., Hudson J.A., Bolick S.C., Wright K.L., Piskurich J.F. Positive regulatory domain I‐binding factor 1 mediates repression of the MHC class II transactivator (CIITA) type IV promoter. Mol. Immunol. 2007;44:1461–1470. doi: 10.1016/j.molimm.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.C., Holtz R., Petroff M.G., Alfaidy N., Murphy S.P. Dampening of IFN‐gamma‐inducible gene expression in human choriocarcinoma cells is due to phosphatase‐mediated inhibition of the JAK/STAT‐1 pathway. J. Immunol. 2007;178:1598–1607. doi: 10.4049/jimmunol.178.3.1598. [DOI] [PubMed] [Google Scholar]
- Chou S.D., Khan A.N., Magner W.J., Tomasi T.B. Histone acetylation regulates the cell type specific CIITA promoters, MHC class II expression and antigen presentation in tumor cells. Int. Immunol. 2005;17:1483–1494. doi: 10.1093/intimm/dxh326. [DOI] [PubMed] [Google Scholar]
- Cresswell P., Ackerman A.L., Giodini A., Peaper D.R., Wearsch P.A. Mechanisms of MHC class I‐restricted antigen processing and cross‐presentation. Immunol. Rev. 2005;207:145–157. doi: 10.1111/j.0105-2896.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- Decker T., Müller M., Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- Dengjel J., Schoor O., Fischer R., Reich M., Kraus M., Muller M., Kreymborg K., Altenberend F., Brandenburg J., Kalbacher H., Brock R., Driessen C. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Rohn W.M., Benveniste E.N. IFN‐gamma regulation of the type IV class II transactivator promoter in astrocytes. J. Immunol. 1999;162:4731–4739. [PubMed] [Google Scholar]
- Dovhey S.E., Ghosh N.S., Wright K.L. Loss of interferon‐gamma inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line. Cancer Res. 2000;60:5789–9576. [PubMed] [Google Scholar]
- Drozina G., Kohoutek J., Jabrane‐Ferrat N., Peterlin B.M. Expression of MHC II genes. Curr. Top. Microbiol. Immunol. 2005;290:147–170. doi: 10.1007/3-540-26363-2_7. [DOI] [PubMed] [Google Scholar]
- Duncan T.J., Rolland P., Deen S., Scott I.V., Liu D.T., Spendlove I., Durrant L.G. Loss of IFN gamma receptor is an independent prognostic factor in ovarian cancer. Clin. Cancer Res. 2007;13:4139–4145. doi: 10.1158/1078-0432.CCR-06-2833. [DOI] [PubMed] [Google Scholar]
- Dunn G.P., Old L.J., Schreiber R.D. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Dunn G.P., Koebel C.M., Schreiber R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- Feinberg J., Fieschi C., Doffinger R., Feinberg M., Leclerc T., Boisson‐Dupuis S., Picard C., Bustamante J., Chapgier A., Filipe‐Santos O., Ku C.L., de Beaucoudrey L. Bacillus Calmette Guerin triggers the IL‐12/IFN‐gamma axis by an IRAK‐4‐ and NEMO‐dependent, non‐cognate interaction between monocytes, NK, and T lymphocytes. Eur. J. Immunol. 2004;34:3276–3284. doi: 10.1002/eji.200425221. [DOI] [PubMed] [Google Scholar]
- Fernandez M.A., Ruiz‐Cabello F., Oliva M.R., Cabrera T., Jimenez P., Lopez Nevot M.A., Garrido F. Beta2‐microglobulin gene mutation is not a common mechanism of HLA class I total loss in human tumors. Int. J. Clin. Lab. Res. 2000;30:87–92. doi: 10.1007/BF02874164. [DOI] [PubMed] [Google Scholar]
- Fojtova M., Boudny V., Kovarik A., Lauerova L., Adamkova L., Souckova K., Jarkovsky J., Kovarik J. Development of IFN‐gamma resistance is associated with attenuation of SOCS genes induction and constitutive expression of SOCS 3 in melanoma cells. Br. J. Cancer. 2007;97:231–237. doi: 10.1038/sj.bjc.6603849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido F., Algarra I. MHC antigens and tumor escape from immune surveillance. Adv. Cancer Res. 2001;83:117–158. doi: 10.1016/s0065-230x(01)83005-0. [DOI] [PubMed] [Google Scholar]
