Abstract
Aptamers are nucleic acid ligands that are generated artificially by in vitro selection and behave similar to antibodies. The development of aptamer-based sensing systems or strategies has been in vogue for the past few decades, because aptamers are smaller in size, stable, cheaper and undergo easier modifications. Owing to these advantages, several facile aptamer-based colorimetric strategies have been created by controlling the assembly and disassembly of aptamers on unmodified gold nanoparticle probes. As these kinds of assay systems are rapid and can be visualized unaided by instruments, they have recently become an attractive method of choice. The formation of purple-colored aggregates (attraction) from the red dispersed (repulsion) state of GNPs in the presence of mono- or divalent ions is the key principle behind this assay. Due to its simplicity and versatility, this assay can be an alternative to existing diagnostic assays. Here, we have investigated the critical elements involved in colorimetric assays, and have screened different proteins and small ligands to evaluate biofouling on GNPs.
Keywords: Gold nanoparticles, Aptamer, Colorimetric, Aggregation, Biofouling
Graphical abstract
Highlights
-
•
Facile aptamer-based colorimetric strategies have been discussed.
-
•
Controlling the assembly and disassembly of aptamers on unmodified gold nanoparticles.
-
•
Rapid assay system and can be visualized unaided by instruments.
-
•
Screened different proteins and small ligands to evaluate biofouling on gold nanoparticles.
-
•
Very much suitable for aptamer interactions with small molecules rather than proteins.
1. Introduction
Nanomaterials can facilitate signal transduction with electroactive tags for sensing and imaging purposes. The benefits obtained from nanotechnology mainly depend on specific tailored materials designed with essential structures at the nanoscale level to achieve a specific goal, thus greatly extending the range of applications, including diagnosis. Among the several nanomaterials that have been fabricated, the gold nanoparticle (GNP) is an ideal material that is widely used in the development of sensors, owing to its unique characteristics, such as easy water dispersal, compatibility with surface functionalization, biological non-reactivity, and ability to be tailored with uniform and different nano-sizes (Lim et al., 2011, Upadhyayula, 2012, Guirgis et al., 2012). GNPs adsorb in the visible light spectrum around 520 nm (green light) due to the excitation of plasmons in the particle and this wavelength can be adapted to several optical sensors (Nagel et al., 2011, Tinguely et al., 2011, Gopinath et al., 2012a, Gopinath et al., 2012b, Gopinath et al., 2013). In the past, several attractive gold-based sensing surfaces have emerged, and the applications of these surfaces have expanded to sensor development in conjunction with various fields (Baldrich et al., 2004, Varma et al., 2004, Guo, 2005, Gronewold et al., 2005, Odenthal and Gooding, 2007, Peng et al., 2007, Wang et al., 2008, Gopinath et al., 2008a, Marquette and Blum, 2008, Song et al., 2008, Giljohann et al., 2010, Iliuk et al., 2011, Zanoli et al., 2012).
Beneficial sensing systems have several characteristics, including ease of use, low cost, rapid process, and results that are easily understandable without prior knowledge and distinguishable by the naked eye. Several sensing systems that exhibit these characteristics have been proposed and commercially established, including color-based sensing on membrane filters (such as immunochromatography tests) or solution-based colorimetric assays (Pavlov et al., 2004, Huang et al., 2005, Liu and Lu, 2006, Wang et al., 2007, Wei et al., 2007, Chen et al., 2008, Zhao et al., 2008a, Li et al., 2009, Lau et al., 2010, Kim et al., 2010, Song et al., 2011, Chávez et al., 2012, Selvakumar and Thakur, 2012). However, the compatibility of the biomolecules used as ligands or analytes must be ascertained. Considering these basic parameters of ideal nanomaterials, i.e., visual interpretation of assay results, and biocompatibility, GNP-based colorimetric assays might be the suitable candidates. GNP-based colorimetric assays can be efficient biosensors, as the molecular recognition event can be transduced visually, obviating the need for any special equipment (Zhao et al., 2008a, Kim and Jurng, 2011). The involvement of interparticle plasmon coupling or local refractive index change-induced plasmon band shift makes this colorimetric assay facile. GNPs are capable of adsorbing small oligonucleotides due to their propensity for electrostatic attractions, hydrophobic absorption, and covalent binding (Zhao et al., 2008a, Lou et al., 2012). Under the category of small oligonucleotides, aptamers have been developed rapidly for use in diagnosis, drugs, molecular validation, etc. (Gopinath, 2011). The features of aptamers including their variety of structural conformations, and their selectivity and specificity, yield excellent matches with target molecules and are ideal for aptasensor development.
Aptamers are artificially evolved molecules against specific targets, from small molecules to whole cells, that are generated by the separation and amplification processes using randomized RNA or DNA libraries. This strategy involves an in vitro selection process, called ‘systematic evolution of ligands by exponential enrichment’ (SELEX). This selection process mimics the natural selection of aptamers as reported in the case of riboswitches (Breaker, 2011). A typical aptamer generation process starts with about 1014 molecules, under stringent conditions. Only high-affinity molecules are retained through successive selection cycles ( Fig. 1). The selected aptamers in the past have possessed significant (>10,000-fold) abilities to discriminate between closely related molecules (Jenison et al., 1994, Geiger et al., 1996). The ability of aptamers to discriminate between molecules has led to the development of high-performance sensing systems. Several functionalization chemistries or modification procedures are available for sensing plates or on aptamers to form the appropriate anchorage and create aptasensors. However, these approaches may have limitations with lower-affinity interactions due to structural changes upon aptamer modification, high cost, and low yield of modified products. The strategy of spontaneous adsorption becomes an easy way to resolve these issues. As demonstrated previously, the GNP-based colorimetric assay is a popular strategy for aptamer–ligand interactions, and does not involve modifying the analyst or ligands (Pavlov et al., 2004, Wei et al., 2007, Wang et al., 2007, Kim et al., 2010, Selvakumar and Thakur, 2012). In the present overview, the colorimetric detection of the controlled assembly and disassembly of GNP-based aptamer–ligand interactions has been described. Further, the suitability of these assays for biomolecular interactions are validated by testing their bio-fouling and non-biofouling characteristics.
Fig. 1.

