Abstract
Recent advances in miniaturization of analytical systems and newly emerging technologies offer platforms with greater automation and multiplexing capabilities than traditional biological binding assays. Multiplexed bioanalytical techniques provide control agencies and food industries with new possibilities for improved, more efficient monitoring of food and environmental contaminants. This review deals with recent developments in planar-array and suspension-array technologies, and their applications in detecting pathogens, food allergens and adulterants, toxins, antibiotics and environmental contaminants.
Keywords: Bioanalytics, Biological binding assay, Environmental contaminant, Environmental monitoring, Food contaminant, Food monitoring, Miniaturization, Multiplex, Planar array, Suspension array
1. Introduction
The presence and the prevalence of diverse, potentially harmful, contaminants in our food and environment require our continual attention [1], [2] The contaminants treated in this review are classified as follows: foodborne pathogens (Salmonella spp., Vibrio spp., Campylobacter spp., Escherichia coli O157 and Listeria monocytogenes), food allergens and proteinaceous adulterants, toxins (mycotoxins, bacterial and plant toxins), residues of antibiotics in food, and environmental contaminants [persistent organic pollutants (POPs), including polychlorinated biphenyls (PCBs) and polycyclic aromatic hydrocarbons (PAHs), pesticides and herbicides, and endocrine-disrupting chemicals (EDCs)].
For successful monitoring of levels and trends of these contaminants in our environment and food, and determination of their significance with regard to public health, powerful analytical methods are applied. These include traditional analytical methods [e.g., gas and liquid chromatography (GC and LC) coupled to mass spectrometry (MS)], classical microbiological culturing methods coupled to biochemical and serological identification, electrophoresis and immunoassays in traditional formats (e.g., immunoblots and radiolabeled immunoassays). The combination of MS detectors with LC-separation techniques is probably the most commonly applied methodology for identifying contaminants in food and environmental samples [3].
Since more and more products nowadays contain multiple and processed ingredients, which are often shipped from different parts of the world, and share common production lines and storage spaces, food safety and environmental monitoring becomes a challenging task. Currently, it is common practice to screen first a large number of samples for possible contamination and then subject suspected samples to further confirmation. Traditional analytical methods require dedicated laboratories, equipment and highly-trained personnel for detection and identification of each type of hazardous agent (e.g., antibiotics, bacteria, and allergens). Screening tools today are therefore based on assays incorporating biological recognition elements (biological binding assays), which offer simpler, more rapid analysis. The most commonly used bioanalytical methods for routine monitoring are enzyme-linked immunosorbent assays (ELISAs) in 96-well plate format [2], [4], [5]. Even though some level of automation has been achieved in the recent years, ELISAs remain laborious, time-consuming and expensive, when multiple targets need to be screened for, so there is a growing need for new multi-analyte screening methods, which will enable rapid, simultaneous detection of contaminants in numerous samples.
This review provides an outlook on the recent developments in bioanalytical multiplex technologies and their applications for food safety and environmental monitoring.
2. Multiplex technologies
2.1. Planar arrays
There is an increasing interest in planar-array technologies for food and environmental analysis, with fluorescent, bioluminescent or chemiluminescent (CL) labels for detection, and direct (label-free) detection. Microarrays and/or multi-channel platforms offer high multiplexing capabilities for the biological binding assays, and they are particularly useful when multi-analyte screening is needed. Short measurement times, automation, reduced sample volumes and high sensitivity are among the main advantages offered by such systems. The most prominent planar-array technologies already applied to food and environmental analysis include the Naval Research Laboratory (NRL) array biosensor based on total internal reflection fluorescence (TIRF), the CL microarray and the Surface Plasmon Resonance (SPR)-based biosensor [6], [7], [8].
2.1.1. The NRL array biosensor
The NRL array biosensor is based on a planar waveguide that directs evanescent excitation light to fluorophores that are bound to the surface. It is composed of three parts: an array of immobilized molecular recognition elements (usually antibodies), an image capturing and processing system, and a fluidics handling unit. Array sensor optics comprise a patterned glass slide, which is placed on a support and is illuminated by launching 635 nm light from a diode laser into one end, a GRIN lens array, which focus the fluorescent patterns, an emission filter, which rejects unwanted laser light and a Peltier-cooled CCD camera which images the array.
Due to the limited penetration depth of the evanescent wave, only fluorescence of the fluorescent probes which are close to the waveguide surface is measured. This approach in combination with fluorescent dyes, which are excited at longer wavelengths (e.g., Cy5 and AlexaFluor 647), significantly reduces interference from the bulk fluid, offering better reproducibility and sensitivity when used for analysis in complex sample matrixes. Optical properties of such fluorophores also allow a small, lightweight source of excitation (e.g., diode laser) and a compact optical detection system (e.g., CCD camera). Multi-analyte detection is achieved through the “bar-code” approach. Biotinylated capture antibodies are immobilized on the avidin-coated waveguide surface in columns, using a polydimethylsiloxane (PDMS) block with several channels. The PDMS block is then oriented perpendicularly and several different samples are passed through the channels. The array biosensor is portable and automated, and thus is very well suited to in-field measurements. Also, its detection principles allow analysis of crude matrixes with minimal sample preparation. Commercial versions of the array biosensor have recently become available [6]. The array biosensor has been used for the detection of small molecules, toxins, proteins, bacteria and viruses in numerous applications in food safety, diagnostics, homeland security and environmental monitoring.
2.1.2. CL-based microarrays
The parallel affinity sensor array (PASA) was the first biosensor for multiplexed detection of antibiotics in milk [9]. The PASA is based on a CL read-out of the microarray via the CCD camera, it includes a flow cell and an integrated fluidic system for reagents handling. The disposable chips, microarrayed with haptens or antibodies, are inserted into the flow cell and placed in the dark chamber. The light is emitted from the microarrays via chemical reaction with assistance of an enzyme label, omitting the need for a light source. Conventional microscope glass slides, modified with (3-glycidyl-oxypropyl)trimethoxysilane, were used as a solid-phase support for the microarray. The haptens were microarrayed using a conventional non-contact spotting technique based on piezoelectric nano-pumps, producing spots of diameter ∼350 μm.
Multi-analyte immunoassays in an indirect competitive ELISA format can be implemented for rapid, automated multiplexed analysis. Usually, the sample is pre-mixed with a specific antibody and injected over a hapten microarray. Free antibody binds to hapten spots and then the secondary antibody, labeled with peroxidase (POD), is introduced. Formation of the immuno-complex is detected by light emission in the presence of a luminol-based POD substrate. The direct immunoassay format was also demonstrated in this system. The microarrays are for multiple uses, and are regenerated between sequential measurement cycles. The analyzed samples do not require enrichment and pretreatment steps, and the target analytes can be detected within 5 min (excluding the regeneration cycle) with sensitivities reaching ng/L concentrations of analyte. The recent version of PASA, which is a self-contained system for fully automated multiplexed immunoanalysis, designated MCR3, was reported by Kloth et al. [7] (Fig. 1 A).
Figure 1.