- Garrido F., Cabrera T., Concha A., Glew S., Ruiz‐Cabello F., Stern P.L. Natural history of HLA expression during tumour development. Immunol. Today. 1993;14:491–499. doi: 10.1016/0167-5699(93)90264-L. [DOI] [PubMed] [Google Scholar]
- Garrido F., Ruiz‐Cabello F., Cabrera T., Perez‐Villar J.J., Lopez‐Botet M., Duggan‐Keen M., Stern P.L. Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol. Today. 1997;18:89–95. doi: 10.1016/s0167-5699(96)10075-x. [DOI] [PubMed] [Google Scholar]
- Gialitakis M., Kretsovali A., Spilianakis C., Kravariti L., Mages J., Hoffmann R., Hatzopoulos A.K., Papamatheakis J. Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by Trichostatin A. Nucleic Acids. Res. 2006;34:765–772. doi: 10.1093/nar/gkj462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannopoulos A., Constantinides C., Fokaeas E., Stravodimos C., Giannopoulou M., Kyroudi A., Gounaris A. The immunomodulating effect of interferon‐gamma intravesical instillations in preventing bladder cancer recurrence. Clin. Cancer Res. 2003;9:5550–5558. [PubMed] [Google Scholar]
- Giroux M., Schmidt M., Descoteaux A. IFN‐gamma‐induced MHC class II expression: Transactivation of class II transactivator promoter IV by IFN regulatory factor‐1 is regulated by protein kinase C‐alpha. J. Immunol. 2003;171:4187–4194. doi: 10.4049/jimmunol.171.8.4187. [DOI] [PubMed] [Google Scholar]
- Gleave M.E., Elhilali M., Fradet Y., Davis I., Venner P., Saad F., Klotz L.H., Moore M.J., Paton V., Bajamonde A. Interferon gamma‐1b compared with placebo in metastatic renal‐cell carcinoma. Canadian Urologic Oncology Group. N. Engl. J. Med. 1998;338:1265–1271. doi: 10.1056/NEJM199804303381804. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Soza A., Kuckelkorn U., Kloetzel P.M. Peptide antigen production by the proteasome: Complexity provides efficiency. Immunol. Today. 1996;17:429–435. doi: 10.1016/0167-5699(96)10051-7. [DOI] [PubMed] [Google Scholar]
- Harada H., Fujita T., Miyamoto M., Kimura Y., Maruyama M., Furia A., Miyata T., Taniguchi T. Structurally similar but functionally distinct factors, IRF‐1 and IRF‐2, bind to the same regulatory elements of IFN and IFN‐inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Kobayashi Y., Kohsaka S., Sano K. The mutation in the ATP‐binding region of JAK1, identified in human uterine leiomyosarcomas, results in defective interferon‐gamma inducibility of TAP1 and LMP2. Oncogene. 2006;25:4016–4026. doi: 10.1038/sj.onc.1209434. [DOI] [PubMed] [Google Scholar]
- Holko M., Williams B.R. Functional annotation of IFN‐alpha‐stimulated gene expression profiles from sensitive and resistant renal cell carcinoma cell lines. J. Interferon Cytokine Res. 2006;26:534–547. doi: 10.1089/jir.2006.26.534. [DOI] [PubMed] [Google Scholar]
- Huang S., Bucana C.D., Van Arsdall M., Fidler I.J. Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene. 2002;21:2504–2512. doi: 10.1038/sj.onc.1205341. [DOI] [PubMed] [Google Scholar]
- Hwang E.S., Kim D.W., Hwang J.H., Jung H.S., Suh J.M., Park Y.J., Chung H.K., Song J.H., Park K.C., Park S.H., Yun H.J., Kim J.M. Regulation of signal transducer and activator of transcription 1 (STAT1) and STAT1‐dependent genes by RET/PTC (rearranged in transformation/papillary thyroid carcinoma) oncogenic tyrosine kinases. Mol. Endocrinol. 2004;18:2672–2684. doi: 10.1210/me.2004-0168. [DOI] [PubMed] [Google Scholar]
- Jensen P.E. Recent advances in antigen processing and presentation. Nat. Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- Jett J.R., Maksymiuk A.W., Su J.Q., Mailliard J.A., Krook J.E., Tschetter L.K., Kardinal C.G., Twito D.I., Levitt R., Gerstner J.B. Phase III trial of recombinant interferon gamma in complete responders with small‐cell lung cancer. J. Clin. Oncol. 1994;12:2321–2326. doi: 10.1200/JCO.1994.12.11.2321. [DOI] [PubMed] [Google Scholar]
