Abstract
The budding yeast SKP1gene, identified as a dosage suppressor of a known kinetochore protein mutant, encodes an intrinsic 22.3 kDa subunit of CBF3, a multiprotein complex that binds centromere DNA in vitro. Temperature-sensitive mutations in SKP1 define two distinct phenotypic classes. skp1-4 mutants arrest predominantly as large budded cells with a G2 DNA content and short mitotic spindle, consistent with a role in kinetochore function. skp1-3 mutants, however, arrest predominantly as multiply budded cells with a G1 DNA content, suggesting an additional role during the G1/S phase. Identification of Skp1p homologs from C. elegans, A. thaliana, and H. sapiens indicates that SKP1 is evolutionarily highly conserved. Skp1p therefore represents an intrinsic kinetochore protein conserved throughout eukaryotic evolution and may be directly involved in linking kinetochore function with the cell cycle-regulatory machinery.
Introduction
Centromeres (also termed kinetochores) are of fundamental importance to chromosomal inheritance in all eukaryotes (reviewed byPluta et al. 1995). These functionally differentiated structures on eukaryotic chromosomes consist of specific centromere DNA sequences and associated proteins that facilitate binding to the spindle microtubules. Kinetochores mediate chromosome movement to ensure the ordered segregation of sister chromatids into daughter cells during mitosis and meiosis. Kinetochore activity must be regulated to coordinate chromosome segregation with other events in the cell cycle and to ensure that bipolar attachment of all chromosomes is achieved before anaphase begins. The structural complexity of centromeres varies widely among higher and lower eukaryotes, as exemplified by the centromeres of animal cells (which span several thousand kilobases of DNA) and budding yeast (which span <0.2 kb of DNA). Despite these apparent organizational differences, centromeres in higher and lower eukaryotes are likely to share evolutionarily conserved components.
The structurally compact centromere of the budding yeast, Saccharomyces cerevisiae, has been analyzed extensively using molecular genetic techniques (reviewed byHegemann and Fleig 1993). The centromere DNA sequence encompasses a 125 bp region that includes three conserved elements, termed CDEI, CDEII, and CDEIII (12, 20). CDEIII (25 bp) is absolutely essential, as evidenced by single point mutations that destroy centromere activity (Hegemann et al. 1988). Lechner and Carbon 1991 described the purification of a 240 kDa multicomponent protein complex, termed CBF3, that specifically binds CDEIII sequences in vitro. The CBF3 complex has three major components that are 110, 64, and 58 kDa in size. The genes encoding p110 (NDC10/CBF2/CTF14) (15, 24, 8), p64 (CEP3/CBF3b) (Strunnikov et al. 1995; Lechner 1994), and p58 (CTF13) (Doheny et al. 1993) have been cloned and characterized. All three proteins are essential for viability, and mutations in any one of them cause a chromosome missegregation phenotype, consistent with a role in kinetochore function. At nonpermissive temperature, the ndc10-1 mutation causes asymmetric segregation of chromosomes in which all chromosomes segregate to one pole following nuclear division. Temperature-sensitive mutations in ctf13 or cep3/cbf3c exhibit G2/M accumulation at nonpermissive temperature with preanaphase short mitotic spindles. Cep3p contains a predicted zinc finger and may therefore represent a DNA-binding component. Ndc10p contains a GTP-binding motif. None of the three CBF3 proteins described to date exhibits extensive homology to any known proteins in the databases.
The coordination of centromere function with cell cycle progression is likely to require regulation of kinetochore protein activity and a signal transduction system that monitors completion of chromosome attachment and relays the status of events to the cell cycle machinery. A direct role for the kinetochore in a checkpoint that monitors completion of metaphase is suggested by studies in both animal and yeast cells. Microinjection of anti–centromere protein antibodies into cultured human cells induces mitotic delay (Bernat et al. 1990). In cultured mammalian cells, the onset of anaphase following nuclear envelope breakdown is delayed even when only a single kinetochore is not attached to microtubules (Rieder et al. 1994). Laser ablation of the unattached kinetochore destroys the delay in anaphase onset, suggesting an inhibitory effect of the unattached kinetochore in cell cycle progression. In yeast cells, a preanaphase delay is induced by the presence of a single mutant centromere DNA (Spencer and Hieter 1992) or by hypomorphic mutations in kinetochore proteins (Doheny et al. 1993). Recently, three groups have presented evidence that the preanaphase delay induced by abnormal kinetochore function requires genes that are involved in a mitotic checkpoint–monitoring spindle assembly (48, 49, 37). Very little is currently known about the molecular components or mechanism that signals defects in kinetochore structure to cell cycle regulators.
Kinetochores execute a variety of activities, including binding of kinetochore protein complexes to centromere DNA, binding of CEN DNA–kinetochore protein complexes to microtubules, movement of chromosomes along the microtubule fibers, and responses to and involvement in cell cycle regulation. Given this complexity, additional structural and regulatory components must be identified to gain a complete understanding of kinetochore function. To accomplish this, we have employed a variety of genetic screens in S. cerevisiae using ctf13-30, a temperature-sensitive mutation in the gene encoding p58 of CBF3, as a starting point. In this paper, we describe the identification of the SKP1 gene as a dosage suppressor of a ctf13-30 mutant. Genetic and biochemical analysis indicates that the 22.3 kDa Skp1 protein is a fourth subunit of CBF3 that was previously unrecognized. Unlike the other CBF3 components, Skp1p is a highly conserved protein found in multicellular eukaryotes. Furthermore, Skp1p has recently been shown to be a component of a cyclin A–cyclin-dependent kinase 2 (CDK2) complex purified from transformed human fibroblasts (Zhang et al. 1995) and to interact directly with Cdc4p and cyclin F through a novel structural motif (Bai et al., 1996 [this issue of Cell]). We conclude that Skp1p is involved in both structural and regulatory functions and may provide a direct link between the kinetochore and the cell cycle machinery.
Results
High Copy Suppression of a Known Kinetochore Component
To identify further additional components of the S. cerevisiae kinetochore, we performed a screen for dosage suppressors of the temperature sensitivity caused by the ctf13-30 mutation. A 2μ genomic library was transformed into a ctf13-30 mutant, and temperature-resistant transformants were selected (see Experimental Procedures). We obtained 12 plasmid clones that fell into two classes of overlapping genomic segments: 7 of 12 corresponded to the CTF13 gene; 5 of 12 contained a new locus. We named the locus SKP1, for suppressor of kinetochore protein 1. In addition to dosage suppression of the temperature sensitivity for growth, increased dosage of SKP1 suppressed the chromosome missegregation phenotype caused by the ctf13-30 mutation at semi-permissive temperature, even when SKP1 was introduced at a modestly increased copy number on a centromere vector (data not shown). Insertion of a DNA fragment containing a prototrophic marker into a unique NsiI site disrupted the ability of the genomic plasmid clone to suppress the temperature-sensitive and chromosome missegregation phenotypes of a ctf13-30 mutant. When the gene disruption was integrated into the genome, dissection of a heterozygous diploid resulted in viability segregating 2+:2− (see Experimental Procedures). The genomic clone was localized to the right arm of chromosome IV using physical mapping methods, indicating that SKP1 is a previously unidentified essential gene in S. cerevisiae.
