Abstract
Nanomedicine, the application of nanotechnology to medicine, encompasses a broad spectrum of fields including molecular detection, diagnostics, drug delivery, gene regulation and protein production. In recent decades, DNA has received considerable attention for its functionality and versatility, allowing it to help bridge the gap between materials science and biological systems. The use of DNA as a structural nanoscale material has opened a new avenue towards the rational design of DNA nanostructures with different polymeric topologies. These topologies, in turn, possess unique characteristics that translate to specific therapeutic and diagnostic strategies within nanomedicine.
Keywords: DNA, Nanomedicine, Polymer topologies, Nano-assembly, DNA nanostructures
1. Introduction
Nanotechnology is a highly multidisciplinary field that has been gaining attention, both from the scientific and political communities, for its exciting breakthroughs and promising developments. Federal funding to the National Nanotechnology Initiative (NNI) in recent years has rocketed from $464 million in 2001 to $1.8 billion in 2011 (http://www.nano.gov/html/about/funding.html), providing an impetus for rapid development. The influence of nanotechnology on the field of medicine has given rise to a new field known as ‘nanomedicine’, which encompasses the utilisation of nanoscale structures and devices for medical treatment and diagnosis. The National Institute of Health (NIH) has incorporated nanomedicine into its roadmap in 2005 with the vision that it will have a beneficial impact on medicine within the first decade. The fundamental challenge in nanomedicine is to design biocompatible and functional nanoscale assemblies with molecular precision and control. There is a need, therefore, to design and explore novel materials that can meet these criteria. DNA has proven to be an exceptional material with a vast number of manipulation options readily available from the molecular biology toolbox, making DNA nanoscale architectures a promising class of materials for nanomedicine.
1.1. DNA: From Genetic Drug to Generic Polymer
In nanomedicine, DNA usually takes on the role of a therapeutic molecule. There has been extensive work to overcome the barriers encountered when delivering DNA to the cell nucleus [1], [2], [3], and the development of new materials that can serve as efficient nucleic acid-delivery vehicles is of key importance. In many of these systems, polymeric materials have been investigated due to their well-understood physical and chemical properties, biocompatibility and apparent increase in delivery efficiency [4]. At the nanoscale, methods for DNA delivery include condensation of DNA by forming complexes with cationic polymers and synthetic peptides [5], [6], lipid–DNA complexes [7] and encapsulation within hydrogel nanoparticles [8]. At larger scales, sustained release of DNA complexes from a polymer matrix has also been reported [9]. It is important to note that, in each of these systems, as with many biomedical systems, the polymer takes on both a functional and structural role. DNA, although often regarded as a drug, is also a polymer that possesses many of the chemical and physical properties that make other polymers desirable for the same therapeutic applications.
There is no question about the importance of the DNA's role in molecular biology since its structure was discovered in the early 1950s. Perhaps it is somewhat unfortunate that, in spite of its polymeric characteristics, the term ‘DNA’ has become synonymous with ‘genetics’. However, within recent decades, DNA has gained significant recognition as an incredibly versatile ‘generic’ material. Due to its remarkable physical and chemical properties, DNA demonstrates great potential as a structural polymer for bottom-up nanoscale synthesis [10]. Mechanically, DNA is surprisingly robust – below its persistence length of ∼ 50 nm, double-stranded DNA (dsDNA) is a relatively rigid molecule [11]. It is also stable at huge aspect ratios and can extend to several hundred micrometres despite its diameter of only ∼ 2 nm. From a chemical perspective, DNA can be engineered and processed efficiently with angstrom-level accuracy using a plethora of biochemical techniques [12]. A unique inventory of thousands of biochemically characterised enzymes, unlike that of any other known natural or synthetic polymer, is also available for the precise manipulation of DNA [13]. In particular, enzymatic cleavage and ligation facilitates the processing of DNA into monodisperse strands with exact lengths and sequences. In single-stranded DNA (ssDNA), each of the four bases – adenine (A), thymine (T), guanine (G) and cytosine (C) – has a specific affinity to its complement (A/T and G/C). These bases act as side-chains that can be engineered in any order. Typically, ssDNA strands containing complementary base sequences can recognise and bind to each other to form a dsDNA duplex (hybridisation). In addition, ssDNA can also self-anneal to form unique tertiary structures that can bind specifically to macromolecules, rivalling the binding affinities of antibodies. Furthermore, the development of recombinant DNA technology and solid-phase synthesis techniques has facilitated the reliable mass production of DNA.