Schematic representation of the process of ‘systematic evolution of ligands by exponential enrichment (SELEX)’. This process involves the selection of high-affinity molecules from a randomized library of molecules by separation and amplification processes, under stringent conditions. Only high affinity molecules are retained with successive selection cycles. After a few to several rounds of selection cycles, molecules with higher affinities are cloned and sequenced to identify the selected candidates.
2. Factors influence GNP-based colorimetric assays
Colorimetry is a solution-based assay that is used to determine the concentration of colored compounds. A colorimeter or standard spectrophotometer can be used to estimate the concentration of materials in the solution by measuring its absorbance at a suitable wavelength. In this assay, the length of the light path through the solutions is measured by absorbance, with the units of optical density. The color or wavelength of the filter chosen for the colorimeter is the critical one, as the wavelength of light that is transmitted by the colorimeter has to be same as that absorbed by the substance being measured. With these basic parameters, several GNP-based colorimetric assays for aptamer–ligand interactions have been developed; these assays also depend on several other parameters as described below.
2.1. Effects of mono- and divalent ions
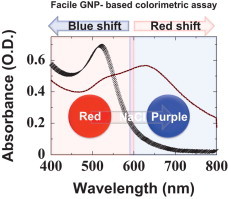
This assay involves the strategy of using balanced interparticle attractive or interparticle repulsive forces to cause aggregation or dispersion, respectively (due to gain or loss of surface charges); van der Waals attractive forces on the surface also cause aggregation (Zhao et al., 2008a). These two states are determined by the presence or absence of ions in the gold colloidal solutions. The as-received gold colloidal solution exhibits strong absorbance of visible light at a wavelength of 520 nm, due to the excitation of plasmons ( Fig. 2a) (Nagel et al., 2011, Tinguely et al., 2011, Gopinath et al., 2012b, Gopinath et al., 2013). Due to the above mechanism, in the presence of salts, GNPs can change color from red to purple (Fig. 2b). The addition of NaCl or other salts caps the repulsion among unmodified negatively charged GNPs, inducing the aggregation of these particles and resulting in a purple or blue solution. To study the dose dependence of these changes on monovalent ions (NaCl), we titrated different concentrations of NaCl from 15 to 120 mM with serial 50% dilutions (15, 30, 60, and 120 mM). As shown in Fig. 2b, with increasing concentrations of NaCl, there were gradual changes in the color of the GNPs from red to purple. At the 15 mM concentration, the color of the solution was reddish-purple because it was in the transition stage, and the color of the solution turned into completely purple at NaCl concentrations >30 mM. Most of these colorimetric assays were designed with an NaCl-induced assembly process (Li and Rothberg, 2004, Wang et al., 2007, Chen et al., 2008, Zhao et al., 2008a, Lou et al., 2012). To examine the effect of divalent ions on these color changes, we tested both CaCl2 and MgCl2, as these 2 candidates are involved in several aptamer–ligand interactions. Based on the NaCl titrations, we chose a concentration of 100 mM for these ions and compared the results with those of the NaCl. These tests revealed that monovalent (NaCl) and divalent (CaCl2 and MgCl2) ions have similar effects; in all cases, the color of the GNPs changed from red to purple (Fig. 2b). The dispersion state of GNPs was evaluated spectrophotometrically and an apparent spectral peak appeared with maximal absorption at nearly 520 nm. When the state was altered using salts, the solution turned purple and drastic changes in the spectral peak could be visualized, with maximal absorbance at nearly 630 nm (Fig. 2c). These changes in the appearance of GNPs under dispersed and aggregate conditions can be visualized under microscope (Fig. 2d). Previously, ions other than NaCl were used for the aggregation of GNPs, as discussed above (Huang et al., 2005, Zhao et al., 2008a). It has been suggested that magnesium ions have a greater affinity to the phosphate groups of the nucleic acid backbone and cause severe effects in neutralizing the molecules (Huang et al., 2005). Because both mono- and divalent ions affect the aggregation of GNPs, an important point to consider is the inclusion of the appropriate ions in addition to NaCl for optimal aptamer performance and ligand binding in GNP-based colorimetric assays.
Fig. 2.
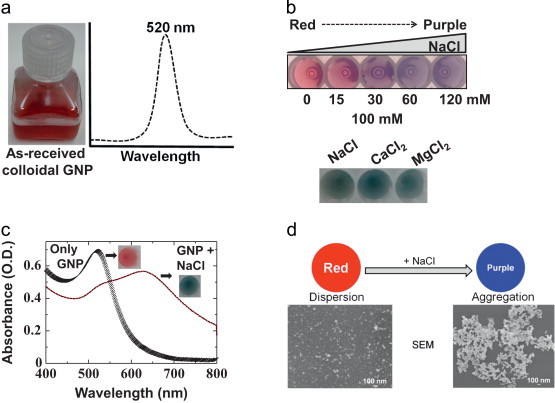
(a) Spectra of as-received GNPs, (b) titrations to evaluate salt-induced aggregation. NaCl concentrations from 15 to 120 mM were used. The color changes from red to purple with increasing salt. Salt induced aggregation of GNPs; 100 mM of different salts were tested. (c) Spectral changes in as-received GNP upon salt-induced aggregation. Color changes from red to purple are indicated. (d) Observation of the assembly and disassembly process of GNPs under salt induction. GNPs were visualized by scanning electron microscopy (SEM; JEOL, JSM-6340F). A scale bar is provided. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. Influence of sizes of GNPs
Apart from the effect of mono- or divalent salts, the sizes of the GNPs may also influence colorimetric assays. Several researchers reported application of GNPs in different sensor platforms using different sizes of GNPs (Lou et al., 2012; references therein). In general, an increase in GNP size, with the concomitant increase in surface area, requires the consumption of greater amounts of aptamers for any given assay. Generally, a GNP size between 5 and 40 nm is ideal for sensor development (Christopher et al., 2005, Lou et al., 2012, Gopinath et al., 2013). Successful fabrication of a label-free biochip based on GNPs, shown that the sizes of diameters in the range of 12–48 nm significantly affects the sensitivity, the detection limit for protein and small molecule with 39-nm-diameter GNPs was 20-fold higher than 13 nm GNPs (Nath and Chilkoti, 2004). Gopinath et al. (2012b) have evaluated the GNPs with different sizes on the waveguide-mode sensing surface modified with varied surface chemistries. In terms of sensitivity, Lou et al. (2012) concluded that larger GNPs are necessary, and demonstrated higher sensitivity using 38-nm GNPs. Smaller GNPs are more stable than larger ones. Larger GNPs will yield more rapid color appearance, but will also undergo self-coagulation. It has been reported that average errors would be negligible for biomolecules attached to larger GNPs, but appreciable with smaller GNPs (Lou et al., 2012) ( Fig. 3a). Medley et al. (2008) screened different sizes of GNPs (5, 20, 50, and 100 nm) and chose 20-nm GNPs for the detection of cancer cells.