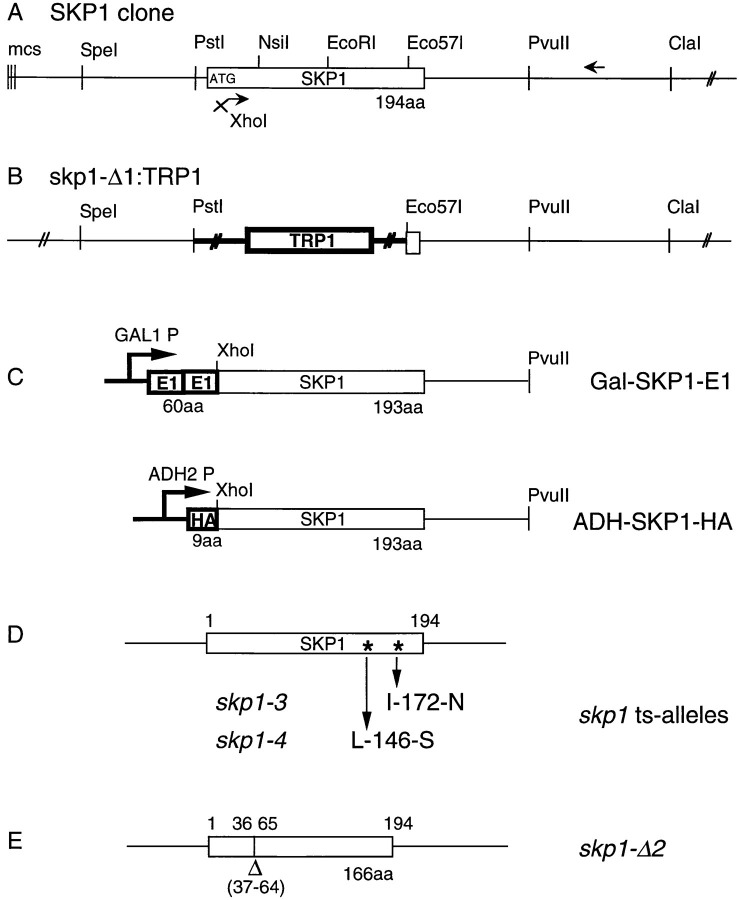
The nucleotide sequence of the SKP1 genomic DNA contains a 582 bp open reading frame (ORF) that encodes a protein of 194 amino acids with a predicted molecular mass of 22.3 kDa (Figure 1A). The restriction maps of genomic SKP1 and derivative clones are shown in Figure 1.
Figure 1.
SKP1 Genomic DNA, Mutant Alleles, and Epitope Tag Fusion Proteins
(A) Restriction map of the cloned SKP1 gene and flanking regions. The vector multiple cloning site (mcs) represents the original left end of the library clone where a genomic Sau3A site was ligated to a BamHI site in the vector.
(B) The skp1-Δ1 allele was constructed by replacement of the 0.73 kb PstI–Eco57I segment with a 1.8 kb TRP1 gene containing fragment (shown in bold).
(C) Schematic structures of the E1 and HA amino-terminal epitope-tagged Skp1p fusion constructs. The E1–Skp1p fusion is under transcriptional control of the GAL1 promotor; the HA–Skp1p fusion is under the transcriptional control of the ADH2 promotor.
(D) Schematic of the skp1 temperature-sensitive alleles used in phenotypic studies.
(E) Schematic of skp1-Δ2. The 28 amino acid in-frame deletion corresponds to amino acid residues 37–64.
SKP1Encodes a Fourth Subunit of CBF3
Lechner and Carbon 1991 described a multiprotein complex (CBF3) present in extracts of S. cerevisiae cells that contains three major protein species 110, 64, and 58 kDa in size. CBF3 binds in vitro to a 100 bp DNA probe that spans CDEIII but lacks CDEI and CDEII (Doheny et al. 1993; see Experimental Procedures). Because of the small size of Skp1p, it was possible that it represented a previously unrecognized subunit of CBF3. To determine whether this was the case, we made two epitope-tagged constructs of Skp1p (see Experimental Procedures). In the first, two tandem copies of the E1 epitope (Pluta et al. 1992), derived from the carboxy-terminal 25 amino acids of avian coronavirus glycoprotein (Machamer and Rose 1987), were placed in-frame at the amino terminus of the SKP1 ORF under the transcriptional control of the GAL1 promoter (Figure 1C). In the second construct, a 9 amino acid epitope derived from the HA1 protein of influenza virus (Field et al. 1988) was placed in-frame at the amino terminus of the SKP1 ORF under the control of the ADH2 promoter (Figure 1C). Both epitope-tagged Skp1p derivatives were able to rescue viability in a skp1-Δ1::TRP1 deletion strain (Figure 1B).
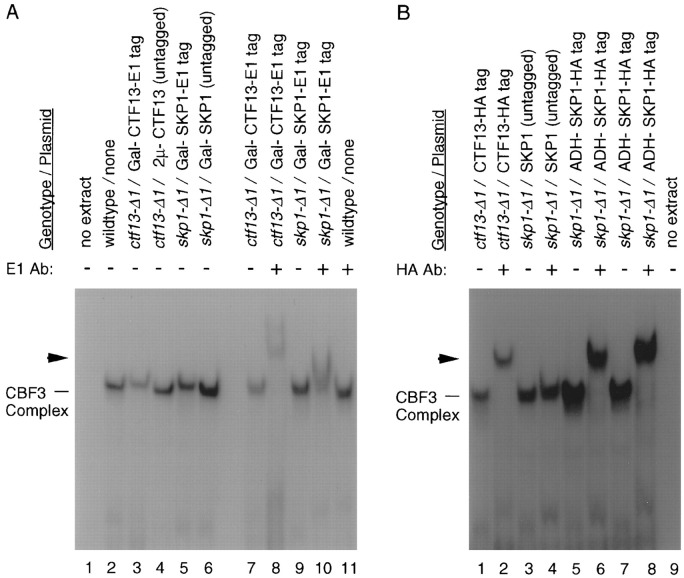
Whole-cell extracts were prepared from a skp1-Δ1 deletion strain carrying either epitope-tagged or untagged copies of Skp1p. As controls, extracts were also prepared from a ctf13-Δ1 deletion strain carrying epitope-tagged Ctf13p. DNA–protein complexes were allowed to form with 32P-labeled CDEIII sequence, and complexes were resolved on a nondenaturing gel (Figure 2). In the absence of anti-E1 antibody, extracts containing the E1 epitope–tagged fusions of Ctf13p and Skp1p show a slight (but clearly observable and reproducible) intrinsic mobility shift up when compared with wild-type and untagged extracts (Figure 2A, lanes 2–6), suggesting that the E1–Skp1p fusion protein is localized to the complex. The magnitude of the intrinsic shift is identical for E1-tagged Ctf13p and E1-tagged Skp1p, indicating equal stoichiometry of these two proteins in the CBF3 complex. When E1 antibody is added to complexes formed using extracts from tagged and untagged Ctf13p and Skp1p strains, a supershift is observed using the Ctf13p-tagged extract (Figure 2A, lane 8) and the E1-tagged Skp1p extract, although the supershift with E1-tagged Skp1p was less pronounced (Figure 2A, lane 10). These results suggested that Skp1p is in the CBF3 complex, as evidenced by both the intrinsic and antibody-induced mobility shift; however, epitope accessibility or antibody binding stability could have compromised a supershift equivalent to that seen with epitope-tagged Ctf13p.
Figure 2.
Mobility Shift Analysis of Skp1p in CBF3–CEN DNA Complexes
DNA–protein complexes formed with 32P-labeled CDEIII probe and whole-cell extracts were analyzed on a nondenaturing polyacrylamide gel (Doheny et al. 1993). Antibodies were added to preformed complexes, and samples were incubated for 20 min at room temperature before gel analysis. Unbound probe was run off the bottom of the gel.
(A) Lanes 1, 2, and 11 are controls. Lanes 3–6, extracts from cells containing E1-tagged or untagged Ctf13p or Skp1p proteins demonstrating an intrinsic mobility shift due to the 60 amino acid epitope fusion in each case. Lanes 7–10, extracts from cells containing E1-tagged Ctf13p or Skp1p subsequently incubated with or without antibodies directed against the E1 epitope as indicated. Lanes 7 and 9 are controls for lanes 8 and 10.