- Jimenez P., Cabrera T., Mendez R., Esparza C., Cozar J.M., Tallada M., Lopez‐Nevot M.A., Ruiz‐Cabello F., Garrido F. A nucleotide insertion in exon 4 is responsible for the absence of expression of an HLA‐A*0301 allele in a prostate carcinoma cell line. Immunogenetics. 2001;53:606–610. doi: 10.1007/s002510100371. [DOI] [PubMed] [Google Scholar]
- Kaplan D.H., Shankaran V., Dighe A.S., Stockert E., Aguet M., Old L.J., Schreiber R.D. Demonstration of an interferon gamma‐dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. USA. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Uddin S., Platanias L.C. The PI3′ kinase pathway in interferon signaling. J. Interferon Cytokine Res. 2005;25:780–787. doi: 10.1089/jir.2005.25.780. [DOI] [PubMed] [Google Scholar]
- Kirkwood J.M., Manola J., Ibrahim J., Sondak V., Ernstoff M.S., Rao U. A pooled analysis of eastern cooperative oncology group and intergroup trials of adjuvant high‐dose interferon for melanoma. Clin. Cancer Res. 2004;10:1670–1677. doi: 10.1158/1078-0432.ccr-1103-3. [DOI] [PubMed] [Google Scholar]
- Kitamura H., Torigoe T., Honma I., Sato E., Asanuma H., Hirohashi Y., Sato N., Tsukamoto T. Effect of human leukocyte antigen class I expression of tumor cells on outcome of intravesical instillation of bacillus calmette‐guerin immunotherapy for bladder cancer. Clin. Cancer Res. 2006;12:4641–4644. doi: 10.1158/1078-0432.CCR-06-0595. [DOI] [PubMed] [Google Scholar]
- Kloetzel P.M. Generation of major histocompatibility complex class I antigens: Functional interplay between proteasomes and TPPII. Nat. Immunol. 2004;5:661–669. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- Komyod W., Bohm M., Metze D., Heinrich P.C., Behrmann I. Constitutive suppressor of cytokine signaling 3 expression confers a growth advantage to a human melanoma cell line. Mol. Cancer Res. 2007;5:271–281. doi: 10.1158/1541-7786.MCR-06-0274. [DOI] [PubMed] [Google Scholar]
- Kwon M.J., Yao Y., Walter M.J., Holtzman M.J., Chang C.H. Role of PKCdelta in IFN‐gamma‐inducible CIITA gene expression. Mol. Immunol. 2007;44:2841–2849. doi: 10.1016/j.molimm.2007.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesinski G.B., Trefry J., Brasdovich M., Kondadasula S.V., Sackey K., Zimmerer J.M., Chaudhury A.R., Yu L., Zhang X., Crespin T.R., Walker M.J., Carson W.E., 3rd. Melanoma cells exhibit variable signal transducer and activator of transcription 1 phosphorylation and a reduced response to IFN‐alpha compared with immune effector cells. Clin. Cancer Res. 2007;13:5010–5019. doi: 10.1158/1078-0432.CCR-06-3092. [DOI] [PubMed] [Google Scholar]
- Levy D.E., Lew D.J., Decker T., Kessler D.S., Darnell J.E., Jr. Synergistic interaction between interferon‐alpha and interferon‐gamma through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Metze D., Nashan D., Muller‐Tidow C., Serve H.L., Poremba C., Luger T.A., Bohm M. Expression of SOCS‐1, suppressor of cytokine signalling‐1, in human melanoma. J. Invest. Dermatol. 2004;123:737–745. doi: 10.1111/j.0022-202X.2004.23408.x. [DOI] [PubMed] [Google Scholar]
- Lienard D., Eggermont A.M., Kroon B.B., Schraffordt Koops H., Lejeune F.J. Isolated limb perfusion in primary and recurrent melanoma: Indications and results. Semin. Surg. Oncol. 1998;14:202–209. doi: 10.1002/(sici)1098-2388(199804/05)14:3<202::aid-ssu3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Logothetis C. Treatment of chemotherapy‐refractory metastatic urothelial tumors. Urol. Clin. North Am. 1992;19:775–777. [PubMed] [Google Scholar]