Fig. 3.
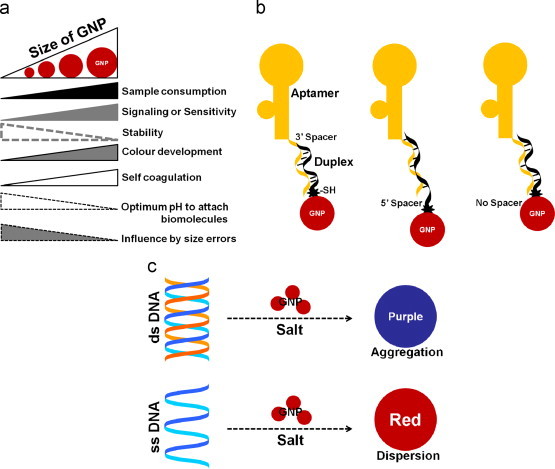
(a) Characteristics of GNPs based on size (Lou et al., 2012). (b) Representation of the effect of spacers on duplex formation. Shown is an extended aptamer tail with a 5′–or 3′-end spacer. (c) Influence of double-stranded and single-stranded DNA on the salt-induced aggregation of GNPs.
Recently, Lou et al. (2012) analyzed antibody attachment and the effect of pH with different sizes (14, 16, 35, and 38 nm) of GNPs. Obviously, a lower amount antibody was required to coat the 16-nm GNP than for GNPs of larger diameter. The optimum pH for the attachment of biomolecules is higher for smaller GNPs than for larger GNPs. However, it was recommended to use acidic pH to immobilize proteins on the gold surfaces (Gopinath, 2010).
2.3. Influence of aptamer strands and lengths
GNPs are mostly investigated for biosensor development due to its noble property and demonstrated for a high sensitivity with oligonucleotide (Guo, 2012). A strategy using aptamer as reversible crosslinkers for ‘sensing aggregates’ with GNPs has been proposed (Chávez et al., 2010). In several instances, sensor surfaces that have been developed for nanoscale biosensors have gold surfaces that use thiolated biomolecules as linkers (Baldrich et al., 2004, Gronewold et al., 2005, Guo, 2005, Odenthal and Gooding, 2007, Marquette and Blum, 2008, Song et al., 2008). This method has no complications to immobilize them on GNPs, forms stable particles with negatively charged ssDNA and further found to stabilize upon binding to the target (Zhao et al., 2008b). Aptamers provide various options with structural variations to bind to their selected target molecules, and these vary with the length of the aptamers (Fig. 3b). Considerations of aptamer lengths are very critical when the aptamer is immobilized on a solid surface (Chávez et al., 2012). In connection with duplex formation, it is also necessary to consider the length of the aptamer or the extension of the aptamer sequence in order to enable complementation with other sequences. Nucleic acids are composed of four nucleotides, adenosine (A), cytidine (C), guanosine (G) and thymine (T), and thymine in DNA, is replaced by uracil (U) in RNA. Canonical complementation occurs between A and T or U, and between G and C. Schreiner et al. (2011) have analyzed the surface-induced weakening and the associated asymmetry in hybridization responses of two strands forming a hairpin stems on gold surfaces. Storhoff et al. (2002) reported that the stability of ssDNA on GNPs is sequence specific, they analyzed with different nucleosides (dA, dG, dC, and dT) and shown different binding affinities to the assemblage. Moreover, dT has a much lower binding affinity to the GNP surface than dG, dC, and dA. In corroboration with these studies, it was reported that thymines are interacting minimally with gold surfaces, whereas adenine interacts well (Schreiner et al., 2011, Chávez et al., 2012).
In the above context, there are differences in electrostatic attractions between double-stranded (ds) or single-stranded (ss) DNA molecules to the GNPs. Upon hybridization, there will be changes in electrostatic, steric repulsion or both (Fujita et al., 2012). In addition, a detailed study with the covalent attachment of GNPs to DNA template was reported by Stevenson et al. (2002). It was also reported that GNPs are capable of adsorbing small oligonucleotides due to hydrophobic absorption in addition to electrostatic attractions and covalent binding (Zhao et al., 2008a, Lou et al., 2012). Peng et al. (2013) have explained the colorimetric detection strategy using ssDNA-modified GNPs binding to the target due to van der Waals force. The advantage of GNPs is that the atoms on the GNP surface can interact with the nitrogenous bases of the ssDNA, enabling their attachment. Usually, ssDNA is flexible and can partially uncoil and expose its bases, leading to greater distances between the phosphate groups, and the molecules therefore have less negative charges. GNP is usually stabilized, for example with citrate, and has a negative charge and repulsion between similarly charged GNPs prevents van der Waals attractions among the GNPs leading to no aggregation. Due to its similar charge, dsDNA cannot attract GNPs and the dispersed GNP will aggregate with NaCl even in the presence of dsDNA (Fig. 3c) (Li and Rothberg, 2004, Wang et al., 2007). Based on this principle, Li and Rothberg (2004) developed a strategy for mismatch analyses on DNA strands. To construct an ideal duplex, a spacer can be created at the 5′ end where the linker is available, or at the 3′ end. The length of the spacer may cause a sliding effect on duplex formation, leading to misalignment. In this case, duplex formation without the spacer regions is highly recommended (Fig. 3b). However, Zhao et al. (2008a) reported that an anti-adenosine aptamer with a thymine-10 (T10) spacer at the 5′ end folded properly and enhanced the stability of the gold colloid more effectively than in the absence of a spacer. Another study with alkanethiol-capped oligonucleotides has been performed using GNPs with poly dT, dA, and dC oligonucleotide sequences of 5–20 bases in length, and the stabilities of the particles were measured. GNPs with poly dT oligonucleotides exhibited a dramatic increase in stability as the length increased from 5 to 20 bases (Storhoff et al., 2002). Recently, it was found that stacking effect with duplex formation of 8 bases to anti-platelet-derived growth factor aptamer, caused significant improvement in the signal-to-noise ratio for GNP-based colorimetric assays (Chang et al., 2012). Chávez et al. (2012) analyzed riboflavin-binding aptamers of different lengths, generated by the addition of bases at the 5′ end. They generated 3 sizes: wild-type (no additional sequences), short-tail (5 random bases at the 5′ end) and long-tail (17 random bases at the 5′ end). Based on their analyses of all these aptamer derivatives, the authors found a tail of 17 random additional bases were more effective than a shorter extension of 5 random bases; however, the response was not as good as no nucleotides were added. The above studies suggested that even though the lengths of the aptamer–GNP conjugates do not cause significant variations, the lengths of the aptamers might play a role in stabilization.