(B) Lanes 1–6, extracts containing either HA-tagged or untagged Ctf13p or Skp1p subsequently incubated with or without an antibody directed against the HA epitopeas indicated. Lanes 7 and 8 are from an independent transformant (repeat of lanes 5 and 6). Lane 9 is a control.
The addition of anti-HA antibody to complexes formed using extracts prepared from untagged and HA-tagged fusions of Ctf13p and Skp1p showed conclusively that Skp1p is present in the CBF3–CEN DNA complex. Two independent extracts containing HA-tagged Skp1p produced a quantitative supershift equivalent to that seen for the HA-tagged Ctf13p (Figure 2B, lanes 2, 6, and 8). We conclude from these data that Skp1p is a fourth subunit of the S. cerevisiae CBF3 complex.
Generation and Analysis of skp1 Temperature-Sensitive Alleles
To facilitate subsequent genetic and phenotypic analysis of mutants defective for SKP1 function, we generated four temperature-sensitive alleles of SKP1 using a method for random mutagenesis of yeast genes described by Muhlrad et al. 1992 that involves mutagenic polymerase chain reaction (PCR) in vitro and gap repair in vivo (see Experimental Procedures). The four alleles were named skp1-1, skp1-2, skp1-3, and skp1-4. The nucleotide sequence of each allele was determined (see Experimental Procedures), and the mutations encoding changes at the protein level were found to be primarily in the carboxyl terminus of the protein (see Table 1).
Table 1.
Mutations in skp1 Temperature-Sensitive Alleles
| Sequence Change |
||||
| Allele | Number of Mutations | Codon | DNA | Protein |
| skp1-1 | 1 | 129 | GAG to GGG | Glu to Gly |
| skp1-2 | 2 | 130 | ATG to GTG | Met to Val |
| 176 | TTC to TCC | Phe to Ser | ||
| skp1-3 | 1 | 172 | ATC to AAC | Ile to Asn |
| skp1-4 | 3 | 146 | TTG to TCG | Leu to Ser |
| 162 | TCT to TCC | No change | ||
| 4 | TCT to TCC | No change | ||
Chromosome missegregation was monitored visually in each skp1 temperature-sensitive strain at semi-permissive temperature using a colony color assay that measures the stability of a marker chromosome fragment (Hieter et al. 1985). By this criterion, the strains fell into two classes. Strains expressing the skp1-1, skp1-2, or skp1-3 alleles exhibited wild-type rates of chromosome missegregation. The strain expressing skp1-4, however, exhibited a dramatically increased rate of chromosome missegregation comparable with that observed for the ctf13-30 mutant. We therefore decided to analyze further strains carrying one mutation in each class (skp1-3 and skp1-4). Our approach was to integrate each mutant allele as a single copy into the genome to avoid potential plasmid copy number effects. Both the skp1-3 and skp1-4 alleles contain single missense mutations (Ile-172 to Asn and Leu-146 to Ser, respectively; Figure 1D). Each mutant allele was integrated at the LEU2 locus in strains containing the skp1-Δ1 deletion mutation (see Experimental Procedures).
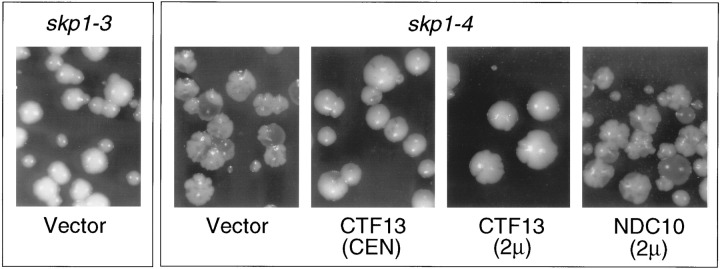
The skp1-3 and skp1-4 mutations were found to cause several allele-specific phenotypes. First, as observed with the plasmid-borne alleles, skp1-3 does not cause increased chromosome missegregation, while skp1-4 causes a dramatic increase in chromosome missegregation (Figure 3, skp1 mutants transformed with vector only). Chromosome transmission fidelity was quantitated in a skp1-4/skp1-4 homozygous diploid using colony color half sector analysis (Koshland and Hieter 1987). The rates of chromosome loss (1:0 segregation) and chromosome nondisjunction (2:0 segregation) in the skp1-4 mutant were 3.9% and 1.7% per cell division, respectively, which represent greater than 100-fold increases relative to wild type. Second, because SKP1 was identified as a high copy suppressor of ctf13-30, it was of interest to test the reciprocal relationship, i.e., whether Ctf13p overexpression could suppress the skp1 temperature-sensitive alleles. Overexpression of Ctf13p rescues the temperature sensitivity of skp1-4, but not that of skp1-3 (data not shown). In addition, at semi-permissive temperature, the chromosome missegregation phenotype caused by the skp1-4 mutation is suppressed by increased dosage of Ctf13p, even when present at one or a few extra copies on a centromere vector (Figure 3). This suppression is kinetochore protein gene specific, since increased dosage of Ndc10p has no effect, suggesting a direct interaction between the Skp1 and Ctf13 proteins. Third, following a 3 hr shift to nonpermissive temperature, skp1-3 causes haploid cells to arrest with a G1 DNA content, while skp1-4 causes haploid cells to arrest with a G2 DNA content (data not shown). These terminal phenotypes were more fully characterized in diploids (see below). Fourth, skp1-3 and skp1-4 mutations exhibit interallelic complementation. Heterozygous and homozygous diploids were created from meiotic segregants of each of the integrated temperature-sensitive alleles. At nonpermissive temperature (37°C), the skp1-3/skp1-4 heteroallelic diploid forms colonies, whereas either homozygous mutant diploid does not. Furthermore, at permissive temperature (25°C), the skp1-3 allele complements the chromosome segregation defect of the skp1-4 allele in the heteroallelic diploid.
Figure 3.
Chromosome Segregation Phenotypes of skp1-3 and skp1-4 Mutants
Yeast strains carrying integrated skp1 temperature-sensitive alleles were tested for chromosome missegregation phenotypes using the colony color sectoring assay (Koshland and Hieter 1987). Loss of the nonessential marker chromosome gives a red sector in a white colony. skp1-3 mutants exhibit a wild-type phenotype (very rarely sectored colonies) when transformed with vector only. skp1-4 mutants exhibit dramatically increased rates of chromosome loss (highly sectored colonies) when transformed with vector only. Introduction of the CTF13 gene on a low copy (CEN) vector or high copy (2μ) vector suppresses the chromosome missegregation phenotype of the skp1-4 mutant. Introduction of the NDC10 gene on a 2μ vector has no effect.
Taken together these data strongly suggest two independent functions for the Skp1 protein: a G2/M function required for kinetochore activity and chromosome segregation and a G1/S function required for cell cycle progression earlier in the cell cycle.