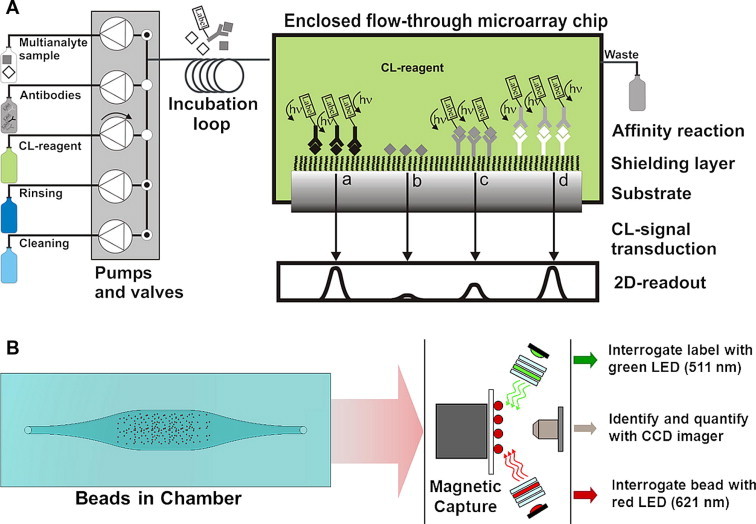
(A) The chemiluminescent (CL) flow-through microarray-platform set-up. CL microarrays measure the light emitted by an enzymatic reaction. The analytes can be quantified via indirect (a and b), direct (c) and sandwich (d) assay formats. This new MCR3 version of the instrument is an automated, stand-alone platform that combines mechanical reagent supply, disposable microarray chip, temperature-controlled flow cell and CCD camera (Reproduced with permission from [7]). (B) Measurement principle of the MAGPIX system (Luminex Corporation) using superparamagnetic MagPlex microspheres and CCD imaging technology. It detects and distinguishes surface reporter fluorescence emissions at 590 nm ± 24 nm on the surface of 1–50 unique xMAP microsphere sets, magnetically captured in a monolayer, from a single sample.
An additional example of a multiplex system using CL readout is based on 96 × 4-well-plate format [10]. This microarray platform comprises a 96-well plate, where each well includes four sub-wells. This approach is limited to the simultaneous screening of four target analytes, but compensates with the amount of samples which can be measured simultaneously in one 96-well plate. Currently it is not automated, but it has a potential to be easily integrated with existing pipetting robots and plate washers.
2.1.3. SPR-based sensors
SPR biosensors do not require the use of reporter elements to generate a signal, which is convenient during assay development and during application by saving labeling steps and washing steps. SPR biosensors are based on optical phenomenon which occurs when the evanescent wave, generated by light, interacts with free electrons in a thin metal film. The intensity of the reflected light at a specific angle is thus reduced. The light angle at which this reduction occurs is called the SPR angle, and it varies with the refractive index of the dielectric medium (usually buffer) close to the metal film (usually gold). When molecules are immobilized on and/or bound to the gold surface, the SPR angle changes, allowing label-free and real-time monitoring [11].
SPR sensors are used in multi-channel and array-based set-ups. The most popular commercially available SPR sensors are the four flow-channel (4FC)-based Biacore systems. Most of the instruments are fully automated with a capacity of 192 samples (two 96-well microtiter plates). Currently, there is a Biacore 4000 SPR system available from GE Healthcare, with the possibility of simultaneous detection of 16 spots in the 4FCs. Many methods for food safety and environmental monitoring have been previously developed on Biacore platforms (see Section 3).
Alternative eight-channel SPR sensor instruments were developed and used for the detection of low-molecular-weight EDCs [12] and an environmental contaminant in a miniaturized, portable format [13]. Taylor et al. reported a custom-built multi-channel SPR sensor for the simultaneous detection of four food pathogens [8]. Several portable SPR devices have been developed for field applications [13], [14], [15]. However, so far, none has demonstrated highly multiplexed analyte detection.
SPR imaging (iSPR) technology takes SPR analysis a step further, offering much higher multiplexing capabilities. There are several commercial iSPR instruments available: SPRi-Plex (Genoptics Bio interactions), ProteOn XPR36 (Bio-Rad Laboratories), SPRimager II ARRAY system (GWC Technologies) and IBIS iSPR (IBIS Technologies B.V.). The instruments differ in optics, fluidics, sample handling and sensor-surface preparations. All of these factors greatly influence sensor output, and the choice of the iSPR instrument is usually made according to the targeted application.
2.1.4. Lateral-flow devices and newly emerging technologies
Lateral-flow devices (LFDs) or dipstick assays are used for qualitative, semi-quantitative and to some extent quantitative monitoring in resource-poor or non-laboratory environments. Multiplexing options are achieved by application of several different test lines [16].
Fenton et al. [16] fabricated paper-based and nitrocellulose-based LFDs that were shaped in two dimensions by a computer-controlled knife. The resulting star, candelabra, and other structures were spotted with multiplex bioassay reagents to produce multiplex lateral-flow assays.
Newly emerging planar technologies include polydiacetylene (PDA) biosensor chips and electrical microarrays. Hee et al. [17] recently reported the development of cross-linked PDA liposome-based chips for multiplex pathogen detection. PDA supramolecules undergo a color change from blue to red under various stimuli (e.g., temperature, pH and mechanical stress), including binding events that take place on the surface. The red state of the PDA also produces fluorescence. For chip production, the PDA liposomes are arrayed using ethylenediamine as an interlinker on amine-covered glass slides by an array spotter and conjugated with different antibodies. Binding to target bacteria can be monitored by the naked eye, due to chromatic transition, or by measuring the fluorescent output (Fig. 2 A). Further research is needed to evaluate the analytical performance of this technology and its applicability to analysis of real samples.
Figure 2.
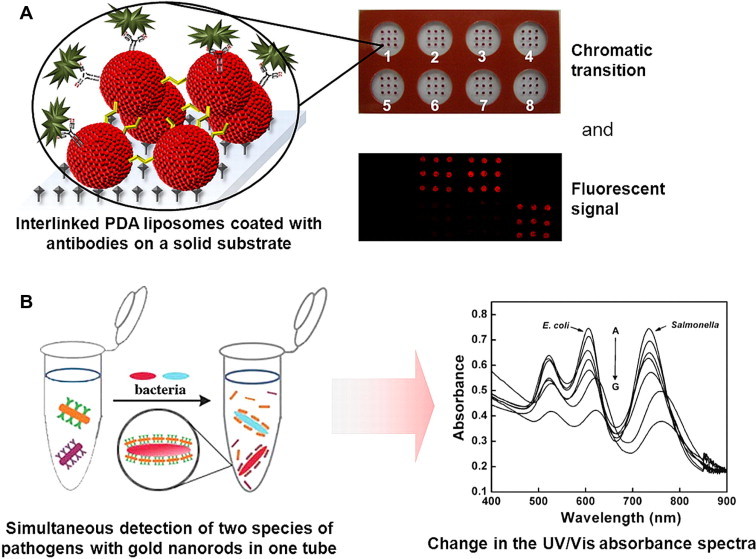
(A) A planar microarray based on interlinked polydiacetylene (PDA) liposomes on a solid substrate. PDA liposomes are coated with different antibodies and arrayed on a glass slide, which is further modified with adhesive silicone to create chambers. In each chamber a different pathogen is detected. Upon binding of the antigen, PDA liposomes undergo a color change to red, visible to the naked eye, and produce fluorescence (Reproduced in modified form with permission from [17]). (B) The simultaneous detection of two species of pathogens in an assay based on gold-nanorod probes. (Left) Two gold nanorods, with different aspect ratios, coated with antibodies form complexes with the target pathogens in one reaction tube. (Right) The change in the UV/Vis absorbance spectra is dose-dependent on the pathogen concentration in the sample, and the two pathogens are distinguished by maximal absorbance peaks at different wavelengths (Reproduced in modified form with permission from [21]).