2.4. Influence of aptamer conformations
Generally, nucleic acids or aptamers show proper folding and form stem and loop regions, called paired and unpaired regions to create appropriate structures. With these proper structures, the aptamer can form the ideal fit and bind tightly to its appropriate ligand. Several software-based or biochemical-based strategies are available to correctly predict aptamer structural information (Gopinath, 2009). Mixtures of canonical and non-canonical conformations, which formed the 4-stranded tetraplex G-quartet structure, were found to increase the charge density on the GNPs, causing the color to remain red in salt-induced ionic conditions even in the presence of the target molecules (Chen et al., 2008). This was also demonstrated for the G-quartet conformation of anti-thrombin aptamers on the GNPs in the presence of the target (Wei et al., 2007).
However, it is necessary to collect information from previous reports about the effects of either folded (properly structured) or unfolded (variations in the structure for the same molecules) aptamers used in the GNP-based colorimetric assays. Several DNA aptamers are exhibiting unique secondary structures, found not to have ability of stabilizing GNPs electrostatically and these aptamers are not suitable for GNP-based colorimetric assays (Zhang et al., 2012). Previously, both properly folded (Zhao et al., 2008a) and improperly folded (Selvakumar and Thakur, 2012) aptamers were used in aptamer-based colorimetric assays. Usually, unfolding happens during the process of aptamer preparation due to the usage of denaturing agents such as urea. However, the proper folding of these unfolded molecules with varied structures can be obtained in this case by denaturing at higher temperature (~92 °C) for 1 or 2 min and cooling to room temperature. In addition, the presence of mono- or divalent ions also assists in proper folding. Selvakumar and Thakur (2012) have reported that even though the anti-vitamin B12 (cyanocobalamin) aptamer was originally selected and used with 1 M lithium chloride (LiCl) for proper folding, the aptamer–target complex formation retains its sensitivity in the absence of LiCl. However, Zhao et al. (2008a) evaluated folded and un-folded DNA aptamers against the target, adenosine, and found that properly folded aptamers showed unique stabilized binding on the GNP that was stable against salt-induced aggregation, whereas unfolded aptamers bound on the GNP were less stable, indicating the importance of properly folded conformations of aptamers for this colorimetric assay. We also carried out the attachment of 3 µM aptamers composed of a 34mer (anti-factor IXa), under heat-denatured and non-heated conditions on the GNP, followed by the addition of 100 mM NaCl, and found that both experiments created stable conditions and prevented aggregation ( Fig. 4). In this case, due to the smaller size of the aptamer, heat-denaturing conditions might not be critical. However, it is commonly expected that a folded aptamer will have higher affinity to the target molecules than an unfolded one. The stability against salt conditions is typical when GNPs and aptamer conjugates exist, which do not aggregate. However, if we add an appropriate target molecule, the GNP in the reaction mix forms aggregates upon the addition of NaCl due to the separation of the attached aptamers from the GNP by aptamer–target complex formation (Fig. 4). However, greater colloidal stabilization (red-colored GNP) was reported with the addition of target to the aptamer-linked GNP (Zhao et al., 2008a). With these strategies, several colorimetric assays have been previously carried out using GNP-aptamer conjugates as the probes. Targets of different molecular sizes have been tested with their aptamer conjugates on the GNP; these strategies are discussed below ( Table 1).
Fig. 4.

Schemes showing the interaction of salt-immobilized GNPs with target molecules. The formation of dispersed and aggregated GNPs under ionic conditions is indicated by the color of the GNPs. The effect of folded and unfolded aptamers on GNP aggregation is shown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Molecules involved in GNP-based colorimetric assays.
| Size of GNP (nm) | Oligo type | Oligo length | Target | Detection limit | UV–visible spectra | References |
|---|---|---|---|---|---|---|
| 12±1 | DNA | – | Thrombin | 20 nM | Red-shift | Pavlov et al. (2004) |
| 15 | DNA | 27 | ATP | 600 nM | Red-shift | Wang et al. (2007) |
| 13 | DNA | 29 | Thrombin | 830 pM | Red-shift | Wei et al. (2007) |
| 20 | DNA | 94 | Cancerous cells | 90 cells | Red-shift | Medley et al. (2008) |
| 14 | DNA | 24 | Mercury | 3 µM | Red-shift | Xue et al. (2008) |
| 13 | DNA | 10 | Mercury | 600 pM | Red-shift | Li et al. (2009) |
| 13 | DNA | 76 | Oxytetracylcine | 25 nM | Red-shift | Kim et al. (2010) |
| 13 | DNA | 21 | Kanamycin | 25 nM | Red-shift | Song et al. (2011) |
| 13 | DNA | 58 | Dopamine | 360 nM | Red-shift | Zheng et al. (2011) |
| 13 | DNA | 33 | Platelet-derived growth factor | 6 nM | Red-shift | Chang et al. (2012) |
| 17±0.6 | DNA | 29 | Riboflavin | 100 nM | Red-shift | Chávez et al. (2012) |
| 15 | RNA | 35 | Vitamin B12 | 0.1 µg/ml | Red shift | Selvakumar and Thakur (2012) |
| 13 | DNA | 19 | Ampicillin | 10 ng/ml | Red shift | Song et al. (2012) |
| 13.3±1.2 | DNA | 27 | Thrombin | 5 pM | Red shift | Peng et al. (2013) |
| 13 | DNA | 22 | Sulfadimethoxine | 50 ng/ml | Red-shift | Chen et al. (2013) |
| 13.3±1.2 | DNA | 35 | PDGF | 25 nM | Blue-shift | Huang et al. (2005) |
| 13 | DNA | 27 | ATP | 10 nM | Blue-shift | Chen et al. (2008) |
| 19.6±0.1 | DNA | 27 | Adenosine | 20 µM | Blue-shift | Zhao et al., 2008a, Zhao et al., 2008b |
3. Detection of small ligands