Cytological Analysis of skp1 Mutants
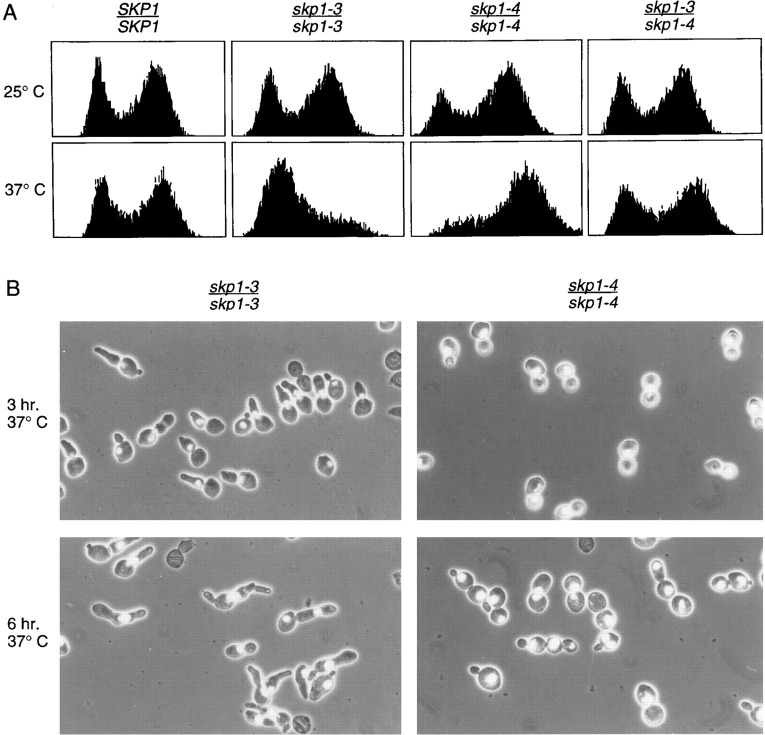
Cytological analysis of skp1 mutants, and accompanying flow cytometric analysis of DNA content per cell, was performed in skp1 homozygous and heteroallelic diploid mutants at permissive and nonpermissive temperatures (Figure 4). As compared with wild type, skp1-3/skp1-3 mutants exhibit typical G1 and G2 profile of DNA content per cell in logarithmically growing cultures at permissive temperature and arrest with a single peak of G1 content DNA after shift to nonpermissive temperature for 3 hr (Figure 4A). In contrast, skp1-4/skp1-4 mutants exhibit a modest accumulation of cells with a G2 DNA content during log phase growth and arrest with a single peak of G2 content DNA after shift to the nonpermissive temperature. As expected from the intragenic complementation data described above, the skp1-3/skp1-4 heteroallelic diploid exhibits DNA content profile similar to wild type in logarithmically growing cells at permissive temperature and after shift to the nonpermissive temperature.
Figure 4.
Phenotypic Analysis of skp1 Temperature-Sensitive Mutants
(A) Shown are G1 arrest (at nonpermissive temperature) of skp1-3 cells and G2/M accumulation (at permissive temperature) and G2/M arrest (at nonpermissive temperature) of skp1-4 cells, as analyzed by flow cytometry (Gerring et al. 1990). The number of cells is depicted on the vertical axis, with fluorescent intensity of emitted light (proportional to DNA content) on the horizontal axis. Logarithmically growing cultures at 25°C were split and incubated at either 25°C or 37°C for 3 hr prior to analysis.
(B) Terminal arrest morphologies of skp1-3 or skp1-4 cells after 3 or 6 hr at 37°C. Cells were stained with 4′,6-diamidino-2-phenylindole and photographed for fluorescence staining and by phase contrast. The fields shown were chosen to emphasize specific phenotypes and are not necessarily representative of overall frequencies.
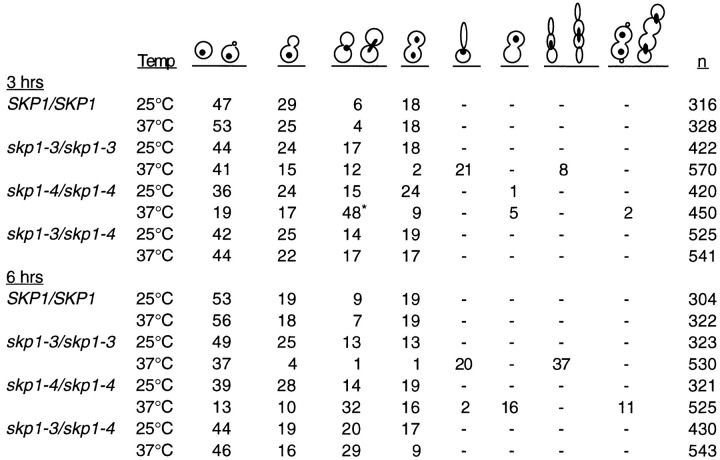
Quantitation of cell and nuclear morphology following growth arrest at high temperature also revealed dramatic differences between the skp1-3 and skp1-4 mutants. After 3 hr at nonpermissive temperature, skp1-3 cells are predominantly unbudded with a single DNA mass. A novel class of cells with abnormally elongated buds (21%) is also seen (Figure 4B and Figure 5). After 6 hr at nonpermissive temperature, 57% of skp1-3 cells exhibit novel cell morphologies, including a multiple budded phenotype accounting for 37% of the cell population. It appears that skp1-3 cells do not replicate their DNA (arrest with a G1 DNA content), but continue the budding process. This arrest phenotype is analogous to that seen in a class of cdc mutants typified by cdc4. In contrast, skp1-4 cells arrest with a predominant cell morphology indicative of the preanaphase G2/M phase of the cell cycle. After 3 hr at the nonpermissive temperature, 48% of cells were large budded with an undivided nucleus positioned at or near the neck between mother and daughter cells. The majority of cells with this morphology were characterized by DNA at or spanning the neck and a short mitotic spindle. Cells with a short spindle spanning the neck accounted for 16% of the cell population, a morphology rarely seen (<1%) in wild-type cells. Interestingly, at 6 hr a proportion of cells (11%) exhibited a novel multiple budded phenotype (Figure 4B and Figure 5). The skp1-4 terminal arrest morphology phenotype is therefore like that of cdc (at medial nuclear division) and is quite similar to that caused by mutations in the 64 kDa (Cep3p) and 58 kDa (Ctf13p) subunits of CBF3.
Figure 5.
Quantitation of Cell and Nuclear Morphology
SKP1/SKP1 wild-type, skp1-3/skp1-3 and skp1-4/skp1-4 homozygous mutants, and skp1-3/skp1-4 heteroallelic diploids were grown to logarithmic phase at 25°C and shifted to 37°C for 3 and 6 hr, and nuclear and bud morphology were scored. The criteria used for each morphologic class scored are shown schematically above the columns. The numbers shown represent percentages of the total cells scored (far right column). Asterisk, in this population of cells, 67% had the nucleus at the neck and 33% had the nucleus spanning the neck (n = 203).
We also tested whether cells grown at nonpermissive temperature could recover when shifted back to the permissive temperature. About 60% of skp1-3 cells were able to recover from the G1 arrest after 6 hr at 37°C. In contrast, only 10% of skp1-4 cells were able to recover from the G2/M arrest (data not shown).
Skp1p Is Highly Conserved in Evolution
To date the components of the CBF3 complex in S. cerevisiae have not shown highly significant amino acid sequence similarities to other proteins currently present in the public databases. However, when a six-way conceptual translation of the database of expressed sequence tags (dbEST) (Boguski et al. 1994) was searched with the Skp1p amino acid sequence, significant matches were found to ORFs encoded in ESTs from Arabidopsis thaliana, Caenorhabditis elegans, and Homo sapiens. Clones corresponding to the EST sequences were obtained in each case, and the complete sequences of the Skp1p homologs in these three organisms were determined. The human Skp1p homolog sequence is identical to the sequence of p19 (Zhang et al. 1995), which is a component of a cyclin A–CDK2 complex purified from transformed human fibroblasts. Coincidentally, the gene encoding human p19 was also named SKP1 (for S phase kinase-associated protein). In addition, a search of the nonredundant protein database (NRDB) revealed three sequences with high similarity to Skp1p: OCP-II (Chen et al. 1995), FP21 (West et al. 1995), and an unknown ORF (Lu et al. 1995). OCP-II is a protein found in the organ of Corti in the guinea pig inner ear, and its role in auditory function is unknown. FP21 is a glycoprotein of unknown function in Dictyostelium and appears to have an unusual glycosylation site. The third match is to a predicted 19 kDa protein that corresponds to an ORF of unknown function encoded in the chlorella virus PBCV-1 genome. In addition, Skp1p exhibits weaker homology to the p15 subunit of a transcription factor called SIII (Garrett et al. 1994), which regulates the activity of the RNA polymerase II elongation complex.