1.2. Polymeric Topologies of DNA
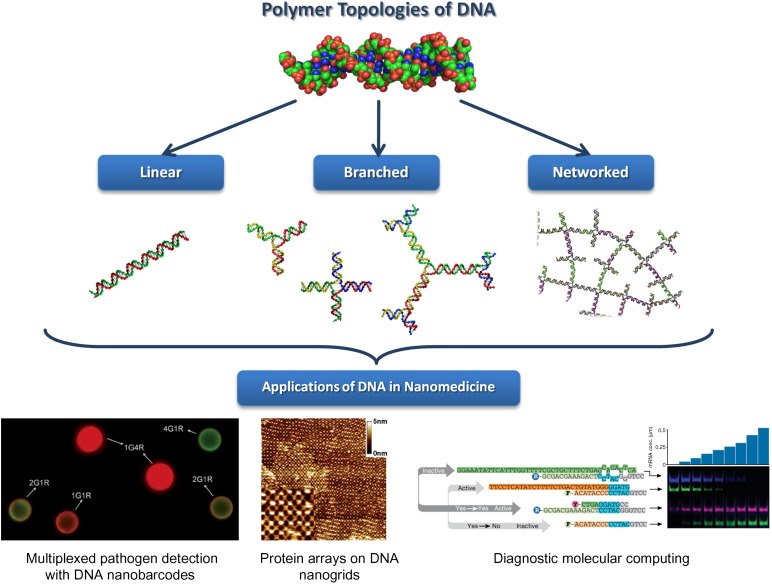
Although naturally occurring DNA is linear, the rational design of single-stranded oligomers has enabled the realisation of three polymer topologies that are relevant to nanomedicine, namely linear, branched and networked (cross-linked) [14]. Each of these topologies is typically associated with properties that could be adapted for specific applications, particularly within the realm of nanomedicine as outlined by Freitas [15], [16]. Linear DNA has traditionally been limited to the fields of genetics and drug delivery, but is being increasingly utilised in molecular recognition and diagnostic systems for its simplicity and distinct binding characteristics. Branched DNA encompasses an extensive library of unique structures that could be tailored individually or collectively for an assortment of potential applications. In particular, DNA dendrimers are unique for their multivalent termini that allow for signal amplification and high-affinity targeting. Finally, networked DNA architectures are structurally stable and provide a three-dimensional scaffold that could be tailored towards controlled-release systems as well as protein-production platforms for therapeutics. There are many excellent reviews that discuss DNA architectures in depth and emphasise their potential use in bottom-up nanoscale synthesis [17], [18], [19], [20], [21], [22], [23], [24], [25]. This article focusses specifically on the application of DNA in nanomedicine. We describe DNA architectures in the context of the three polymer topologies and how their properties can be tailored towards particular applications (Fig. 1 and Table 1 ).
Fig. 1.
Schematic illustrating the assembly of DNA as a polymer with different topologies – linear, branched and networked – and how these topologies translate to applications in nanomedicine. Examples include multiplexed pathogen detection based on branched DNA (Figure reproduced from reference [119]), protein arrays assembled on DNA nanogrids (Figure reproduced from reference [137] with permission), and diagnostic molecular computing (Figure reproduced from reference [152] with permission).
Table 1.
Applications of DNA polymeric topologies in nanomedicine.
| Systems | Potential Applications | References | |
|---|---|---|---|
| Linear | DNA-nanoparticle conjugates | Gene regulation, DNA/mRNA/protein/molecular sensing, multiplexed biomolecular detection | [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58] |
| DNA aptamers | Targeted drug delivery, molecular sensing and quantification, protein arrays, therapeutic use | [68], [69], [75], [76], [80], [81], [82], [83], [84] | |
| DNA-responsive hydrogels | Controlled drug release, DNA sensing, protein capture and release | [97], [98], [99], [100], [101], [102], [103] | |
| Active constructions | Molecular modification and manipulation, gene regulation, diagnostic molecular computing | [141], [144], [145], [146], [148], [149], [150], [152] | |
| Branched | Multi-arm DNA | Biosensing | [116], [117] |
| Dendrimer-like DNA | Targeted drug delivery, multiplexed biomolecular detection | [119], [120], [122] | |
| Active constructions | Molecular modification and manipulation, molecular switches | [142], [143], [147], [151] | |
| Networked | DNA films | Toxin filtration, antibacterial materials, antigen detection | [126], [128], [129] |
| DNA hydrogels | Cell encapsulation, controlled drug release, protein production | [127], [138], [139], [140] | |
| DNA lattices | Protein arrays | [136], [137] |
2. Linear DNA
The specificity of hybridisation makes DNA ideal for nanoscale systems that require molecular recognition. There are a variety of systems that take advantage of the hybridisation of linear DNA for nanomedicine applications, including oligonucleotide sensing, multiplexed pathogen detection, aptameric drug delivery and diagnostics, as well as stimuli-responsive hydrogels.