- Marincola F.M., White D.E., Wise A.P., Rosenberg S.A. Combination therapy with interferon alfa‐2a and interleukin‐2 for the treatment of metastatic cancer. J. Clin. Oncol. 1995;13:1110–1122. doi: 10.1200/JCO.1995.13.5.1110. [DOI] [PubMed] [Google Scholar]
- Maleno I., Cabrera C.M., Cabrera T., Paco L., Lopez‐Nevot M.A., Collado A., Ferron A., Garrido F. Distribution of HLA class I altered phenotypes in colorectal carcinomas: High frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Immunogenetics. 2004;56:244–253. doi: 10.1007/s00251-004-0692-z. [DOI] [PubMed] [Google Scholar]
- Marques L., Brucet M., Lloberas J., Celada A. STAT1 regulates lipopolysaccharide‐ and TNF‐alpha‐dependent expression of transporter associated with antigen processing 1 and low molecular mass polypeptide 2 genes in macrophages by distinct mechanisms. J. Immunol. 2004;173:1103–1110. doi: 10.4049/jimmunol.173.2.1103. [DOI] [PubMed] [Google Scholar]
- Muhlethaler‐Mottet A., Di Berardino W., Otten L.A., Mach B. Activation of the MHC class II transactivator CIITA by interferon‐gamma requires cooperative interaction between Stat1 and USF‐1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- Oesterlund P.I., Pietilä T.E., Veckman V., Kotenko S.V., Julkunen I. IFN Regulatory factor family members differentially regulate the expression of type III IFN (IFN‐λ) genes. J. Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- Paschen A., Mendez R.M., Jimenez P., Sucker A., Ruiz‐Cabello F., Song M., Garrido F., Schadendorf D. Complete loss of HLA class I antigen expression on melanoma cells: A result of successive mutational events. Int. J. Cancer. 2003;103:759–767. doi: 10.1002/ijc.10906. [DOI] [PubMed] [Google Scholar]
- Perez B., Benitez R., Fernandez M.A., Oliva M.R., Soto J.L., Serrano S., Lopez Nevot M.A., Garrido F. A new beta 2 microglobulin mutation found in a melanoma tumor cell line. Tissue Antigens. 1999;53:569–572. doi: 10.1034/j.1399-0039.1999.530607.x. [DOI] [PubMed] [Google Scholar]
- Pestka S. The human interferon alpha species and receptors. Biopolymers. 2000;55:254–287. doi: 10.1002/1097-0282(2000)55:4<254::AID-BIP1001>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Pestka S., Krause C.D., Walter M.R. Interferons, interferon‐like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Platanias L.C. Mechanisms of type‐I‐ and type‐II‐interferon‐mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Quesada J.R., Rios A., Swanson D., Trown P., Gutterman J.U. Antitumor activity of recombinant‐derived interferon alpha in metastatic renal cell carcinoma. J. Clin. Oncol. 1985;3:1522–1528. doi: 10.1200/JCO.1985.3.11.1522. [DOI] [PubMed] [Google Scholar]
- Rahat M.A., Chernichovski I., Lahat N. Increased binding of IFN regulating factor 1 mediates the synergistic induction of CIITA by IFN‐gamma and tumor necrosis factor‐alpha in human thyroid carcinoma cells. Int. Immunol. 2001;13:1423–1432. doi: 10.1093/intimm/13.11.1423. [DOI] [PubMed] [Google Scholar]
- Ramal L.M., Maleno I., Cabrera T., Collado A., Ferron A., Lopez‐Nevot M.A., Garrido F. Molecular strategies to define HLA haplotype loss in microdissected tumor cells. Hum. Immunol. 2000;61:1001–1012. doi: 10.1016/s0198-8859(00)00171-3. [DOI] [PubMed] [Google Scholar]