The protein sequences of the seven Skp1p homologs were aligned and a consensus generated (Figure 6). A remarkable conservation of this protein has been preserved over the kingdoms, with the highest similarity toward the carboxyl terminus of this protein family. The temperature-sensitive mutations in the skp1-3 and skp1-4 alleles are in completely conserved amino acids.
Figure 6.
Multiple Protein Sequence Alignments of Skp1p Homologs
The seven Skp1p homologs, from the six species indicated and the chlorella virus genome (PBCV-1), are aligned and ordered by decreasing similarity to the human protein. The consensus is derived from positions that have a 5 out of 7 identical match or better. Hs, H. sapiens; Cp, C. porcellus; Dd, D. discoideum; At, A. thaliana; Sc, S. cerevisiae; CV, PBCV-1 chlorella virus; Ce, C. elegans. Asterisks mark the positions of temperature-sensitive mutations.
S. cerevisiae Skp1p has an apparent 28 amino acid insertion when aligned with the other homologs (residues 37–64). It was possible that these additional residues were important for the specific functions of Skp1p in yeast. An in-frame deletion of the 28 amino acids was constructed (skp1-Δ2; see Figure 1E) and shown to rescue a null mutation of SKP1. We conclude that the S. cerevisiae protein is not a functionally distinct member of the protein family.
Discussion
CBF3, a multisubunit protein complex that binds the essential CDEIII sequence in centromere DNA, was previously shown to contain three major subunits (58, 64, and 110 kDa in size). We describe the identification of a fourth intrinsic subunit of CBF3, Skp1p, which was isolated by screening for dosage suppressors of a known kinetochore protein mutant. The discovery of Skp1p is of broad significance for several reasons. In addition to its essential structural role at the kinetochore, analysis of skp1 mutants suggest a role in cell cycle progression at both G2/M and G1/S. Skp1p represents an intrinsic kinetochore protein that is highly conserved in evolutionarily diverse eukaryotes, including yeast and mammals. Yeast Skp1p (suppressor of kinetochore protein) is the homolog of human SKP1 (S phase kinase-associated protein), which is a subunit of a cyclin A–CDK2 complex purified from cultured mammalian cells. Skp1p may therefore provide a molecular link that facilitates direct interaction between the kinetochore complex and the cell cycle–regulatory machinery.
Skp1p Is an Intrinsic Kinetochore Component
Several lines of evidence indicate that Skp1p is a 22.3 kDa subunit of the CBF3 complex. First, increased expression of SKP1 suppresses the temperature-sensitive lethality and chromosome missegregation phenotype caused by ctf13-30, a missense mutation in the 58 kDa subunit of CBF3. Second, SKP1 is an essential gene, and the temperature-sensitive mutation skp1-4 causes phenotypes similar to those observed in a ctf13 mutant, including chromosome missegregation at semi-permissive temperature and a terminal phenotype at nonpermissive temperature indicative of a defect in the G2/M phase of the cell cycle. Finally, antibodies recognizing an epitope-tagged Skp1 protein decrease the electrophoretic mobility of a CBF3–CEN DNA–protein complex formed in vitro.
Within the CBF3 complex, the stoichiometry of Skp1p is apparently equal to Ctf13p. CBF3 has an apparent native molecular mass of 240 kDa, consistent with the original hypothesis that it is a heterotrimeric complex containing one molecule each of the 58, 64, and 110 kDa subunits. The intrinsic mobility shift data (Figure 2A) indicate that Skp1p and Ctf13p are present at an equimolar ratio. Addition of a 60 amino acid segment to either Skp1p or Ctf13p results in an identical mobility change for the CBF3–CEN DNA complex in the absence of added anti-epitope antibody. Exact comigration of these complexes has been confirmed in extended electrophoresis runs (data not shown). This “intrinsic” shift is not subject to variation of epitope accessibility to antibody binding, as might be encountered in a standard supershift assay (as in Figure 2B). CBF3 therefore contains a minimum of four subunits, which are likely to be in an equimolar ratio. The DNase I footprint of CBF3 exhibits asymmetry around the center of the dyad and encompasses 56 bp of DNA (Jehn et al. 1991). It is possible that multiple CBF3 complexes bind to a single CDEIII sequence, especially given the dyad symmetry within the 25 bp CDEIII consensus sequence.
Stemmann and Lechner 1996 have independently identified Skp1p (termed Cbf3d) as a protein that copurifies with Ctf13p (termed Cbf3c) and is required for reconstitution of CBF3 in vitro from purified components. These biochemical data strongly support the conclusion that Skp1p is an essential component of CBF3. Furthermore, as these authors point out, the copurification of Skp1p (Cbf3d) with Ctf13p (Cbf3c) through a series of chromatographic steps provides evidence that these proteins directly interact. Our genetic suppression data provide in vivo evidence for protein–protein interaction: increased expression of SKP1 suppresses a ctf13 mutation and, reciprocally, increased expression of CTF13 suppresses the skp1-4 mutation.
Skp1p Is an Evolutionarily Conserved Kinetochore Component
The identification and sequence analysis of homologs of the S. cerevisiae SKP1 gene from A. thaliana, C. elegans, and H. sapiens indicate that this protein is highly conserved (>60% identity in pairwise combinations) throughout the eukaryotic phyla. The human SKP1 is likely encoded at a single locus, since all 20 examples of SKP1-related human EST sequences in dbEST appear to represent overlapping segments of a single mRNA species. Recently, a mouse gene encoding a Skp1p homolog was cloned and related sequences were mapped to two locations in the mouse genome, one corresponding to the cloned gene and the other to a cross-hybridizing sequence (Chen et al. 1995). It is possible that one of these represents a pseudogene, although this point needs clarification. In C. elegans, however, SKP1-related ESTs were derived from two nonidentical mRNA sequences (only one of which was sequenced to completion and reported here). The human genome may therefore contain a single functional SKP1 gene or, alternatively, two functional genes, one of which is expressed at low levels and not represented in the cDNA libraries used for EST sequencing. Skp1p homologs present in the public databases include OCP-II (Cavia porcellus), FP21 (Dictyostelium discoideum), and an uncharacterized ORF from the chlorella virus (PBCV-1) genome; however, very little is known about the function of these three proteins in their respective organisms. The human Skp1p homolog protein sequence is identical to a sequence reported by Zhang et al. 1995 for p19SKP1, a protein that copurifies with a p9–p45SKP2–cyclin A–CDK2 complex derived from transformed fibroblasts. Although addition of p45 or p45 and p19 to cyclin A–CDK2 complexes had no effect on kinase activity in vitro, microinjection of anti-p45 antibody into cultured cells prevented entry into S phase. Microinjection of anti-p19 antibody had no effect, so there is currently no direct evidence that p19SKP1 modulates CDK activity either in vitro or in vivo. The G1/S arrest observed in S. cerevisiae skp1-3 mutants reported here supports the hypothesis that p19 is an essential element of the cyclin A–CDK2 S phase kinase.
Skp1p Is Required for Cell Cycle Progression at Both G1/S and G2/M
The distinct sets of phenotypes caused by different temperature-sensitive alleles of SKP1 suggest two independent functions for the Skp1 protein: one acting at G2/M and the other at G1/S. This is reminiscent of allele-specific G1 or G2/M arrest points caused by different cdc28 mutant alleles (seeNasmyth 1993). The G2/M accumulation and chromosome missegregation phenotypes in skp1-4 mutants are similar to those observed in ctf13 and cep3/cbf3c mutants, both of which carry mutations in CBF3 components. The G2/M delay in cell cycle progression could be due to a physical impediment to chromosome separation or, alternatively, to activation of a cell cycle checkpoint. The identification of mad and bub mutants that bypass a cell cycle arrest induced by defects in spindle assembly provides strong evidence for a surveillance mechanism that monitors spindle integrity in yeast (21, 28). Two groups have recently shown that these same functions are required for the G2/M delay observed in response to kinetochore defects caused by mutations in ctf13 or CEN DNA (48, 37). It will therefore be of interest to determine the effects of mad and bub mutants on the skp1-4-mediated G2/M delay.