Observations of small-molecule interactions are difficult to analyze by several currently available sensors. Aptamer and small molecule interactions are important because small molecules are mainly involved in drug development. Several currently available drug or chemical libraries used in the drug-discovery process is mainly composed of small molecules. Wang et al. (2007) tested the effect of dsDNA or ssDNA aptamer binding to ATP using this assay; sequences complementary to the aptamer were liberated into the solution upon the addition of ATP and caused concomitant changes in the color of the solution. Chen et al. (2008) analyzed urinary adenosine samples with a G-quartet aptamer structure and found that the red-wine color of the GNPs with higher salt conditions persisted even in the presence of target molecules (Chen et al., 2008). With respect to folded and unfolded aptamers, Zhao et al. (2008a) further analyzed aptamer–adenosine interactions; the GNP-aptamer conjugates were stabilized further upon the addition of the target. In these cases, the wavelength of the absorbed light is blue-shifted rather than red-shifted, as reported in other cases. Aptamer-based colorimetric detection of a small molecule, Hg2+, on unmodified gold nanoparticle (UGNP) as the colorimetric probe was performed by Li et al. (2009). As discussed above, this assay was carried out based on differential salt-induced aggregation of UGNP in the presence or absence of the target molecule Hg2+. In the presence of Hg2+, 2 T-rich single-stranded oligonucleotide DNA aptamers form a duplex through the T–Hg2+–T complex, are thus unable to bind UGNP, and fail to stabilize the particles. In the absence of Hg2+, ssDNA oligonucleotide aptamers bind UGNP via the nitrogen atoms of the bases. This stabilizes the nanoparticles, which exhibit resistance to salt-induced aggregation, giving the solution a red appearance. The corresponding changes in UV–visible spectra were used to quantitatively analyze Hg2+. In this study, at 5 min incubation, the absorption ratio (A 670/A 520) of the assay demonstrated a linear correlation to the target concentration (from 1×10−4 M to 1×10−9 M). The sensitivity of GNPs is due to the high molar absorptivity in the visible region (as is evident from the higher absorption ratio with increasing Hg2+ concentration). This assay achieved a picomolar detection limit (600 pM). Similarly, using DNA/nanoparticle conjugates, Xue et al. (2008) developed detection systems with mercury, although they obtained a higher detection limit (in the order of micromolar concentrations). In a further advance in colorimetric-based assays, Ogawa and Maeda (2009) designed logic gates, based on aptazymes with small ligands. To formulate a universal strategy, Xia et al. (2010) designed colorimetric detection assays for DNA, proteins, small molecules, and ions on the GNP and conjugated polyelectrolytes. High sensitivity detection of an anti-bacterial agent (oxytetracycline) by aptamer-coated GNPs was demonstrated for small molecule analyses (Kim et al., 2010). Another DNA-aptamer based detection assay has been developed for kanamycin using a similar strategy (Song et al., 2011). Chen et al. (2013) made critical analyses with aptamer-immobilized GNPs against the target sulfadimethoxine and determined the pH value, aptamer concentration and salt concentration as 8.0, 2 µM and 2 M, respectively, for the sensitive detection. Most colorimetric-based detection systems have been used DNA–aptamers, however, a detection system has been developed for vitamin B12 using a 2′-fluoro modified RNA aptamer immobilized on GNPs (Selvakumar and Thakur, 2012). The reported GNP-based colorimetric assays rely on small molecules and aptamer interactions in most cases, including those discussed in this section (Table 1).
4. Detection of macromolecules
In the above section, we have discussed GNP-based colorimetric assays for small molecules; similar detection systems have also been proposed for macromolecules such as proteins and whole cells (Table 1). Among several molecules that have been tested with GNP-based colorimetric strategies, thrombin is one of the predominant targets (Pavlov et al., 2004, Lu and Liu, 2006, Wei et al., 2007, Jian and Huang, 2011, Chen et al., 2012, Peng et al., 2013). Pavlov et al. (2004) reported the use of an aptamer-functionalized GNP, using thrombin as the target molecule, with a detection sensitivity limit of 20 nM. Confirming these results, the addition of thrombin to other control nucleic acid did not change the color with the addition of salt; moreover, bovine serum albumin (BSA) and human IgG maintained the stability as these are negative samples for anti-thrombin aptamer. Similarly, Wei et al. (2007) demonstrated this assay with the anti-thrombin–thrombin interaction and discussed proper aptamer folding in the presence of thrombin. Recently, Peng et al. (2013) demonstrated the colorimetric assay with thrombin and attained the sensitivity as low as 5 pM. Huang et al. (2005) designed different strategies for evaluating the interaction between platelet-derived growth factor (PDGF) and the PDGF receptor using GNPs modified with thiolated aptamers as probes with a view to develop a colorimetric sensing assay for cancer diagnosis. They found that aptamer-immobilized GNPs were dispersed at lower and higher concentrations of the target, and aggregated at medium concentrations, due to the formation of cross-links between the aptamer-modified GNPs. The addition of 0.1% BSA to the anti-PDGF-modified GNPs does not allow the formation of aggregates in NaCl concentrations up to 3 M. Chang et al. (2012) also demonstrated specific interactions between the target PDGF and the anti-PDGF aptamer with a different strategy. Apart from proteins, the development of sensors for the detection of whole cells, such as those causing cancer, is very important in terms of medical diagnosis. Gold materials-based detection has been used in the development of tumor sensors, drug-delivery agents, and as enhancers in plasmonic photothermal therapy to treat cancer (Lim et al., 2011). Using aptamer and GNP conjugates, a colorimetric assay for diseased (cancer) cell detection that discriminates against normal cells has been demonstrated by Medley et al. (2008).