- Ramana C.V., Gil M.P., Schreiber R.D., Stark G.R. Stat1‐dependent and ‐independent pathways in IFN‐gamma‐dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- Rock K.L., York I.A., Goldberg A.L. Post‐proteasomal antigen processing for major histocompatibility complex class I presentation. Nat. Immunol. 2004;5:670–677. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- Rodriguez T., Mendez R., Del Campo A., Aptsiauri N., Martin J., Orozco G., Pawelec G., Schadendorf D., Ruiz‐Cabello F., Garrido F. Patterns of constitutive and IFN‐gamma inducible expression of HLA class II molecules in human melanoma cell lines. Immunogenetics. 2007;59:123–133. doi: 10.1007/s00251-006-0171-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez T., Mendez R., Del Campo A., Jimenez P., Aptsiauri N., Garrido F., Ruiz‐Cabello F. Distinct mechanisms of loss of IFN‐gamma mediated HLA class I inducibility in two melanoma cell lines. BMC Cancer. 2007;7:34. doi: 10.1186/1471-2407-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saban M.R., Hellmich H.L., Simpson C., Davis C.A., Lang M.L., Ihnat M.A., O'Donnell M.A., Wu X.R., Saban R. Repeated BCG treatment of mouse bladder selectively stimulates small GTPases and HLA antigens and inhibits single‐spanning uroplakins. BMC Cancer. 2007;7:204. doi: 10.1186/1471-2407-7-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A., Toyota M., Ikeda H., Morimoto Y., Akino K., Mita H., Suzuki H., Sasaki Y., Kanaseki T., Takamura Y., Soejima H., Urano T. Epigenetic inactivation of class II transactivator (CIITA) is associated with the absence of interferon‐gamma‐induced HLA‐DR expression in colorectal and gastric cancer cells. Oncogene. 2004;23:8876–8886. doi: 10.1038/sj.onc.1208144. [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Munz C. Antigen‐loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C., Levy D.E., Decker T. JAK‐STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Seliger B., Hammers S., Hohne A., Zeidler R., Knuth A., Gerharz C.D., Huber C. IFN‐gamma‐mediated coordinated transcriptional regulation of the human TAP‐1 and LMP‐2 genes in human renal cell carcinoma. Clin. Cancer Res. 1997;3:573–578. [PubMed] [Google Scholar]
- Seliger B., Ritz U., Ferrone S. Molecular mechanisms of HLA class I antigen abnormalities following viral infection and transformation. Int. J. Cancer. 2006;118:129–138. doi: 10.1002/ijc.21312. [DOI] [PubMed] [Google Scholar]
- Sexl V., Kovacic B., Piekorz R., Moriggl R., Stoiber D., Hoffmeyer A., Liebminger R., Kudlacek O., Weisz E., Rothammer K., Ihle J.N. Jak1 deficiency leads to enhanced Abelson‐induced B‐cell tumor formation. Blood. 2003;101:4937–4943. doi: 10.1182/blood-2001-11-0142. [DOI] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D. IL‐28, IL‐29 and their class II cytokine receptor IL‐28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Shin‐Ya M., Hirai H., Satoh E., Kishida T., Asada H., Aoki F., Tsukamoto M., Imanishi J., Mazda O. Intracellular interferon triggers Jak/Stat signaling cascade and induces p53‐dependent antiviral protection. Biochem. Biophys. Res. Commun. 2005;329:1139–1146. doi: 10.1016/j.bbrc.2005.02.088. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Tajima K., Ito Y., Demachi A., Nishida K., Akatsuka Y., Tsujimura K., Hida T., Morishima Y., Kuwano H., Mitsudomi T., Takahashi T., Kuzushima K. Interferon‐gamma differentially regulates susceptibility of lung cancer cells to telomerase‐specific cytotoxic T lymphocytes. Int. J. Cancer. 2004;110:403–412. doi: 10.1002/ijc.20139. [DOI] [PubMed] [Google Scholar]
- Takaoka A., Yanai H. Interferon signalling network in innate defence. Cell. Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Uddin S., Lekmine F., Sassano A., Rui H., Fish E.N., Platanias L.C. Role of Stat5 in type I interferon‐signaling and transcriptional regulation. Biochem. Biophys. Res. Commun. 2003;308:325–330. doi: 10.1016/s0006-291x(03)01382-2. [DOI] [PubMed] [Google Scholar]