The allele-specific G1/S arrest observed in skp1-3 mutants appears to be distinct from the G1 arrest observed in cdc28 mutants. The skp1-3-mediated arrest is similar to those exhibited by mutants later in G1. In particular, the multiple budded phenotype is strikingly similar to cdc4, cdc34, and cdc53 mutants, which arrest at the G1/S boundary. As noted above, the homology of Skp1p to human p19, which may associate with a cyclin A–CDK2 complex in mammalian cells, is consistent with a regulatory role for entry into S phase. In addition, Bai et al. 1996 provide strong evidence that Skp1p plays a critical role in the transition from G1 to S phase in yeast and mammals. Combined genetic and biochemical experiments show that Skp1p directly interacts with Cdc4p and cyclin F through a novel structural motif. These authors suggest that Skp1p is a component required for ubiquitin-mediated proteolysis of cyclins and CDK inhibitors.
Skp1p and Cell Cycle Progression
Skp1p is an intrinsic subunit of the kinetochore complex required for assembly of CBF3 on CEN DNA (Stemmann and Lechner 1996; this work). Skp1p also apparently plays an essential regulatory role for entry into S phase (Bai et al. 1996; this work). Temperature-sensitive mutations cause phenotypes consistent with independent roles in kinetochore function at G2/M and cell cycle transition regulation at G1/S (although there is no direct evidence bearing on one or more execution points). Interallelic complementation supports this idea. What is the relationship between these apparently disparate functions?
One possibility is that defects in Skp1p localized within the kinetochore complex can induce a cell cycle checkpoint at either G2/M or G1/S. Defects in CBF3 structure in skp1-4 mutants could lead to aberrant or delayed attachment of kinetochores to the mitotic spindle, which in turn would delay the onset of anaphase. Defects in CBF3 structure in skp1-3 mutants could lead to the G1/S arrest observed by induction of a checkpoint that monitors events at the kinetochore taking place in early S phase. It is known, for example, that centromeres are the earliest-replicating sequences on yeast chromosomes (Ferguson and Fangman 1992). If CEN DNA replication depends on kinetochore structure, failure to complete CEN DNA synthesis in a skp1 mutant could, in theory, preclude subsequent replication of the chromosome.
Alternatively, Skp1p may play spatially distinct but related regulatory roles: one at the kinetochore for progression through mitosis and another elsewhere for entry into S phase. The presence of Skp1p at the kinetochore could provide a direct molecular link to protein complexes (e.g., cyclin–CDKs or other regulatory complexes) whose activities are critical to the metaphase/anaphase transition. Analysis in yeast and higher eukaryotes has provided strong evidence that kinetochores act dynamically to regulate cell cycle progression through mitosis (reviewed byGorbsky 1995). A phospho-epitope in animal cells that is present on unattached kinetochores but is extinguished when bipolar attachment is achieved may be a component or target of a kinetochore checkpoint (Campbell and Gorbsky 1995). As suggested previously (29, 16), eventual achievement of bipolar attachment could, through mechanical tension on the kinetochore complex, induce a conformational change in a key regulatory protein and thereby regulate the activity of an associated kinase. Though speculative, it is possible that Skp1p may function at the kinetochore to signal completion of metaphase. For example, Skp1p could facilitate docking of a cyclin–CDK complex (or proteins involved in regulating a kinase) at the kinetochore. This specific localization of a regulatory kinase (or other protein complex) may act as a determinant in its activity upon a kinetochore-localized substrate. Perhaps the regulatory capacity of Skp1p has been recruited to the kinetochore by its direct incorporation within the CBF3–CEN DNA complex.
Experimental Procedures
Yeast Strains and Media
The ctf13-30 allele (YPH972) has been previously described (Doheny et al. 1993). The yeast strains and their genotypes used in this study are described in Table 2. The high copy suppression screen was done in YPH972R, a derivative of YPH972 lacking the chromosome fragment. Methods and media for growth and sporulation were as described previously (Rose et al. 1990). Yeast transformations were done by the method described by Ito et al. 1983.
Table 2.
Strains Used in This Study
| S. cerevisiae Strain S288C Derivatives | Genotype | Reference |
| YPH972Ra | Matαura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 ctf13-30 | Doheny et al. 1993 |
| YPH975Ra | Matαura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 ctf13Δ1::HIS3pRS316/CTF13 | Doheny et al. 1993 |
| YPH985 | Mata/Matαura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his-Δ200 leu2-Δ1/leu2-Δ1 CFIII (CEN3.L.YPH 983) HIS3 SUP11 | This study |
| YPH1015 | Mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CFIII (CEN3.L.YPH983) HIS3 SUP11 | This study |
| YPH1096 | Mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CFIII (CEN3.L.YPH983) HIS3 SUP11pRS316SKP1 | This study |
| YPH1099 | Mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 skp1Δ1::TRP1 CFIII (CEN3.L.YPH983) HIS3 SUP11pRS316SKP1 | This study |
| YPH1161 | Mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 skp1Δ1::TRP1 skp1-4::LEU2 CFIII(CEN3.L.YPH983) HIS3 SUP11 | This study |
| YPH1172 | Mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 skp1Δ1::TRP1 skp1-3::LEU2 CFIII(CEN3.L.YPH983) HIS3 SUP11 | This study |
| YPH1197 | Mata/Matαura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 skp1-3::LEU2/skp1-3::LEU2 CFIII (CEN3.L.YPH983) HIS3 SUP11 | This study |
| YPH1201 | Mata/Matαura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 skp1-4::LEU2/skp1-4::LEU2 CFIII (CEN3.L.YPH983) HIS3 SUP11 | This study |
| YPH1208 | Mata/Matαura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 skp1-3::LEU2/skp1-4::LEU2 CFIII (CEN3.L.YPH983) HIS3 SUP11 | This study |
These strains are mitotic segregants that have lost the marker chromosome fragment (and therefore form red colonies).
High Copy Suppression
A 2μ yeast genomic library was prepared from YPH1 (Sikorski and Hieter 1989) genomic DNA. After partial Sau3A digestion, 6–8 kb of genomic DNA segments was ligated into BamHI–BglII-restricted pRS202, a derivative of pRS306 that contains, first, the 2μ origin and REP3 sequences from Yep24 (HindIII to PstI) inserted into the AatII site and, second, BglII linkers inserted at the SmaI site of the polylinker. A sample of the library base of 19,500 transformants showed that 32 out of 36 plasmids had average insert sizes of 6–8 kb. All of the recombinants were pooled, and DNA was prepared for transformation into yeast.
Library DNA was transformed into YPH972R, and after 18 hr at room temperature the transformants were shifted to the nonpermissive temperature at 37°C selecting for uracil prototrophy. Approximately 18,300 independent transformants were screened at the nonpermissive temperature, yielding 15 temperature-resistant colonies after 5 days of growth. The library clones were recovered from the yeast strains and retested for the suppression phenotype. Restriction site and Southern blot analysis of the plasmid DNAs indicated that 7 of 15 contained CTF13 sequences, 3 of 15 contained vector only or an insert that failed to retest, and 5 of 15 contained overlapping segments. The latter class of plasmids were able to suppress the temperature sensitivity of ctf13-30, although not as well as wild-type Ctf13p.