2.1. DNA–Nanoparticle Conjugates for Detection and Gene Regulation
In the past decade, there has been considerable interest in using hybrid nanoparticles consisting of oligonucleotides covalently tethered to gold nanoparticles (AuNPs) as a tool for molecular diagnostics and gene regulation [26], [27]. Compared with conventional probes used in molecular diagnostics, oligonucleotide-functionalised AuNPs have achieved higher sensitivity and selectivity, lower cost and greater ease of detection [28]. From a materials perspective, AuNPs are relatively easy to synthesise and are biocompatible [29]. In addition to diagnostics, they have also been utilised in the investigation of gene function and treatment of disease [30], [31].
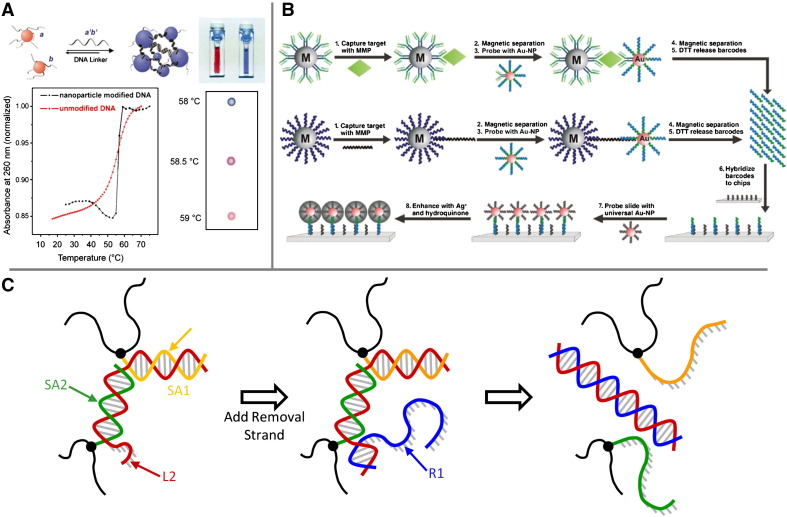
Mirkin pioneered the development of DNA–AuNP conjugates for molecular diagnostics and gene regulation. As early as 1996, Mirkin and co-workers described a method of assembling AuNPs into macroscopic aggregates using DNA as linkers through thiol bonds [32]. Subsequently, Mirkin's group employed DNA-modified AuNPs as a scaffold to develop a highly selective, colorimetric polynucleotide-detection technique [26], [33] (Fig. 2A). AuNPs aggregated in the presence of complementary target ssDNA in solution as a result of DNA hybridisation. The aggregation induced a red-to-purple colour change, which enabled the precise detection of as little as 10 femtomoles of the target sequence [34]. This colorimetric method was also demonstrated for the highly selective and sensitive detection of mercuric ions and cysteine in aqueous solution [35], [36]. Colorimetric biosensing is particularly promising since it eliminates the necessity of expensive and complicated instruments for detection [37].
Fig. 2.
Application of linear DNA polymeric topologies in molecular diagnosis and gene regulation. (A) Oligonucleotide detection based on aggregation of DNA-modified AuNPs. Introduction of a target ssDNA into a solution containing AuNPs functionalized with complementary DNA sequences resulted in the formation of an aggregated network of nanoparticles with a corresponding red-to-purple color change. Reproduced from reference [33] with permission. (B) DNA-AuNP conjugates as biobarcodes. Each target molecule is coded by ssDNA with specific sequences and lengths. The target molecule binds specifically to a monoclonal antibody on a magnetic microparticle and is sandwiched by the DNA-AuNP containing the corresponding polyclonal antibody and the ssDNA barcodes. The ssDNA barcodes are subsequently released for specific identification of the target molecule. Reproduced from reference [47] with permission. (C) DNA-mediated reversible sol-gel transitions. Polyacrylamide modified with DNA sequences SA1 and SA2 crosslinks with the addition of a target sequence, L2, partially complementary to both SA1 and SA2. Upon addition of a removal strand that hybridizes completely with L2, the crosslinking is disrupted and the gel reverts to a liquid state.