5. Molecular screening on GNP – ‘biofouling’
In the studies discussed above, the issue of biofouling, which involves non-specific binding of target molecules on UGNP, remains unclear. In the past, especially in sensor development, several attempts have been made to prevent non-specific binding and improve the specificity of the sensing system (Nagasaki, 2011). Even though several natural polymers are available, the use of synthesized polymers is an efficient way to reduce such non-specific interactions (Otsuka et al., 2000, Nagasaki et al., 2007, Kamimura et al., 2008, Yoshimoto et al., 2010, Xue et al., 2008, Xia et al., 2010). These studies have significantly improved the quality and sensitivity of several sensing strategies. Non-specific bio-fouling remains one of the key issues in the development of higher sensitivity sensors; therefore, in the development of colorimetric assays, we screen different macro- and small molecules for specific attachment. In GNP-based colorimetric assays, aptamers are attached to the GNP by physical adsorption and are considered basic molecules. However, the attachment of the target to the GNP, irrespective of its interaction with the aptamers, is not investigated in many instances. To answer this issue, molecular screening should be performed in these studies with different commonly used molecules.
5.1. Macromolecules (proteins)
Non-specificity is commonly seen in the case of macromolecules such as proteins in several sensing systems, and is drastically reduced or nullified with small molecules. To address the question of specific interactions, we initially screened different classes of important proteins that are involved in several applications. The screened proteins were human clotting factor IX (FIX), erythropoietin (EPO), hemagglutinin (HA), severe acute respiratory syndrome (SARS) envelope-GST, rabbit IgG, streptavidin, protein A, and BSA. These proteins were directly attached on the as-received GNPs (dispersed state) followed by the addition of NaCl with the expectations of forming aggregates. As reported recently (Chang et al., 2012), 60 mM NaCl was noticed as an enough ionic condition to change the color of the GNPs from red to purple in the absence of protein. Initially, we used FIX at a concentration of 100 nM for adsorption on the GNPs; with the addition of 60 mM NaCl there was no change in the reddish color of the GNPs and they remained in the dispersed state. This state was unchanged even with the addition of 10-fold higher NaCl (600 mM) and remained constant even under such strong ionic conditions ( Fig. 5). This indicates that the FIX protein has a strong affinity for GNPs and prevents the formations of aggregates. To test whether this effect was seen only for FIX or with other proteins too, we screened 2 more proteins (EPO and HA), and the results showed similar effects of the physical adsorption of EPO and HA on the GNPs. Further screening with other proteins (SAR envelope-GST, rabbit IgG, streptavidin, protein A and BSA) with 100 nM concentration and the addition of 100 mM NaCl was also yielded the similar results (Fig. 5). Based on these results, it can be generalized that most proteins can adsorb physically to GNPs and may not be suitable for the evaluation on GNP-aptamer conjugates without reducing non-specificity. Tsai et al. (2012) have also shown the strong physical adsorption of BSA on GNPs with their displacement strategy using dithiothreitol. In reporting aptamer-protein interactions using GNP-based colorimetric assays, the selection of the appropriate negative control proteins is mandatory (to void non-specific binding). It is wise to test the desired target molecules for non-specificity before performing the main interactive analyses that rely on colorimetric assays with GNP-aptamer conjugates.
Fig. 5.
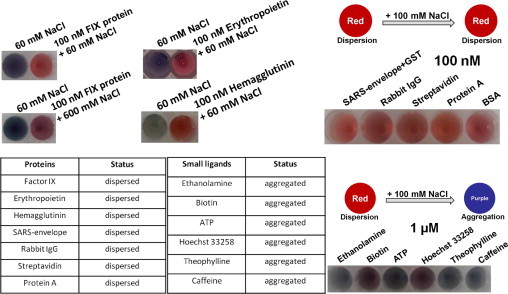
Screening of macro- and micro-molecules for salt-induced aggregation. Biofouling and non-biofouling characteristics are determined by the direct attachment of these molecules on the GNP and NaCl was added to visualize GNP aggregation.
5.2. Small molecules
Based on the above analyses, it is clear that compatibility is an important key point for interactive analyses using GNPs, especially with protein. Other than these macromolecules, as stated above, small ligands are generally considered to exhibit little or no lower non-specific interactions on GNPs. To evaluate this, we screened different small molecules that have been routinely used in the past for different sensing purposes. The screened small molecules included ethanolamine, biotin, ATP, Hoechst 33258, theophylline, and caffeine; bearing in mind the necessity for higher amounts of small molecules compared to proteins, in this study we used 1 μM of molecules to be tested. Among these molecules, ethanolamine, ATP, theophylline, and caffeine showed no interactions with GNPs, as confirmed by the addition of 100 mM NaCl, which yielded a clear purple or blue color, indicating the formation of aggregates under ionic conditions. In general, amine groups are expected to have non-specific interactions on GNPs, as in the above cases where the proteins have amine groups on their surfaces. However, ethanolamine, which was used in the small molecule screening has amine groups and is a common blocking reagent, especially for SiO2 surfaces (Gopinath et al., 2012b, Gopinath et al., 2013), although it did not result any non-specificity on the GNPs used here. At the same concentration (100 mM) of NaCl, biotin and Hoechst 33258 yield a reddish-purple color and in this case, it may be necessary to use higher concentrations of NaCl (>100 mM). Based on these screening studies with both macromolecules and small molecules, it can be concluded that small molecules are well suited for GNP-based colorimetric assays.
6. Perspectives
GNPs are capable of attracting small oligonucleotides, paving the way for the development of colorimetric assays for different biomolecular interactions. This label-free approach is fast, robust, and dependent only on routine equipmental tools, abrogating the need for additional steps such as modification of the aptamer and separation of the bound and unbound aptamer–target complexes. An overall comparison of reports made in the past revealed that GNP sizes between 10 and 20 nm are suitable and ideal for the detection of small molecules. In terms of sensitivity this method may not able to reach femtomolar concentrations. In addition, in most cases, DNA aptamers/oligos rather than RNA aptamers are used to develop colorimetric assays, indicating their suitability or cost-effect (Table 1). Although several strategies for the detection of aptamer–ligand interactions using colorimetric-based assays have been developed, this convenient assay is highly reliable for molecules that exhibit greater non-fouling on GNPs. The GNP-based colorimetric assay is very convenient for evaluating aptamer–ligand interactions, and these assays for molecular screening can be performed without prior experience. Moreover, considering the suitability of this assay for smaller ligands, this method is recommendable for the small molecule screening of chemical or drug libraries against aptamer. Further, GNP-based controlled assembly and disassembly strategies are promising for biological applications due to their optical properties and can be applied to the design of GNP-based nanoassembly methods.
Acknowledgments
This study was supported by the Industrial Technology Research Grant Program 2009 from the New Energy and Industrial Technology Development Organization (NEDO) of Japan. Authors would like to thank Dr. Makoto Fujimaki, National Institute of Advanced Industrial Science and Technology, for his comments.