Molecular Characterization of SKP1
The library clone (3B) chosen for further characterization contained a 6.3 kb insert of genomic DNA (Figure 1A). A 1.68 kb SpeI–PvuII fragment was subcloned and shown to suppress the temperature sensitivity of a ctf13-30 mutant. Fill-in of a unique NsiI site with T4DNA polymerase (New England Biolabs) abolished the suppressing activity. A series of subclones and deletion derivatives were sequenced by standard methods (42, 17, 44, 32). CEN vector derivatives containing the 1.68 kb SpeI–PvuII region were constructed by insertion into the SpeI and SmaI polylinker sites of pRS vectors (Sikorski and Hieter 1989), resulting in two constructs, pRS315SKP and pRS316SKP. These CEN vector recombinants were able to suppress the temperature sensitivity of ctf13-30, but not to the extent of the 2μ library clone 3B. Insertion into the SKP1 ORF of a 1.8 kb BglII fragment containing the TRP1 gene was accomplished by blunt-end ligation into the NsiI site. One-step gene replacement was accomplished by excision of a NotI–XhoI fragment and transformation into YPH985 selecting for Trp+ transformants. The integration of the TRP1 marker into the genome was confirmed by Southern blot analysis of restricted genomic DNA. Trp+ diploids were sporulated, and 20 tetrads dissected resulted in 2+:2− segregation for viability. All viable spores were Trp−, demonstrating that SKP1 was an essential gene.
SKP1 was physically mapped to chromosome IV by hybridization of a radioactive 32P-labeled (Feinberg and Vogelstein 1984) EcoRI–NotI (polylinker) fragment isolated from 3B to filters containing overlapping λ and cosmid clones of the S. cerevisiae genome (L. Riles and M. Olson). This placed SKP1 on the right arm of chromosome IV, between gcn2 and trp4.
The skp1-Δ1::TRP1 allele is a deletion of the entire SKP1 ORF between the PstI and Eco57I sites and replacement with vector and TRP1 sequences using one-step gene replacement (Figure 1B). This deletion extends from 100 bp upstream of the start of SKP1 to 20 bp upstream of the stop codon. The integration vector, pRS304 ΔSKP1, was constructed by inserting the ∼1.3 kb PstI (polylinker)–PstI fragment and the 1.16 kb Eco57I–ClaI fragment from the original library clone 3B into the PstI site and the ClaI–KpnI (blunt) site of pRS304 (Sikorski and Hieter 1989), respectively. pRS304ΔSKP1 was linearized with EcoRI and transformed into YPH1096 selecting for Trp+ transformants. Transformants were streaked on 5-fluoroorotic acid (FOA) (Boeke et al. 1987) to determine whether the integration took place in the genome or on the Ura+ plasmid, pRS316SKP. Transformants that failed to grow on FOA were selected, and Southern blot analysis of genomic DNA was performed to confirm the integration and removal of the SKP1 ORF. YPH1099 is the skp1 null strain generated in this way.
Biochemical Analysis
The plasmid containing the GAL1 promotor–E1 epitope fused to SKP1, p415GEU1SKP, was constructed from the plasmid p415GEU1, a derivative of p414GEU1 (Doheny et al. 1993), which replaced the TRP1 gene with the LEU2 gene using PvuI–PvuI sites in the vector sequence (Sikorski and Hieter 1989). A PCR strategy was used to place the E1 epitope in-frame with the second codon of the SKP1 ORF using an engineered XhoI cloning site and a downstream genomic EcoRI site (see Figure 1A). To avoid possible PCR errors, wild-type genomic sequence from NsiI to PvuII replaced the PCR-generated sequence. When transformed into the skp1 deletion strain, p415GEU1SKP rescued growth on galactose-containing, but not dextrose-containing, media. The untagged construct was prepared by cloning the XhoI–PvuII fragment from p415GEU1SKP into p415GEU2 (J. Kroll, unpublished data) at the XhoI–SmaI sites in the polylinker. When transformed into the skp1 deletion strain, p415GEU2SKP rescued growth on both galactose- and dextrose-containing media, presumably because of low level expression under glucose-repressing conditions.
The HA epitope was fused to SKP1 in pAD5 (Neiman et al. 1993), which is a 2μ–LEU2 plasmid that carries an ADH1 promoter, an ATG, and the Lerner HA epitope (Field et al. 1988) followed by a SalI cloning site. A 1.3 kb XhoI–SacI fragment from p415GEU1SKP was cloned into the SalI–SacI site in the polylinker of pAD5, resulting in pAD5SKP. This HA-tagged construct was able to rescue viability in the skp1 deletion strain.
Extracts were prepared for biochemical analysis of CBF3–CEN DNA complexes according to the methods described in Doheny et al. 1993. The DNA fragment containing CDEIII used in the gel mobility shift assay was the same as reported previously (Doheny et al. 1993) except it was labeled by a different method. The plasmid pSF262a (a gift from P. Sorger) allows excision of the CDEIII region with HindIII–EcoRI, resulting in a 100 bp fragment that was labeled using Taq polymerase according to the method described previously (Niedenthal and Hegemann 1993).
Generation and Integration of Temperature-Sensitive Alleles
Synthetic oligonucleotides were synthesized corresponding to sequences approximately 250 bp on either side of the polylinker of pRS316. They served as primer for a PCR using pRS316SKP as a template following conditions for mutagenesis as described by Muhlrad et al. 1992. Approximately 10 ng of template was amplified under standard PCR conditions (Perkin Elmer) except the concentration of MgCl2 was increased to 3 mM and the dNTP pool was reduced to 60 μM. Approximately 500 ng of mutagenized PCR product was cotransformed into YPH1099 with 500 ng of XhoI-linearized pRS315 and 5 μg of herring sperm DNA (Sigma). Transformants were replica plated twice to medium containing FOA to ensure eviction of pRS316SKP. Of the 1300 transformants screened by this method, 16% failed to grow on FOA, suggesting knockouts of SKP1 or reclosing of the gapped vector. There was an approximately 4-fold stimulation of transformants with the addition of PCR product as compared with gapped vector only. It was also observed that approximately 80% of the gapped vector only transformants failed to grow on FOA. Transformants that grew presumably recombined with pRS316SKP. The transformants were then replica plated in parallel and grown at 25°C and 37°C. Four temperature-sensitive alleles, skp1-1, skp1-2, skp1-3, and skp1-4, were isolated, and the mutagenized plasmids were recovered and retested for sensitivity at 37°C. The nucleotide sequence of each allele was done to identify the mutations (44, 32), and an alignment of the sequences revealed that the mutations fell mostly in the carboxyl terminus (see Table 1).
Two of the temperature-sensitive alleles, skp1-3 and skp1-4, were integrated into the genome at leu2-Δ1 on chromosome III using a LEU2 derivative of p679 (Doheny et al. 1993). The mutagenized plasmid DNA was excised from the pRS315SKP derivative as a 1.6 kb SpeI (blunt)–HindIII (polylinker) and inserted into a SmaI–HindIII polylinker site of p679LEU2, resulting in BPH650 (skp1-3) and BPH651 (skp1-4). Transformation of these two constructs linearized with NotI directed the γ-integration of the temperature-sensitive alleles to leu2-Δ1 located either on chromosome III or the artifical chromosome III fragment in YPH1099. Independent transformants were grown in the presence of FOA to select against the wild-type SKP1 and then tested for temperature sensitivity. Transformants that grew at the permissive temperature but failed to grow at the nonpermissive temperature were prepared for analysis by orthogonal field alteration gel electrophoresis (OFAGE) according to the methods described by Carle and Olson 1984. The genomic location of the temperature-sensitive allele was accomplished by hybridization of a 1.3 kb PstI (genomic)–PstI (polylinker) containing SKP1 from pRS315SKP to a Southern blot of the OFAGE gel (data not shown). The genomic integration of skp1-3 was confirmed in YPH1172 and that of skp1-4 in YPH1161.
Homozygous and heterozygous skp1 diploids were generated from meiotic sister spores obtained from dissection of tetrads from backcrosses of YPH1172 and YPH1161 to YPH500 (Sikorski and Hieter 1989; see Table 2).