Electrical microarrays employ an electrical signal readout from an array of microelectrodes. For example, Elsholz et al. [18] described an electrical oligonucleotide microarray for identification and detection of multiple pathogens via RNA hybridization. The signal was generated by alkaline-phosphatase-mediated conversion of p-aminophenol to its electrically-active phosphate derivative and enhanced by redox cycling. This system was reported to be fast and easy to use, and did not require PCR amplification, but it has not yet been applied to analysis of food or environmental samples. Additional examples of analytical microarray systems, which have not yet been applied to food and environmental monitoring, can be found in the review by Seidel et al. [9].
2.2. Suspension arrays
2.2.1. Flow cytometry
In fluorescence-based flow cytometry, cells or particles are aligned in a flow stream and optically interrogated. Size, density, and fluorescence at multiple wavelengths can be quantified creating suspension microarrays.
The Multi Analyte Profiling (xMAP) technology from Luminex Corporation is the most prominent suspension microarray commercially available. This technology employs small carboxylated polystyrene microspheres (5.6 μm), which are internally dyed with a red fluorophore and an infrared fluorophore. Up to 100 different color-coded bead sets can be distinguished by varying the ratio of the two fluorophores. Each bead set can be coupled to a different biological probe, allowing simultaneous measurements of up to 100 different biomolecular interactions in a single well. The carboxylated bead surface allows simple chemical coupling of capture reagents (e.g., antibodies or drug-protein conjugates).
Increased multiplexing capability can be obtained with the FlexMAP 3D system of Luminex, using a third fluorophore, which offers 500-plex and runs three times faster than the Luminex 100 or 200 systems.
Recently, a MagPix instrument was launched by Luminex, offering a low-cost, compact, rugged, alternative for multi-analyte diagnostic and environmental testing. It moves away from a flow-cytometry-based system to an instrument based on magnetic bead array analyzed on a magnet in a 2D readout with light emitting diodes (LEDs) and a CCD camera, offering a more robust system suitable for in-field measurements (Fig. 1B). Several microbead-based suspension microarrays, which may be run on standard flow cytometers, are commercially available: QuantumPlex (Bangs Laboratories, Inc.); FlowCytomix (Bender MedSystems); and, Cyto-Plex (Duke Scientific Corp). Some are already dedicated to environmental and food analysis [e.g., Fungi-PLEX5 (Soft Flow, Inc.) and Sal Plex (RnA, Utrecht, The Netherlands)].
Recently, a very compact, robust and simple-to-fabricate microflow cytometer was developed by Kim et al. [19]. Having capabilities for point-of-care and on-site analysis, it has great potential for applications in food and environmental monitoring.
2.2.2. Nanoparticles
Implementation of new nano-tools [e.g., gold nanoparticles (NPs) and quantum dots (QDs)] to analytical applications promises increased sensitivity, multiplexing capabilities, and reduced costs. Moreover, NP-based devices offer simplicity of optical configuration, ease of fabrication, great potential for miniaturization, simple handling and short assay times.
For example, fluorescent NPs, known as QDs, offer multiplex detection by conjugating these semiconductor nanocrystals with biorecognition molecules. Their broad absorption spectra and narrow emission spectra make them ideal for optical multiplexing [20]. Other properties of QDs include negligible photobleaching and fairly high quantum yields. Even though QDs offer many advantages over conventional fluorophores, their synthesis is considered difficult, and commercial variants are still limited in number and expensive.
As an alternative to fluorescently-labeled NPs, gold-nanorod probes have been proposed, applying localized SPR (LSPR) detection principles (Fig. 2B) [21]. The optical properties of gold nanorods depend on shape and are affected by changes in the dielectric constant in the vicinity of the nanorod surface, a phenomenon that is also known as LSPR. The elongated NPs provide higher sensitivity to the local dielectric environment than spherical NPs of the same size. Gold nanorods can be easily fabricated at different aspect ratios, offering multiplexing possibilities.
3. Applications
3.1. Foodborne pathogens
Proper detection methods of foodborne pathogens are vital for maintenance of food safety, and rapid detection of microorganism is necessary to curb outbreaks that can affect large populations. Screening each sample for a single pathogen using commonly applied techniques (e.g., culturing and molecular techniques) is time-consuming, labor-intensive and costly. Multiplex technologies offer parallel analysis of several pathogens in a single experimental run, reducing assay time, labor and costs. There are numerous applications of molecular methods for pathogen detection. Here, we report on the multiplexed techniques based on bioassays that enable simultaneous detection of at least two pathogens.
The NRL array biosensor, in combination with antimicrobial peptides for the detection of E. coli and Salmonella with sensitivities in the ranges of 0.5–5 × 105 and 0.1–5 × 106 cells/mL, respectively, was demonstrated by Kulagina et al. [22], [23].
Hee et al. [17] reported a polydiacetylene (PDA) liposome-based biosensor for multiplex pathogen detection. Proof of concept was demonstrated using two mixtures, one containing Cryptosporidium parvum and E. coli O157 and another containing Giardia lamblia, S. typhimurium and Encephalitozoon intestinalis at concentrations of 106 CFU/mL. Within 30 min, the presence of pathogens could be detected qualitatively by the chromatic transition of PDA to red or quantitatively by the fluorescence signal.
A 96-well microtiter plate-based antibody microarray was developed by Gehring et al. [24]. Within 2.5 h, E. coli O157:H7 and S. typhimurium were detected at concentrations of 106 and 107 CFU/mL, respectively, in buffer and ground beef. Alongside, chicken IgG was detected at ng/mL levels, as a model for proteinaceous toxin. Even though the sensitivity achieved with this method was less than the sensitivity reported for the NRL array biosensor, microarrays in a multi-well plate offer automatic sample handling during multiple steps, enabling screening of large sample sets.
A flow-through microarray coupled to a CL readout was described by Wolter et al. [25]. With this method, multiple bacteria were simultaneously detected in water samples within 13 min. CL-based detection enabled optical readout of the microarray with high sensitivity and without an external light source. Polyethylene glycol-modified glass was used as a support platform for a sandwich immunoassay for the parallel detection of E. coli O157:H7, S. typhimurium and Legionella pneumophila, with limits of detection (LODs) of 3 × 103, 3 × 106 and 1 × 105 cells/mL, respectively.
Karsunke et al. [26] modified this method by developing a disposable plastic multichannel version, using an acrylonitrile-butadiene-styrene copolymer. The chip contained six flow-through microchannels, which enabled calibration and measurement in one experiment, reducing the total assay time to 18 min. For monitoring drinking water supply, both systems should be used after bacteria-enrichment steps (e.g., microfiltration or immunomagnetic concentration).
Immunoassays combined with a CL readout have also been implemented in a microtiter-plate format. Magliulo et al. [10] developed a CL-EIA for multiplex detection of E. coli O157:H7, S. typhimurium, Yersinia enterocolitica and L. monocytogenes. Using a new polystyrene plate design, where each of the 96 wells contained four sub-wells at the bottom, the four bacteria were simultaneously detected in meat and fecal samples with 0.1–1 × 105 CFU/mL sensitivity. A portable point-of-care device, based on electrochemical detection, was also reported for the simultaneous measurements of E. coli and B. subtilis DNA, utilizing a silicon glass-based micro chamber [27].
Label-free, quantitative and simultaneous detection of E. coli O157, Salmonella choleraesuis typhimurium, L. monocytogenes and C. jejuni was achieved by Taylor et al. [8] using an eight-channel SPR sensor. They reported sensitivities of 0.034–1.2 × 105 CFU/mL in apple juice. A sandwich immunoassay was performed, in order to amplify the direct response obtained with bacteria binding to the sensor-chip surface. The LODs obtained with this sensor were comparable to those obtained with the NRL array biosensor, but the assay time was longer (approximately 100 min in the SPR sensor). A direct SPR-based immunoassay, using monoclonal antibodies spotted on a Protein G-modified gold-sensor chip, was reported for the detection of E. coli, S. typhimurium, L. pneumophila, and Y. enterocolitica [28].