Mirkin's group developed several other highly sensitive and selective detection strategies. These include chip-based detection methods that rely upon either light scattering or silver staining [38], [39], Raman dye-labelled oligonucleotide-gold probes for multiplexed detection of DNA targets [40], [41], an electrical detection method for target DNA which is captured in the gap between two electrodes [42] as well as a real-time DNA-detection method [43]. Notably, a nanoparticle-based biobarcode assay was developed for the ultrasensitive detection of biomolecules, such as proteins [44], [45], [46], [47], [48], messenger RNA (mRNA) [49] and the human immunodeficiency virus (HIV)-1 antigen [50], as well as for the multiplexed detection with high selectivity but without the need for enzymatic target amplification and labelling [28], [47] (Fig. 2B).
For gene regulation, Mirkin's group reported a method that employed DNA-modified AuNPs for cellular transfection and targeted gene silencing [27]. This ‘antisense nanoparticle’ was constructed by attaching multiple strands of antisense DNA to the surface of an AuNP. It was observed that the AuNP-immobilised antisense DNA became more stable and could bind to the target mRNA more effectively than the free antisense DNA strands. This technique possesses many advantages compared to antisense DNA complexed with commercial agents, such as LipofectamineTM and CytofectinTM; in particular, more effective gene knockdown, less susceptibility to degradation, lower toxicity and enhanced cellular absorption were noted for these AuNP–DNA conjugates. Interestingly, the large number of proteins (from serum) that adsorbed onto the AuNP surface appeared to enhance cellular uptake of the negatively charged antisense nanoparticles [31]. In addition, a higher surface density of oligonucleotides bound more proteins, which ultimately increased the uptake efficiency.
In addition to Mirkin's extensive work, there are many other groups that have used DNA-modified nanoparticles for sensing and diagnostics. For example, a molecular ruler created by tethering double-stranded DNA to a single AuNP enabled label-free and real-time measurements of protein-DNA interactions at an extremely high resolution [51]. Storhoff developed a ‘spot-and-read’ colorimetric detection method for detecting zeptomole quantities of a nucleic acid target without any target or signal amplification [52]. More recently, Qin and co-workers reported a novel AuNP-based quantitative DNA assay with single-nucleotide polymorphism (SNP) discrimination selectivity [53]. The precise quantification is due to the formation of defined DNA–AuNP conjugate groupings in the presence of target/linker DNA, which were then characterised and quantified by gel electrophoresis. Niemeyer and co-workers have also utilised linear DNA as an intermediate linker for generating protein–nanoparticle conjugates that are ideal for immunological diagnostics [54], [55]. Niemeyer's group also demonstrated binding and dissociation of proteins from the surfaces of DNA–nanoparticle conjugates under physiological conditions, which ensures that the biological activity of the protein is not compromised [56]. In addition to metallic nanoparticle-based systems, quantum dots (QDs) conjugated to ssDNA have also been used for ultra-sensitive detection of DNA in a separation-free format [57]. Alivisatos and co-workers also reported that DNA–QD conjugates, when used in conjunction with microarrays, enabled the rapid detection of SNPs and single-base deletions [58].
2.2. DNA Aptamers for Drug Delivery and Diagnostic Detection
Owing to its high chain flexibility, ssDNA is capable of self-annealing and folding into unique tertiary structures. Aptamers are defined as short nucleic acid ligands that can bind to target molecules with high specificity and affinity [59], [60], [61]. Both the concept of aptamers and the procedure for aptamer generation were brought about in the early 1990s [62], [63]. Over the past 17 years, aptamers have been generated against a wide array of molecular targets, including many known proteins, carbohydrates, lipids, nucleotides and other small molecules, as well as highly complex structures such as viruses [64], [65], [66]. Aptamers are typically 15–40 nucleotides in length [60] and are generally produced through the systematic evolution of ligands by exponential enrichment (SELEX) process [62], [67], an iterative, in vitro evolutionary process based on identification of a tight binding sequence followed by amplification.