Internal Deletion of SKP1 ORF
The alignment of S. cerevisiae to conserved homologs in other species (see Figure 6) defined a region of 28 amino acids unique to S. cerevisiae. Fortunately, a genomic NsiI site was at the junction. This region of nonhomology was removed from the SKP1-coding region by a PCR strategy using a synthetic oligonucleotide, PH139, which included a unique NsiI cloning site in-frame with the sequence 5′-GGTGACGACGACGATGAGGATG-3′ located at the end of the 28 amino acid region to be removed. An ∼980 bp PCR product generated from PH139 and a primer in the vector sequence of pRS316SKP was restricted with NsiI–XhoI and cloned into the NsiI (genomic)–Xho (polylinker) of pRS315SKP, resulting in pRS315 skp1-Δ2. This construct rescued the deletion strain YPH1099.
Acknowledgements
Correspondence should be addressed to P. H. We are particularly grateful to O. Stemmann, J. Lechner, C. Bai, and S. Elledge for communicating results prior to publication. We thank F. Spencer, R. Skibbens, and J. Lamb for comments on the manuscript; M. Basrai for contributions to experimental design; G. Merkulov for construction of the E1-tagged SKP1 expression vector; S. Tugendreich for help in protein sequence alignments; and members of the Hieter lab for helpful discussions and ideas. This work was supported by a National Institutes of Health grant (CA16519) to P. H.
References
- 1.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J.W, Elledge S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell, this issue. 1996 doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Bernat R.L, Borisy G.G, Rothfield N.F, Earnshaw W.C. Injection of anticentromere antibodies in interphase disrupts events required for chromosome movement in mitosis. J. Cell Biol. 1990;111:1519–1533. doi: 10.1083/jcb.111.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke J, Truehart J, Natsoulis G, Fink G. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Meth. Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 4.Boguski M, Tolstochev C, Bassett D. Gene discovery in dbEST. Science. 1994;265:1993–1994. doi: 10.1126/science.8091218. [DOI] [PubMed] [Google Scholar]
- 5.Campbell M.S, Gorbsky G.J. Microinjection of mitotic cells with the 3F3/2 anti-phospoepitope antibody delays the onset of anaphase. J. Cell Biol. 1995;129:1195–1204. doi: 10.1083/jcb.129.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carle G.F, Olson M. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alteration gel electrophoresis. Nucl. Acids Res. 1984;12:5647–5665. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Thalmann I, Adams J.C, Avraham K.B, Copeland N.G, Jenkins N.A, Beier D.R, Corey D.P, Thalmann R, Duyk G.M. cDNA cloning, tissue distribution, and chromosomal localization of Ocp2, a gene encoding a putative transcription-associated factor predominantly expressed in the auditory organs. Genomics. 1995;27:389–398. doi: 10.1006/geno.1995.1068. [DOI] [PubMed] [Google Scholar]
- 8.Doheny K.F, Sorger P.K, Hyman A.A, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg A.P, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson B.M, Fangman W.L. A position effect on the time of replication origin activation in yeast. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- 11.Field J.J, Nikawa D, Broek B, MacDonald L, Rogers L, Wilson I, Lerner R, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparison and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 13.Garrett K, Tan S, Bradsher J, Lane W, Conaway J, Conaway R. Molecular cloning of an essential subunit of RNA polymerase II elongation factor SIII. Proc. Natl. Acad. Sci. USA. 1994;91:5237–5241. doi: 10.1073/pnas.91.12.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerring S.L, Spencer F, Hieter P. The CHL1(CTF1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goh P.-Y, Kilmartin J.V. NDC10: a gene involved in chromosome segregation in S. cerevisiae. J. Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbsky G.J. Kinetochores, microtubules and the metaphase checkpoint. Trends Cell. Biol. 1995;5:143–148. doi: 10.1016/s0962-8924(00)88968-0. [DOI] [PubMed] [Google Scholar]
- 17.Hattori M, Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal. Biochem. 1986;152:232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- 18.Hegemann J, Fleig U. The centromere of budding yeast. Bioessay. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- 19.Hegemann J, Shero J, Cottarel G, Philippsen P, Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hieter P, Mann C, Snyder M, Davis R. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 21.Hoyt M.A, Totis L, Roberts B.T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 22.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jehn B, Niedenthal R, Hegemann J. In vivo analysis of the Saccharomyces cerevisiae centromere CDEIII sequence: requirements for mitotic chromosome segregation. Mol. Cell. Biol. 1991;11:5212–5221. doi: 10.1128/mcb.11.10.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2) specifying an essential protein component of the budding yeast kinetochore. J. Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koshland D, Hieter P. Visual assay for chromosome ploidy. Meth. Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- 26.Lechner J. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, CBF3. EMBO J. 1994;13:5203–5211. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lechner J, Carbon J. A 240 kD multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 28.Li R, Murray A.W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Nicklas R.B. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 30.Lu Z, Li Y, Zhang Y, Kutish G.F, Rock D.L, Van Etten J.L. Analysis of 45 kb of DNA located at the left end of the chlorella virus PBCV-1 genome. Virology. 1995;206:339–352. doi: 10.1016/s0042-6822(95)80049-2. [DOI] [PubMed] [Google Scholar]
- 31.Machamer C.E, Rose J.K. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 1987;105:1205–1214. doi: 10.1083/jcb.105.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCombie W.R, Heiner C, Kelly J.M, Fitzgerald M.G, Gocayne J.D. Rapid and reliable fluorescent cycle sequencing of double stranded templates. DNA Seq. 1992;2:289–296. doi: 10.3109/10425179209030961. [DOI] [PubMed] [Google Scholar]
- 33.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 34.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr. Opin. Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 35.Neiman A.M, Stevenson B.J, Xu H.-P, Sprague G.F, Herkowitz I, Wigler M, Marcus S. Functional homology of protein kinases required for sexual differentiation in Schizosaccharomyces pombe and Saccharomyces cerevisiae suggests a conserved signal transduction module in eukaryotic organisms. Mol. Biol. Cell. 1993;4:107–120. doi: 10.1091/mbc.4.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niedenthal R.K, Hegemann J.H. An efficient method to generate phosphatase insensitive 3′ labelled DNA probes using Taq polymerase. Nucl. Acids Res. 1993;21:4413. doi: 10.1093/nar/21.18.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol. Biol. Cell, in press. 1996 doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pluta A.F, Saitoh N, Goldberg I, Earnshaw W.C. Identification of a subdomain of CENP-B that is necessary and sufficient for localization to the human centromere. J. Cell Biol. 1992;116:1081–1093. doi: 10.1083/jcb.116.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pluta A.F, Mackay A.M, Ainsztein A.M, Goldberg I.G, Earnshaw W.C. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1595. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 40.Rieder C, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell. Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose M.D, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1990. [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sikorski R.S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith L.M, Sander J.Z, Kaiser R.J, Hughes P, Dodd C, Connel C.R, Heiner C, Kent S.B, Hood L.E. Fluoresence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 45.Spencer F, Hieter P. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1992;89:8908–8912. doi: 10.1073/pnas.89.19.8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stemmann O, Lechner J. The S. cerevisiae kinetochore contains a cyclin-CDK complexing homolog, as identified by in vitro reconstitution. EMBO J., in press. 1996 [PMC free article] [PubMed] [Google Scholar]
- 47.Strunnikov A.V, Kingsbury J, Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Burke D.J. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells W.A.E, Murray A.W. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West C.M, Kozarov E, van der Wel H, Field M, Gritzali M, Brown R.D., Jr. Characterization of FP21, a cytosolic glycoprotein from Dictyostelium. J. Biol. Chem. 1995;270:3022–3030. doi: 10.1074/jbc.270.7.3022. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A–CDK2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
Further reading
The accession numbers for the SKP1 cDNA sequences reported in this paper are U43179 (S. cerevisiae), U60980 (C. elegans), and U60981 (A. thaliana).