The use of beads with varying properties in biological binding assays offers many multiplexing possibilities. There are examples of such systems applied to detection of foodborne pathogens.
A multiplexed, bead-based, mesofluidic system (BMS) was developed for the simultaneous detection of eight major foodborne pathogens {i.e. Salmonella enterica, S. aureus, L. monocytogenes, V. parahaemolyticus, Shigella sonnei, Enterobacter sakazakii, E. coli O157:H7 and C. jejuni [29]}. Glass microbeads, coated with specific nucleotide probes, were arranged in microchannels in a predetermined order. Fluorescently-labeled PCR products of pathogenic amplicons were infused into the microchannels, where they were captured by corresponding probes. LODs obtained for the eight tested pathogens from pure cultures were in the range 0.5–6 × 103 CFU/mL. The tested pathogens were correctly detected and identified in 184 endogenously infected food samples, including eggs, pork, chicken, shellfish, ice cream and milk powder.
Fluorescent NPs incorporating different ratios of three dyes, were suggested for the simultaneous detection of multiple bacteria by Wang et al. [30]. Proof of concept was demonstrated by coating the NPs with polyclonal antibodies against E. coli, S. typhimurium and S. aureus. Confocal imaging of the target bacteria showed specific coverage with the fluorescent NPs. Despite promising multiplexing possibilities, the analytical capabilities of this method are yet to be evaluated.
Another study reported the use of commercially-available QDs for parallel detection of E. coli and S. typhimurium coupled with immuno-magnetic bead separation [31]. The authors reported an LOD of approximately 104 CFU/mL, requiring a total assay time of 2 h. Multiplexing capacity in this assay was rather low (up to four species) due to the limited availability of commercial QDs.
Simultaneous detection of E. coli and S. typhimurium was also achieved using gold nanorods with different aspect ratios, coated with polyclonal antibodies against the two pathogens [21]. Within 30 min, the target pathogens were simultaneously detected at concentrations less than 102 CFU/mL.
Superparamagnetic particles were also employed for multi-analyte detection of food pathogens. Koets et al. [32] reported a giant magneto-resistance (GMR)-based biosensor for simultaneous detection of four antibiotic resistant genes of Salmonella with pM sensitivity.
3.2. Food allergens and proteinaceous adulterants
Food adulteration refers to the situation when the food product fails to meet safety and quality standards determined in the relevant legislation. Even though the outcome of most food-adulteration cases is economical, there are additional concerns that include possible allergic reactions to adulterants and offence to religious beliefs [33]. Certain forms of adulteration can also be unintentional (e.g., cross-contamination due to shared manufacturing and storing facilities), so adequate monitoring techniques are of an interest to both legal authorities and the food industry.
The next generation of protein-detection techniques is based on miniaturized planar and suspension arrays and enables multiplexed protein analysis. For many adulterants, the fastest and the easiest detection method so far has proved to be ELISA, which is most commonly used in a singleplex format.
As an exception, a multi-allergen ELISA in competitive, indirect format was described for the simultaneous determination of peanut and several tree-nut allergens in chocolate with LODs below 1 μg/g protein for each allergenic food [4]. They assembled multi-allergen microtiter plates by combining eight-well strips coated with proteins from each of the five allergenic foods.
An optical resonance-enhanced absorption (REA)-based near-field biosensor immunoassay was proposed as a novel platform for allergen detection by Maier et al. [34]. In this study, gold NPs were used as probes for signal generation in a distance-dependent interferometric set-up in planar-chip format. Aluminum discs coated with poly(styrene-methyl methacrylate) and a specific polyclonal antibody were used to detect ovalbumin and ovomucoid with a sensitivity of 1 ng/mL. The main advantage of this approach is that the signal is visible to the naked eye and thus has minimal technical requirements.
For the rapid, simultaneous detection of several allergens, Rebe Raz et al. [35] constructed a reusable antibody microarray directed against 12 major food allergens and applied it to label-free and direct allergen detection in food using an angle-scanning iSPR system. Each measurement cycle produced quantitative data on the concentration of 12 allergens within 12 min. The sensitivity of the on-chip allergen detection was reported to be in the low-μg/g range for cookies and dark chocolates, which is adequately compatible with food-allergen analysis and comparable to most commercially available ELISAs.
Haasnoot et al. [36] developed a triplex for the detection of plant proteins in milk powders, using a competitive immunoassay in combination with the xMAP technology. This was realized by coupling soluble wheat proteins and proteins from soy and pea to three different microsphere sets. A mixture of these microsphere sets was incubated with a mixture of three affinity-purified polyclonal antibodies raised against these proteins and labeled with a fluorophore. The sensitivities of the three assays were determined as 0.5–0.6 μg/mL at 50% binding.
The microsphere-based technology was also applied for the detection of Cry1Ab protein in genetically modified maize with an LOD of 0.018% (weight (GMO)/weight), and was described as the first application of a quantitative high-throughput immunoassay in GMO analysis with multiplex options by Fantozzi et al. [37].
Recently, Bremer et al. [38] reported application of the microsphere-based technology for the indirect detection of recombinant bovine somatotropins (rbST) via changes in multiple rbST-dependent biomarkers in cow serum. rbST enhances growth and lactating performances of livestock, but its use is banned in the European Union. The simultaneous detection of total insulin-like growth factor 1 (IGF1) and one of its binding proteins was demonstrated with sensitivities in the low-ng/mL range.
3.3. Toxins
Due to the diversity of toxin structures, which vary from low-molecular-weight compounds to proteins, it is fairly impossible to use one analytical technique for their detection. Since bioanalytical techniques utilize an interaction with a biological recognition element for detection, they enable the possibility of simultaneous screening of multiple toxins from a single matrix in a rapid, cost-effective manner.
Microchannel and SPR-based biosensors have been described for several mycotoxins [39]. Such rapid SPR immunoassay was developed for the combined detection of T2-toxin and HT2-toxin in naturally contaminated cereals and maize-based baby food [40].
A microarray of immobilized antigens on a plastic probe tray, in combination with polyclonal antibodies and an enzyme-labeled second antibody, was described for the simultaneous detection of aflatoxin B1 (AFB1) and fumonisin B1 (FB1) with LODs in standard solutions of 3 ng/mL and 43 ng/mL, respectively [41].
Applications of the NRL array biosensor for the rapid, simultaneous detection of multiple toxins were reviewed by Taitt et al. [42]. This sensor was used for the detection of ochratoxin A (OTA), deoxynivalenol (DON) and AFB1 individually and in combinations in various food matrices. This system was also applied for the detection of two large protein toxins [botulinum toxoid A (BotA) and staphylococcal enterotoxin B (SEB)] in the sandwich-immunoassay format in various food matrices.
Mak et al. [43] combined the specificity of immunoassays with the sensitivity and the simplicity of magnetic detection to develop a novel multiplex magnetic nanotag (MNT)-based detection platform for mycotoxins that functions on a sub-pM concentration level. Unlike fluorescent labels, MNTs can be detected with inexpensive giant magnetoresistive sensors (e.g., spin-valve sensors). They reported the simultaneous detection of AFB1, zearalenone and HT2-toxin with LODs at the pg/mL level.
Another approach to the simultaneous detection of SEB and immunodominant antigen A homologue of S. epidermidis used electrical protein-array-chip technology [44]. This procedure is based on an enzyme-linked sandwich immunoassay in which the detection is achieved by measuring the electrical current generated by redox recycling of an enzymatically-released substance. The toxins could be detected in milk and urine at a concentration of 1 ng/mL in less than 23 min.