References
- Baldrich E., Restrepo A., O’Sullivan C.K. Analytical Chemistry. 2004;76:7053–7063. doi: 10.1021/ac049258o. [DOI] [PubMed] [Google Scholar]
- Breaker R.R. Molecular Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Wei S., Wu T., Lee C., Lin C. Biosensors & Bioelectronics. 2012;42:119–123. doi: 10.1016/j.bios.2012.10.072. [DOI] [PubMed] [Google Scholar]
- Chávez J.L., Lyon W., Kelly-Loughnane N., Stone M.O. Biosensors & Bioelectronics. 2010;26:23–28. doi: 10.1016/j.bios.2010.04.049. [DOI] [PubMed] [Google Scholar]
- Chávez J.L., MacCuspie R.I., Stone M.O., Kelly-Loughnane N. Journal of Nanoparticle Research. 2012;14:1166–1177. [Google Scholar]
- Chen S., Huang Y., Huang C., Lee K, Lin Z., Chang H. Biosensors & Bioelectronics. 2008;23:1749–1753. doi: 10.1016/j.bios.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Chen A., Jiang X., Zhang W., Chen G., Zhao Y., Tunio M.T., Liu J., Lv Z., Li C., Yang S. Biosensors & Bioelectronics. 2013;42:419–425. doi: 10.1016/j.bios.2012.10.059. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Tseng, C., Chang, H., Hung, Y., Huang, C., 2012. Biosensors & Bioelectronics 26, 3160–3166. [DOI] [PubMed]
- Christopher P., Robinson N., Shaw M.K. Forensic Science and Medicine, Drugs of Abuse: Body Fluid Testing. 2005. p. 87. [Google Scholar]
- Fujita M., Katafuchi Y., Ito K., Kanayama N., Takarada T., Maeda M. Journal of Colloid and Interface Science. 2012;368:629–635. doi: 10.1016/j.jcis.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Geiger A., Burgstaller P., Eltz H.V., Roeder A., Famulok M. Nucleic Acids Research. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giljohann D.A., Seferos D.S., Daniel W.L., Massich M.D., Patel P.C., Mirkin C.A. Angewandte Chemie International Edition. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath S.C.B. Aptamers. In: Meyers R.A., editor. Encyclopedia of Analytical Chemistry. John Wiley; Chichester: 2011. pp. 1–27. (Published on 15th March) [Google Scholar]
- Gopinath S.C.B. Sensors and Actuators B: Chemical. 2010;150:722–733. [Google Scholar]
- Gopinath S.C.B. Analytica Chimica Acta. 2009;636:117–128. doi: 10.1016/j.aca.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Gopinath S.C.B., Awazu K., Fujimaki M. Sensors. 2012;12:2136–2151. doi: 10.3390/s120202136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath S.C.B., Awazu K., Fujimaki M., Shimizu K., Mizutani W., Tsukagoshi K. Analyst. 2012;137:3520–3527. doi: 10.1039/c2an35159e. [DOI] [PubMed] [Google Scholar]
- Gopinath S.C.B., Awazu K., Fujimaki M., Shimizu K. Acta Biomaterialia. 2013;9:5080–5087. doi: 10.1016/j.actbio.2012.09.027. [DOI] [PubMed] [Google Scholar]
- Gopinath S.C.B., Awazu K., Kumar P.K.R., Tominaga J. ACS Nano. 2008;2:1885–1895. doi: 10.1021/nn800285p. [DOI] [PubMed] [Google Scholar]
- Gronewold T.M.A., Glass S., Quandt E., Famulok M. Biosensors & Bioelectronics. 2005;20:2044–2052. doi: 10.1016/j.bios.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Guirgis B.S., Cunha Sá E, Gomes C., Cavadas I., Silva M., Doria I., G. Analytical and Bioanalytical Chemistry. 2012;402:1019–1027. doi: 10.1007/s00216-011-5489-y. [DOI] [PubMed] [Google Scholar]
- Guo P. Journal of Nanoscience and Nanotechnology. 2005;5:1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. Journal of Biophotonics. 2012;5:483–501. doi: 10.1002/jbio.201200015. [DOI] [PubMed] [Google Scholar]
- Huang C., Huang Y., Cao Z., Tan W., Chang H. Analytical Chemistry. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- Iliuk A.B., Hu L., Tao W.A. Analytical Chemistry. 2011;83:4440–4452. doi: 10.1021/ac201057w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenison R.D., Gill S.C., Pardi A., Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Jian J.W., Huang C.C. Chemistry. 2011;17:2374–2380. doi: 10.1002/chem.201002825. [DOI] [PubMed] [Google Scholar]
- Kamimura M., Miyamoto D., Saito Y., Soga K., Nagasaki Y. Langmuir. 2008;24:8864–8870. doi: 10.1021/la801056c. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Jurng J. Analyst. 2011;136:3720–3724. doi: 10.1039/c1an15261k. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Kim J.H., Kim I.A., Lee S.J., Jurng J., Gu M.B. Biosensors & Bioelectronics. 2010;26:1644–1649. doi: 10.1016/j.bios.2010.08.046. [DOI] [PubMed] [Google Scholar]
- Lau P.S., Coombes B.K., Li Y. Angewandte Chemie International Edition. 2010;49:7938–7942. doi: 10.1002/anie.201002621. [DOI] [PubMed] [Google Scholar]
- Li L., Li B., Qi Y., Jin Y. Analytical and Bioanalytical Chemistry. 2009;393:2051–2057. doi: 10.1007/s00216-009-2640-0. [DOI] [PubMed] [Google Scholar]
- Li H., Rothberg L. Proceedings of the National Academy of Sciences USA. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Z., Li J., Ng C., Yung L.L., Bay B. Acta Pharmacologica Sinica. 2011;32:983–990. doi: 10.1038/aps.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lu Y. Nature protocols 1. 2006:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- Lou S., Ye J., Li K., Wu A. Analyst. 2012;137:1174–1181. doi: 10.1039/c2an15844b. [DOI] [PubMed] [Google Scholar]