- Umeda T., Niijima T. Phase II study of alpha interferon on renal cell carcinoma. Summary of three collaborative trials. Cancer. 1986;58:1231–1235. doi: 10.1002/1097-0142(19860915)58:6<1231::aid-cncr2820580610>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Uze G., Monneron D. IL‐28 and IL‐29: Newcomers to the interferon family. Biochimie. 2007;89:729–734. doi: 10.1016/j.biochi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- van den Elsen P.J., Holling T.M., Kuipers H.F., van der Stoep N. Transcriptional regulation of antigen presentation. Curr. Opin. Immunol. 2004;16:67–75. doi: 10.1016/j.coi.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Waiboci L.W., Ahmed C.M., Mujtaba M.G., Flowers L.O., Martin J.P., Haider M.I., Johnson H.M. Both the suppressor of cytokine signaling 1 (SOCS‐1) kinase inhibitory region and SOCS‐1 mimetic bind to JAK2 autophosphorylation site: Implications for the development of a SOCS‐1 antagonist. J. Immunol. 2007;178:5058–5068. doi: 10.4049/jimmunol.178.8.5058. [DOI] [PubMed] [Google Scholar]
- Wang W., Edington H.D., Rao U.N., Jukic D.M., Land S.R., Ferrone S., Kirkwood J.M. Modulation of signal transducers and activators of transcription 1 and 3 signaling in melanoma by high‐dose IFNalpha2b. Clin. Cancer Res. 2007;13:1523–1531. doi: 10.1158/1078-0432.CCR-06-1387. [DOI] [PubMed] [Google Scholar]
- Wellbrock C., Weisser C., Hassel J.C., Fischer P., Becker J., Vetter C.S., Behrmann I., Kortylewski M., Heinrich P.C., Schartl M. STAT5 contributes to interferon resistance of melanoma cells. Curr. Biol. 2005;15:1629–1639. doi: 10.1016/j.cub.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Wiesenfeld M., O'Connell M.J., Wieand H.S., Gonchoroff N.J., Donohue J.H., Fitzgibbons R.J., Jr., Krook J.E., Mailliard J.A., Gerstner J.B., Pazdur R. Controlled clinical trial of interferon‐gamma as postoperative surgical adjuvant therapy for colon cancer. J. Clin. Oncol. 1995;13:2324–2329. doi: 10.1200/JCO.1995.13.9.2324. [DOI] [PubMed] [Google Scholar]
- Windbichler G.H., Hausmaninger H., Stummvoll W., Graf A.H., Kainz C., Lahodny J., Denison U., Muller‐Holzner E., Marth C. Interferon‐gamma in the first‐line therapy of ovarian cancer: A randomized phase III trial. Br. J. Cancer. 2000;82:1138–1144. doi: 10.1054/bjoc.1999.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.H., Krauer K.G., Hatzinisiriou I., Estcourt M.J., Hersey P., Tam N.D., Edmondson S., Devenish R.J., Ralph S.J. Interferon‐resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48‐ISGF3gamma. J. Biol. Chem. 1997;272:28779–28785. doi: 10.1074/jbc.272.45.28779. [DOI] [PubMed] [Google Scholar]
- Wright K.L., White L.C., Kelly A., Beck S., Trowsdale J., Ting J.P. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med. 1995;181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi S., Dyer K.F., Kimak M., Zhang Q., Gooding W.E., Chaillet J.R., Chai R.L., Ferrell R.E., Zamboni B., Hunt J., Grandis J.R. Decreased STAT1 expression by promoter methylation in squamous cell carcinogenesis. J. Natl. Cancer Inst. 2006;98:181–189. doi: 10.1093/jnci/djj020. [DOI] [PubMed] [Google Scholar]
- Yang D., Stewart T.J., Smith K.K., Georgi D., Abrams S.I., Liu K. Downregulation of IFN‐gammaR in association with loss of Fas function is linked to tumor progression. Int. J. Cancer. 2008;122:350–362. doi: 10.1002/ijc.23090. [DOI] [PubMed] [Google Scholar]
- Yu H., Jove R. The STATs of cancer ‐ new molecular targets come of age. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
Further reading
- Isaacs A., Lindenmann J. Virus interference. I. The interferon. J. Interferon Res. 1987;7:429–438. doi: 10.1089/jir.1987.7.429. By A. Isaacs and J. Lindenmann, 1957. [DOI] [PubMed] [Google Scholar]