Pauly et al. [45] developed a multiplexed immunoassay for the simultaneous quantification of five bacterial and plant toxins in complex matrices using the xMAP technology. Sandwich immunoassays were combined for proteotoxins ricin, abrin, botulinum neurotoxins type A and B and SEB, and excellent sensitivities of 2–546 ng/L were obtained in a minimal sample volume of 50 μL. Advancing existing bead-array technology, the novel magnetic and fluorescent microbeads were introduced for an enrichment step, which further increased the sensitivity of the assay to 0.3–85 ng/L, enabling analysis in a 500-μL sample volume. The method was successfully applied to simultaneous identification of the target toxins in complex food matrices (e.g., milk, baby food and yoghurt). Another example of implementation of this xMap technology for multiplex detection of toxins was reported for B. anthracis spore, Y. pestis, SARS-CoV, SEB and ricin [46]. In our group, this magnetic bead-based technology was used to develop a multiplexed competitive immunoassay for the detection of several mycotoxins in feed extract [47].
Kim et al. [19] applied the microflow cytometer to multiplexed detection of bacteria and toxins, demonstrating assay performance close to that of a commercial bench-top flow cytometer.
3.4. Antibiotics
Widespread use of veterinary antibiotics in feeding animals has the potential to provoke development of antibiotic-resistant bacteria. Microbial inhibition tests and immuno-based or receptor-based assays are the two main applied screening techniques for detection of antibiotics in food and related products. Due to their high cost effectiveness and broad spectrum characteristics, microbial inhibition methods are preferred for large-scale monitoring programs on veterinary-drug residues. However, since these methods rely on growth inhibition of a susceptible bacterium in the presence of the antibiotic compound, they require rather lengthy incubation times of 4–18 h [48].
A faster alternative is a whole-cell-based bioassay, also named whole-cell biosensor, which has been described for the detection of tetracyclines [49]. This assay is based on a genetically-engineered luminescent bacterial strain that contains the regulation unit of tetracycline-resistance factor (tetracycline-responsive element) to control the expression of the luciferase operon, resulting in tetracycline-dependent light production. The assay is performed in a 96-well microtiter plate format, allowing simultaneous analysis of several samples within 4 h and with little preparation. With addition of membrane-permeabilizing and chelating agents, sensitivities of 5 ng/g for doxycycline, 7.5 ng/g for chlortetracycline and 25 ng/g of tetracycline were reached. Whole-cell biosensors have the potential to displace growth-inhibition assays as the favored method for tetracycline-residue screening, since they are better suited to high-throughput analysis and achieve similar sensitivities. However, such bioassays have not yet been described for other kinds of antibiotics. Currently, whole-cell biosensors incorporating various microbial reporters are widely used in pharmaceutical-drug discovery [50] and for monitoring environmental chemical contaminants [51]. Their application range will most probably extend in the near future to include detection of antibiotics in food.
The most frequently-used immunochemical method for antibiotics detection is the ELISA in 96-well microtiter-plate format, and many ELISA kits to detect specific antibiotic compounds are commercially available. In general, they are sensitive and easy to use, have high specificity, require minimal sample preparations, and are therefore suitable for the screening of a large number of samples in a short time (about 2–3 h). These tests can be used within food-producing facilities. The use of generic structures for the development of group-specific antibodies enables detection of groups of compounds, widening the screening range [52], [53]. Penicillin-binding protein was also used for the detection of the antibiotics from the beta-lactam group in different food matrices [54]. It was immobilized to a microplate and the amount of a bifunctional reagent (with ampicillin and digoxigenin as functional groups), measured with anti-digoxigenin conjugated with horseradish peroxidase, was used to quantify the amount of beta-lactams present in the sample extracts. A multi-analyte screening ELISA for sulfonamides, fluoroquinolones and beta-lactam antibiotics in milk, using three class-selective bioreceptors in a planar microarray configuration, was also recently described [5].
LFDs were described for the detection of several (group) specific antibiotics {e.g., sulfonamides in eggs and chicken muscles [55] and cephems [56] in milk}. Examples of commercially-available products are the Rapid One Step Assay (ROSA) tests for β-lactams, tetracyclines, enrofloxacin and sulfadimethoxine/sulfamethazine of Charm Sciences Inc. (Lawrence, MA, USA). Unisensor (Angleur, Belgium) developed a receptor-based assay in dipstick format (Twin sensor) for simultaneous, rapid detection of β-lactams and tetracyclines in raw milk. In general, the commercial availability of these rapid qualitative tests is still limited to a few antibiotics.
While bioassays or whole-cell biosensors utilize the response of entire cells to detect biologically-active agents, biosensor instruments use biological recognition elements (e.g., antibodies, enzymes, lectins, receptors and nucleic acids) coupled to transducers [51]. For the detection of antibiotics, electrochemical and optical biosensors are most frequently applied.
Zacco et al. [57] developed a novel electrochemical immunosensing strategy for the detection of sulfonamide antibiotics in raw, full-cream milk with an LOD of 1.4 μg/L based on magnetic beads coated with class-specific anti-sulfonamide antibodies and sulfonamide peroxidase as tracer.
Even though Biacore Q kit-based assays for veterinary drug tests in foodstuffs, including antibiotics, β-agonists, and antiparasitic drugs, have become commercially available, the Biacore systems are considered expensive and offer limited multiplexing possibilities. Also, the antibodies are too specific for the simultaneous detection of antibiotics from different groups and the systems are therefore less suitable for control agencies and food industries. To provide increased, more efficient control of antibiotics in the food chain, cheaper options and more extensive multiplex systems are needed.
Rebe Raz et al. [58] used the IBIS iSPR for the simultaneous detection of seven antibiotics in milk. By multiplexing seven immunoassays in a competitive format, they were able to measure all the target compounds at parts per billion (ppb) levels in diluted skimmed milk, within 10 min.
The automated microarray PASA system was one of the first immunochemical biosensor platforms with the potential to test for numerous antibiotics in parallel in less than 5 min and with LODs in milk in the range 0.1–32 μg/L [59].
Another interesting automated multiplex biochip array technology (Evidence from Randox) has 25 immunoassays that can be measured simultaneously using chemiluminescent signals, and spots are measured with a CCD camera. Randox supplies arrays for growth-promoters and antimicrobials, but a major disadvantage is that it is a closed system, which is not suitable for assay development.
Next to these commercially available systems, Chen et al. [60] recently developed a simple, practical biochip system with a drug-protein conjugate array spotted onto activated agarose surface-modified glass slides and fluorescent-labeled antibodies for the simultaneous detection of eight antibiotics in six sample extracts using a laser confocal scanner.
The suspension-array technology in the Luminex flow cytometer was successfully applied for the simultaneous detection of aminoglycosides and sulfonamides in milk and blood serum at relevant levels and with an assay time of about 2 h [61]. Liu et al. [62] also used it for the simultaneous detection of chloramphenicol, clenbuterol and 17-beta-estradiol, and they considered this technology high throughput and simple to operate at high sensitivity and low cost.
Peng et al. [20] demonstrated simultaneous determination of five drug residues (dexamethason, gentamicin, clonazepam, medroxyprogesterone acetate and ceftiofur) in one well of a microplate using a mixture of five antibody-coated QDs in an indirect competition fluorescent-linked immunosorbent assay. They described this technology as being less time-consuming than ELISA and sufficiently flexible to be used in other systems for simultaneous multicolor detection of drugs.