- Lu Y., Liu J. Current Opinion in Biotechnology. 2006;17:580–588. doi: 10.1016/j.copbio.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Marquette C.A., Blum L.J. Analytical and Bioanalytical Chemistry. 2008;390:155–168. doi: 10.1007/s00216-007-1631-2. [DOI] [PubMed] [Google Scholar]
- Medley C.D., Smith J.E., Tang Z., Wu Y., Bamrungsap S., Tan W. Analytical Chemistry. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- Nagasaki Y. Polymer Journal. 2011;43:949–958. [Google Scholar]
- Nagasaki Y., Kobayashi H., Katsuyama Y., Jomura T., Sakura T. Journal of Colloid and Interface Science. 2007;309:524–530. doi: 10.1016/j.jcis.2006.12.079. [DOI] [PubMed] [Google Scholar]
- Nagel J., Chunsod P., Zimmerer C., Simon F., Janke A., Heinrich G. Materials Chemistry Physics. 2011;129:599–604. [Google Scholar]
- Nath N., Chilkoti A. Analytical Chemistry. 2004;76:5370–5378. doi: 10.1021/ac049741z. [DOI] [PubMed] [Google Scholar]
- Odenthal K.J., Gooding J. Analyst. 2007;132:603–610. doi: 10.1039/b701816a. [DOI] [PubMed] [Google Scholar]
- Ogawa A., Maeda M. Chemical Communications. 2009;31:4666–4668. doi: 10.1039/b910288d. [DOI] [PubMed] [Google Scholar]
- Otsuka H., Nagasaki Y., Kataoka K. Biomacromolecules. 2000;1:39–48. doi: 10.1021/bm990005s. [DOI] [PubMed] [Google Scholar]
- Pavlov V., Xiao Y., Shlyahovsky B., Willner I. Journal of the American Chemical Society. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- Peng Y., Li L., Mu X., Guo L. Sensors and Actuators B. Chemical. 2013;177:818–825. [Google Scholar]
- Peng L., Varma M.M., Cho W., Regnier F.E., Nolte D.D. Applied Optics. 2007;46:5384–5395. doi: 10.1364/ao.46.005384. [DOI] [PubMed] [Google Scholar]
- Schreiner S.M., Hatch A.L., Shudy D.F., Howard D.V., Howell C., Zhao J. Analytical Chemistry. 2011;83:4288–4295. doi: 10.1021/ac200814y. [DOI] [PubMed] [Google Scholar]
- Selvakumar L.S., Thakur M.S. Analytical Biochemistry. 2012;427:151–157. doi: 10.1016/j.ab.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Song K., Cho M., Jo H., Min K., Jeon S.H., Kim T. Analytical Biochemistry. 2011;415:175–181. doi: 10.1016/j.ab.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Song K., Jeong E., Jeon W., Cho M., Ban C. Analytical and Bioanalytical Chemistry. 2012;402:2153–2161. doi: 10.1007/s00216-011-5662-3. [DOI] [PubMed] [Google Scholar]
- Song S., Wang L., Li J., Zhao J., Fan C. Trends in Analytical Chemistry. 2008;27:108–117. [Google Scholar]
- Stevenson K.A., Muralidharan G., Maya L., Wells J.C., Barhen J., Thundat T. Journal of Nanoscience and Nanotechnology. 2002;2:397–404. doi: 10.1166/jnn.2002.110. [DOI] [PubMed] [Google Scholar]
- Storhoff J.J., Elghanian R., Mirkin C.A., Letsinger R.L. Langmuir. 2002;18:6666–6670. [Google Scholar]
- Tinguely J., Sow I., Leiner C., Grand J., Hohenau A., Felidj N. BioNanoScience. 2011;1:128–135. [Google Scholar]
- Tsai D., Shelton M.P., DelRio F.W., Elzey S., Guha S., Zachariah M.R. Analytical and Bioanalytical Chemistry. 2012;404:3015–3023. doi: 10.1007/s00216-012-6418-4. [DOI] [PubMed] [Google Scholar]
- Upadhyayula V.K. Analytica Chimica Acta. 2012;715:1–18. doi: 10.1016/j.aca.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Varma M.M., Nolte D.D., Inerowicz H.D., Regnier F.E. Optics Letters. 2004;29:950–952. doi: 10.1364/ol.29.000950. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang L., Liu X., Liang Z., Song S., Li W. Advanced Materials. 2007;19:3943–3946. [Google Scholar]
- Wang X., Zhao M., Nolte D.D. Applied Optics. 2008;47:2779–2789. doi: 10.1364/ao.47.002779. [DOI] [PubMed] [Google Scholar]
- Wei H., Li B., Li J., Wang E., Dong S. Chemical Communications. 2007;36:3735–3737. doi: 10.1039/b707642h. [DOI] [PubMed] [Google Scholar]
- Xia F., Zuo X., Yang R., Xiao Y., Kang D., Vallée-Bélisle A. Proceedings of the National Academy of Sciences USA. 2010;107:10837–10841. doi: 10.1073/pnas.1005632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Wang F., Liu X. Journal of the American Chemical Society. 2008;130:3244–3245. doi: 10.1021/ja076716c. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Nishio M., Sugasawa H., Nagasaki Y. Journal of the American Chemical Society. 2010;132:7982–7989. doi: 10.1021/ja910372e. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Nozawa M., Matsumoto S., Echigo T., Nemoto S., Hatta T. Langmuir. 2009;25:12243–12249. doi: 10.1021/la901143h. [DOI] [PubMed] [Google Scholar]
- Yuan X., Yoshimoto K., Nagasaki Y. Analytical Chemistry. 2009;81:1549–1556. doi: 10.1021/ac802282c. [DOI] [PubMed] [Google Scholar]
- Zanoli L.M., D′Agata R., Spoto G. Analytical and Bioanalytical Chemistry. 2012;402:1759–1771. doi: 10.1007/s00216-011-5318-3. [DOI] [PubMed] [Google Scholar]
- Zhang X., Servos M.R., Liu J. Langmuir. 2012;28:3896–3902. doi: 10.1021/la205036p. [DOI] [PubMed] [Google Scholar]
- Zhao W., Brook M.A., Li Y. ChemBioChem. 2008;9:2363–2371. doi: 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- Zhao W., Chiuman W., Lam J.C.F., McManus S.A., Chen W., Chi Y., Pelton R., Brook M.A., Li Y. Journal of the American Chemical Society. 2008;130:3610–3618. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wang Y., Yang X. Sensors and Actuators B. Chemical. 2011;156:95–99. doi: 10.1016/j.snb.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]