3.5. Environmental contaminants
Because of their integrated nature, biosensors are ideal for environmental monitoring and detection, as they can be portable and provide selective, sensitive and rapid responses in real time. However, most of them are single-analyte detectors, and that is a major disadvantage because of the possible presence for instance of many pesticides.
Mauriz et al. [15] described multi-analyte SPR immunoassays for environmental biosensing of the pesticides DDT, chlorpyrifos and carbaryl in a two-channel biosensor with sensitivities of 18–50 ng/L. Based on recent advances in NP engineering, Nichkova et al. [63] described the application of two commercially-available QDs as labels in an immunoassay microarray for the simultaneous microscopic detection of two biomarkers of exposure to two major classes of compounds: pyrethroid insecticides and triazine herbicides.
Guo et al. [64] developed a lateral-flow strip test for the simultaneous detection of carbofuran and triazophos using two specific gold-labeled monoclonal antibodies as detector reagents with LODs in spiked water at 32 μg/L and 4 μg/L, respectively.
The NRL biosensor was also utilized for simultaneous measurements of up to 30 contaminants. This resulted in an automated water analyzer computer-supported system (AWACSS) that could measure several organic pollutants at the low-ppt level in a single analysis within a few min and without any prior sample pre-treatment steps [9].
Weller et al. [65] described the application of the PASA system for environmental contaminants in water. However, they only demonstrated this miniaturized sensor with a few analytes [i.e. trinitrotoluene (TNT), 2,4-D and triazines (atrazine and terbuthylazine)], for which the lowest LOD (20 ng/L in water) was obtained with terbuthylazine.
Since immunoassays tend to be very specific, other biomolecules (enzymes, receptors or transport proteins) or whole cells are often applied to achieve the broader detection of compounds or bio effect related detections. Several biosensors incorporate enzymes, using biocatalytic reactions to detect contaminants. For example, enzyme-based biosensors have been applied for the detection of phenolic estrogens (e.g., phenol, catechol, bisphenol A, genistein, quercetin, nonylphenol, and diethylstilbestrol) using the ability of tyrosinase to catalyze the oxidation of the phenolic estrogens to o-diphenol and o-quinone [66]. Enzymes (e.g., horseradish peroxidase, alkaline phosphatase, oxidases, urease, L-cysteine desulfhydrolase and invertase) have been utilized in the detection of various metals (e.g., arsenic, silver, mercury, cadmium, lead, copper and zinc) [66]. Lack of selectivity is described as the major disadvantage of enzyme-activity-inhibition assays, as some enzymes are inhibited by several metals and even some anions and pesticides. Metal determination by enzyme activation is considered to be more selective, because fewer metal ions can activate a particular enzyme.
The binding of estrogen receptors (ERs) to EDCs has also been used to fabricate biosensors. A good example is the determination of estrogenic compounds in water by SPR using ER dimerization [67]. The ligand-activated ER dimer was detected by its interaction with a specific DNA consensus sequence estrogen-response element. Estrogenic compounds (e.g., 17β-estradiol, estriol, estrone, and ethynyl estradiol) activated the dimerization process at different concentration levels.
Habauzit et al. [68] used an SPR sensor for the determination of estrogenic compounds in water using the ER-dimerization properties. They demonstrated the direct detection of 17β-estradiol at concentrations above 1.4 μg/L and concluded that this method could be a good way to measure the estrogenic potency of compounds and their presence in water.
Such an SPR biosensor was also used for the detection of chemicals that may interfere with the thyroid system. There, inhibition assays with the two main thyroid hormone transport proteins, T4 binding globulin (TBG) and transthyretin (TTR), were used in combination with a T4-coated biosensor chip [69] and the most potent binding was observed with hydroxylated metabolites of the brominated diphenyl ethers (BDEs).
Whole-cell biosensors provide one of the newest tools used in environmental monitoring. They are particularly useful for assaying contaminant toxicity and bioavailability, and that makes them suitable for the detection of unknown agents. A recombinant yeast cell-based estrogen bioassay, expressing human ERα and yeast-enhanced green fluorescent protein in response to estrogens, was recently developed and applied to screening estrogenic activity in animal feed [70].
4. Concluding remarks
The choice of the multiplex bioanalytical technique is essentially dictated by the particular application in mind. The target analyte is not the only decision-driving force, but also the environment where the analysis needs to be performed and the implications of the results obtained. Our outlook, based on this review, suggests that the need for multiplexed analysis in food and environmental safety is met by rapidly developing biological binding-assay-based technologies (Fig. 3 ). The multi-analyte methods described feature versatile, innovative, technological platforms and implement a range of bio-recognition elements.
Figure 3.
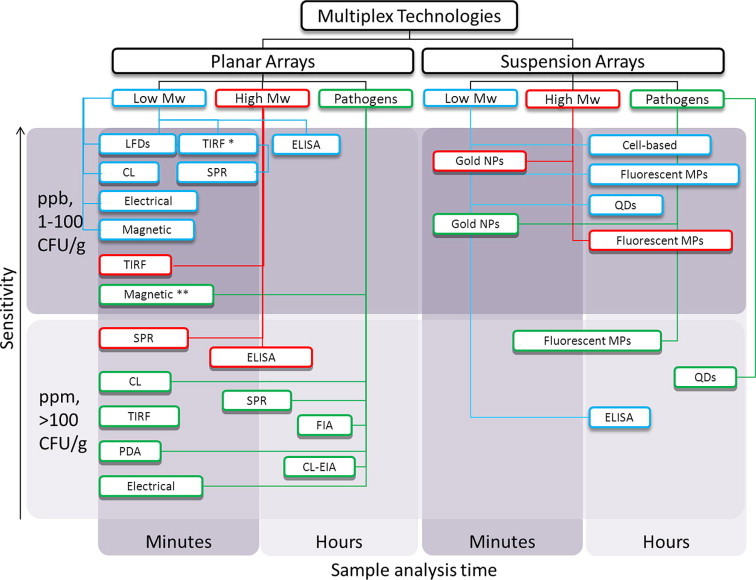
Overview of multiplex bioanalytical technologies currently applied for food and environmental analysis. The methods are distinguished first accordingly to the array platform (planar or suspension), and classified according to application (low/ high molecular weight analytes and pathogen detection). They are then classified with regard to sensitivity [parts per billion (ppb) and parts per million (ppm) for low and high molecular weight analytes, and colony-forming units (CFU) for pathogens; in gram food or mL buffer (depending on the assay)] and with regard to assay time (min or h). Abbreviations: CL, Chemiluminescence; TIRF, Total internal reflection fluorescence; ELISA, Enzyme-linked immunosorbent assay; LFD, Lateral flow device; SPR, Surface Plasmon Resonance, PDA, Polydiacetylene-based microarray; FIA, Fluorescence-based immunoassay; CL-EIA, Chemiluminescence-based enzyme-linked immunoassay; NP, Nanoparticle; MP, Microparticle; QD, Quantum dot. ∗Parts per trillion sensitivity. ∗∗pM sensitivity of PCR products
However, only a few novel technological platforms have been thoroughly studied with regards to application to real-life samples and even fewer validated. This might be the reason for the limited dissemination and application of these technologies to routine analysis.
Most probably, conventional analytical techniques in routine food safety and environment monitoring will not be completely replaced, since they are essential for confirmation purposes. However, suspension-array-based technologies have already become common screening techniques and portable planar-array-based biosensors will probably dominate on-site contaminant detection in the near future.
Acknowledgements
We thank Dr. Mariel G. Pikkemaat for her critical reading and remarks on the food pathogens section. We thank Professors Michael Seidel, Sang Jun Sim and Joseph Irudayaraj, and Jan van Gils for providing us with the original figures for use in this manuscript. We thank The Dutch Technology Foundation (STW) for funding this study (Project TMF 6635: “Multi-analyte screening with μfluidic biochips“).
References
- 1.European Environmental Agency (EEA), The EEA and UNEP Annual Message on the State of Europes’s Environment, 1998.
- 2.Bock S.A., Muñoz-Furlong A., Sampson H.A. J. Allergy Clin. Immunol. 2007;119:1016. doi: 10.1016/j.jaci.2006.12.622. [DOI] [PubMed] [Google Scholar]
- 3.Malik A.K., Blasco C., Picó Y. J. Chromatogr., A. 2010;1217:4018. doi: 10.1016/j.chroma.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Ben Rejeb S., Abbott M., Davies D., Claroux C., Delahaut P. Food Addit. Contam. 2005;22:709. doi: 10.1080/02652030500158450. [DOI] [PubMed] [Google Scholar]
- 5.Adrian J., Pinacho D.G., Granier B., Diserens J.M., Sanchez-Baeza F., Marco M.P. Anal. Bioanal. Chem. 2008;391:1703. doi: 10.1007/s00216-008-2106-9. [DOI] [PubMed] [Google Scholar]
- 6.Ligler F.S., Sapsford K.E., Golden J.P., Shriver-Lake L.C., Taitt C.R., Dyer M.A., Barone S., Myatt C.J. Anal. Sci. 2007;23:5. doi: 10.2116/analsci.23.5. [DOI] [PubMed] [Google Scholar]
- 7.Kloth K., Niessner R., Seidel M. Biosens. Bioelectron. 2009;24:2106. doi: 10.1016/j.bios.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Taylor A.D., Ladd J., Yu Q., Chen S., Homola J., Jiang S. Biosens. Bioelectron. 2006;22:752. doi: 10.1016/j.bios.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Seidel M., Niessner R. Anal. Bioanal. Chem. 2008;391:1521. doi: 10.1007/s00216-008-2039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magliulo M., Simoni P., Guardigli M., Michelini E., Luciani M., Lelli R., Roda A. J. Agric. Food Chem. 2007;55:4933. doi: 10.1021/jf063600b. [DOI] [PubMed] [Google Scholar]
- 11.Homola J., Yee S.S., Myszka D. In: Surface Plasmon Resonance Biosensors. Ligler F.S., Taitt C.A.R., editors. Elsevier Science; Amsterdam, The Netherlands: 2002. p. 7. [Google Scholar]
- 12.Dostálek J., Přibyl J., Homola J., Skládal P. Anal. Bioanal. Chem. 2007;389:1841. doi: 10.1007/s00216-007-1536-0. [DOI] [PubMed] [Google Scholar]
- 13.Kim S.J., Gobi K.V., Iwasaka H., Tanaka H., Miura N. Biosens. Bioelectron. 2007;23:701. doi: 10.1016/j.bios.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima H., Harada Y., Asano Y., Nakagama T., Uchiyama K., Imato T., Soh N., Hemmi A. Talanta. 2006;70:419. doi: 10.1016/j.talanta.2006.02.068. [DOI] [PubMed] [Google Scholar]
- 15.Mauriz E., Calle A., Manclús J., Montoya A., Lechuga L. Anal. Bioanal. Chem. 2007;387:1449. doi: 10.1007/s00216-006-0800-z. [DOI] [PubMed] [Google Scholar]
- 16.Fenton E.M., Mascarenas M.R., López G.P., Sibbett S.S. ACS Appl. Mater. Interfaces. 2008;1:124. doi: 10.1021/am800043z. [DOI] [PubMed] [Google Scholar]
- 17.Hee P.C., Pyo K.J., Wook L.S., Li J.N., Pil J.Y., Jun S.S. Adv. Funct. Mater. 2009;19:3703. [Google Scholar]
- 18.Elsholz B., Worl R., Blohm L., Albers J., Feucht H., Grunwald T., Jurgen B., Schweder T., Hintsche R. Anal. Chem. 2006;78:4794. doi: 10.1021/ac0600914. [DOI] [PubMed] [Google Scholar]
- 19.Kim J.S., Anderson G.P., Erickson J.S., Golden J.P., Nasir M., Ligler F.S. Anal. Chem. 2009;81:5426. doi: 10.1021/ac9005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng C., Li Z., Zhu Y., Chen W., Yuan Y., Liu L., Li Q., Xu D., Qiao R., Wang L., Zhu S., Jin Z., Xu C. Biosens. Bioelectron. 2009;24:3657. doi: 10.1016/j.bios.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 21.Chungang W., Joseph I. Small. 2008;4:2204. [Google Scholar]
- 22.Kulagina N.V., Lassman M.E., Ligler F.S., Taitt C.R. Anal. Chem. 2005;77:6504. doi: 10.1021/ac050639r. [DOI] [PubMed] [Google Scholar]
- 23.Kulagina N.V., Shaffer K.M., Anderson G.P., Ligler F.S., Taitt C.R. Anal. Chim. Acta. 2006;575:9. doi: 10.1016/j.aca.2006.05.082. [DOI] [PubMed] [Google Scholar]
- 24.Gehring A., Albin D., Reed S., Tu S.-I., Brewster J. Anal. Bioanal. Chem. 2008;391:497. doi: 10.1007/s00216-008-2044-6. [DOI] [PubMed] [Google Scholar]
- 25.Wolter A., Niessner R., Seidel M. Anal. Chem. 2008;80:5854. doi: 10.1021/ac800318b. [DOI] [PubMed] [Google Scholar]
- 26.Karsunke X., Niessner R., Seidel M. Anal. Bioanal. Chem. 2009;395:1623. doi: 10.1007/s00216-009-2905-7. [DOI] [PubMed] [Google Scholar]
- 27.Yeung S.W., Lee T.M.H., Cai H., Hsing I.M. Nucleic Acids Res. 2006;34:e118. doi: 10.1093/nar/gkl702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh B.-K., Lee W., Chun B.S., Bae Y.M., Lee W.H., Choi J.-W. Biosens. Bioelectron. 2005;20:1847. doi: 10.1016/j.bios.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Jin S.Q., Yin B.C., Ye B.C. Appl. Environ. Microbiol. 2009;75:6647. doi: 10.1128/AEM.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L., Zhao W., O’Donoghu M.B., Tan W. Bioconjug. Chem. 2007;18:297. doi: 10.1021/bc060255n. [DOI] [PubMed] [Google Scholar]
- 31.Yang L., Li Y. Analyst (Cambridge, UK) 2006;131:394. doi: 10.1039/b510888h. [DOI] [PubMed] [Google Scholar]
- 32.Koets M., Van Der Wijk T., Van Eemeren J.T.W.M., Van Amerongen A., Prins M.W.J. Biosens. Bioelectron. 2009;24:1893. doi: 10.1016/j.bios.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 33.Ballin N.Z., Vogensen F.K., Karlsson A.H. Meat Sci. 2009;83:165. doi: 10.1016/j.meatsci.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Maier I., Morgan M.R.A., Lindner W., Pittner F. Anal. Chem. 2008;80:2694. doi: 10.1021/ac702107k. [DOI] [PubMed] [Google Scholar]
- 35.Raz S.R., Liu H., Norde W., Bremer M.G.E.G. Anal. Chem. 2010;82:8485. doi: 10.1021/ac101819g. [DOI] [PubMed] [Google Scholar]
- 36.Haasnoot W., Du Pre J.G. J. Agric. Food Chem. 2007;55:3771. doi: 10.1021/jf063281o. [DOI] [PubMed] [Google Scholar]
- 37.Fantozzi A., Ermolli M., Marini M., Scotti D., Balla B., Querci M., Langrell S.R.H., Van Den Eede G. J. Agric. Food Chem. 2007;55:1071. doi: 10.1021/jf061506p. [DOI] [PubMed] [Google Scholar]
- 38.Bremer M.G.E.G., Smits N.G.E., Haasnoot W., Nielen M.W.F. Analyst (Cambridge, UK) 2010;135:1147. doi: 10.1039/b925372f. [DOI] [PubMed] [Google Scholar]
- 39.Maragos C.M. World Mycotoxin J. 2009;2:221. [Google Scholar]
- 40.Meneely J.P., Sulyok M., Baumgartner S., Krska R., Elliott C.T. Talanta. 2010;81:630. doi: 10.1016/j.talanta.2009.12.055. [DOI] [PubMed] [Google Scholar]
- 41.Lamberti I., Tanzarella C., Solinas I., Padula C., Mosiello L. Mycotoxin Res. 2009;25:193. doi: 10.1007/s12550-009-0028-9. [DOI] [PubMed] [Google Scholar]
- 42.Taitt C.R., Shriver-Lake L.C., Ngundi M.M., Ligler F.S. Sensors. 2008;8:8361. doi: 10.3390/s8128361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mak A.C., Osterfeld S.J., Yu H., Wang S.X., Davis R.W., Jejelowo O.A., Pourmand N. Biosens. Bioelectron. 2011;26:4503. doi: 10.1016/j.bios.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quiel A., Jurgen B., Piechotta G., Le Foll A.P., Ziebandt A.K., Kohler C., Koster D., Engelmann S., Erck C., Hintsche R., Wehland J., Hecker M., Schweder T. Appl. Microbiol. Biotechnol. 2010;85:1619. doi: 10.1007/s00253-009-2347-3. [DOI] [PubMed] [Google Scholar]
- 45.Pauly D., Kirchner S., Stoermann B., Schreiber T., Kaulfuss S., Schade R., Zbinden R., Avondet M.-A., Dorner M.B., Dorner B.G. Analyst (Cambridge, UK) 2009;134:2028. doi: 10.1039/b911525k. [DOI] [PubMed] [Google Scholar]
- 46.Wang J., Yang Y., Zhou L., Wang J., Jiang Y., Hu K., Sun X., Hou Y., Zhu Z., Guo Z., Ding Y., Yang R. Immunopharmacol. Immunotoxicol. 2009;31:417. doi: 10.1080/08923970902740837. [DOI] [PubMed] [Google Scholar]
- 47.Peters J., Bienenmann-Ploum M., Rijk De T., Haasnoot W. Mycotoxin Res. 2011;27:63. doi: 10.1007/s12550-010-0077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider M.J., Lehotay S.J. Anal. Bioanal. Chem. 2008;390:1775. doi: 10.1007/s00216-008-1918-y. [DOI] [PubMed] [Google Scholar]
- 49.Virolainen N.E., Pikkemaat M.G., Elferink J.W.A., Karp M.T. J. Agric. Food Chem. 2008;56:11065. doi: 10.1021/jf801797z. [DOI] [PubMed] [Google Scholar]
- 50.Urban A., Eckermann S., Fast B., Metzger S., Gehling M., Ziegelbauer K., Rabsamen-Waigmann H., Freiberg C. Appl. Environ. Microbiol. 2007;73:6436. doi: 10.1128/AEM.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel P.D. J. AOAC Int. 2006;89:805. [PubMed] [Google Scholar]
- 52.Huet A.C., Charlier C., Tittlemier S.A., Singh G., Benrejeb S., Delahaut P. J. Agric. Food Chem. 2006;54:2822. doi: 10.1021/jf052445i. [DOI] [PubMed] [Google Scholar]
- 53.Yue N., Wu L., Li L., Xu C. Food Agric. Immunol. 2009;20:281. [Google Scholar]
- 54.Lamar J., Petz M. Anal. Chim. Acta. 2007;586:296. doi: 10.1016/j.aca.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 55.Wang X., Li K., Shi D., Xiong N., Jin X., Yi J., Bi D. J. Agric. Food Chem. 2007;55:2072. doi: 10.1021/jf062523h. [DOI] [PubMed] [Google Scholar]
- 56.Xie H., Ma W., Liu L., Chen W., Peng C., Xu C., Wang L. Anal. Chim. Acta. 2009;634:129. doi: 10.1016/j.aca.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 57.Zacco E., Adrian J., Galve R., Marco M.P., Alegret S., Pividori M.I. Biosens. Bioelectron. 2007;22:2184. doi: 10.1016/j.bios.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 58.Raz S.R., Bremer M.G.E.G., Haasnoot W., Norde W. Anal. Chem. 2009;81:7743. doi: 10.1021/ac901230v. [DOI] [PubMed] [Google Scholar]
- 59.Knecht B.G., Strasser A., Dietrich R., Martlbauer E., Niessner R., Weller M.G. Anal. Chem. 2003;76:646. doi: 10.1021/ac035028i. [DOI] [PubMed] [Google Scholar]
- 60.Chen A., Wang G., Cao Q., Wang Y., Zhang Z., Sun Y., Wang H., Xu C., Zhou Q., Han P., Liu M., Yang Y., Xing W., Mitchelson K.R., Cheng J. J. Forensic Sci. 2009;54:953. doi: 10.1111/j.1556-4029.2009.01049.x. [DOI] [PubMed] [Google Scholar]
- 61.W. Haasnoot, H.D.L.M. Eekelen Van, M.E. Bienenmann-Ploum, H. Gercek, M.W.F. Nielen, Proc. EuroResidue VI Conf., 19–21 May, Egmond aan Zee, The Netherlands, 2008, p. 5.
- 62.Liu N., Su P., Gao Z., Zhu M., Yang Z., Pan X., Fang Y., Chao F. Anal. Chim. Acta. 2009;632:128. doi: 10.1016/j.aca.2008.10.061. [DOI] [PubMed] [Google Scholar]
- 63.Nichkova M., Dosev D., Davies A.E., Gee S.J., Kennedy I.M., Hammock B.D. Anal. Lett. 2007;40:1423. doi: 10.1080/00032710701327088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo Y.R., Liu S.Y., Gui W.J., Zhu G.N. Anal. Biochem. 2009;389:32. doi: 10.1016/j.ab.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 65.Weller M.G., Schuetz A.J., Winklmair M., Niessner R. Anal. Chim. Acta. 1999;393:29. [Google Scholar]
- 66.Wanekaya A.K., Chen W., Mulchandani A. J. Environ. Monit. 2008;10:703. doi: 10.1039/b806830p. [DOI] [PubMed] [Google Scholar]
- 67.Badihi-Mossberg M., Buchner V., Rishpon J. Electroanalysis (NY) 2007;19:2015. [Google Scholar]
- 68.Habauzit D., Armengaud J., Roig B., Chopineau J. Anal. Bioanal. Chem. 2008;390:873. doi: 10.1007/s00216-007-1725-x. [DOI] [PubMed] [Google Scholar]
- 69.Marchesini G.R., Meimaridou A., Haasnoot W., Meulenberg E., Albertus F., Mizuguchi M., Takeuchi M., Irth H., Murk A.J. Toxicol. Appl. Pharmacol. 2008;232:150. doi: 10.1016/j.taap.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 70.Bovee T.F.H., Bor G., Heskamp H.H., Hoogenboom R.L.A.P., Nielen M.W.F. Food Addit. Contam. 2006;23:556. doi: 10.1080/02652030600557163. [DOI] [PubMed] [Google Scholar]