Aptamers behave like drugs, analogous to monoclonal antibodies that function through high-affinity binding to a target molecule [68], distinguishing them from antisense oligonucleotides and ribozymes [69]. With their high specificity and strong affinity for their targets, aptamers are very powerful tools for diagnostic and therapeutic applications [64], [65], [66], [70]. They are potentially useful in the treatment of a wide variety of human maladies, including infectious diseases, cancer and cardiovascular disease [71]. As a result, there has been much interest in developing aptamers for cancer therapy [72], [73], [74], [75], [76]. The most advanced aptamer and the first to enter clinical trials is AS1411 [77], a 26-mer unmodified guanosine-rich oligonucleotide that has been shown to be effective against human tumour xenografts in vivo. Aptamers have also been conjugated to QDs and used in conjunction with DNA-intercalating drugs for the simultaneous targeting, imaging and treatment of cancer [75], [76]. This approach proved to be highly effective in targeting cancer cells and decreasing tumour size with fewer side effects on healthy cells in comparison to current methods of chemotherapy.
Aptamers can be adapted to produce many different types of signals, such as electrochemical, acoustic, fluorescence and piezoelectric [59], making them excellent candidates for disease diagnostics and biosensing [65], [66], [78], [79]. Currently, most diagnostic applications of aptamers rely on ligand-induced conformational changes, which can be detected by the corresponding changes of these signals [65]. Such applications include western blot analyses, oligonucleotide-precipitation assays, flow-cytometry and use as affinity reagents coupled to solid matrices for protein purification [66], [78]. Aptamers have also been successfully utilised in a procedure similar to the enzyme-linked immunosorbent assay (ELISA) to detect vascular endothelial growth factor (VEGF) and thrombin [80]. In addition, fluorescence-labelled aptamers have also been used to quantify immunoglobulin E (IgE) and thrombin based on capillary electrophoresis/laser-induced fluorescence (CE/LIF), in which detection as low as 50 pM was reported [81]. A cell-based aptamer-selection method to generate aptamers that can specifically recognise target cancer cells has also been reported [82], [83], [84]. This method shows great promise for developing specific molecular probes for cancer diagnosis and cancer biomarker discovery. In addition to the diagnostic and therapeutic applications, aptamers have also been frequently used for target validation and drug discovery [65], [70], [85], [86].
2.3. Stimuli Response in DNA-Based Hydrogels
For the purposes of our discussion, we make a distinction between ‘DNA-based hydrogels’ and ‘DNA hydrogels’. We define a DNA-based hydrogel as one in which DNA takes on the role of a cross-linking agent or is chemically grafted into the hydrogel matrix. Since the function of these structures is mediated by the hybridisation of linear DNA, we categorise DNA-based hydrogels under the linear DNA topology. In contrast, we regard a ‘DNA hydrogel’ as a hydrogel in which DNA acts as the primary structural unit. The resulting structures are categorised under the network topology (see the section titled ‘DNA networks’).
Hydrogels have been used in a wide range of biomedical applications for the past several decades, ranging from contact lenses to drug-delivery systems [87]. In the most basic description, a hydrogel is a water-swollen, cross-linked polymer network in which water accounts for most of the structure's mass. The degree of swelling is mediated by a balance of the force associated with solvent mixing with the retractive force of the polymer chains, and is dependent on the amount of cross-linking and the solvent conditions [88], [89], [90]. ‘Smart’ hydrogels can be designed to respond to environmental stimuli, such as chemicals, temperature, pH, electrical signals and light [91], which has stirred much interest in their use as biomaterials [92]. Consequently, a variety of applications have been demonstrated with environmentally responsive hydrogels, including chemical sensors [93], microfluidic actuators [94], microlenses [95], [96] and controlled-drug-release systems [97], [98], [99]. The incorporation of DNA into hydrogels has also led to novel systems that exhibit a bulk response triggered by the presence of target ssDNA [100], [101], [102], [103].
DNA-based hydrogels are often constructed by first chemically modifying the terminus of ssDNA with a polymerisable group (such as an acryl group) and then co-polymerising with other monomers. Co-polymerisation of single-terminus modified ssDNA results in grafted polymers, in which the ssDNA can be treated as a functional side-chain. Langrana and co-workers have used grafted ssDNA as a means to control the cross-linking of polyacrylamide [100], [101]. Two different sequences (SA1 and SA2) were modified with Acrydite (Integrated DNA Technologies, Inc.) at their 5′ ends and co-polymerised separately with acrylamide. A target sequence, L2, was designed to be partially complementary to SA1 and SA2. When L2 was added to the mixture of the polymer batches, it was able to cross-link adjacent SA1 and SA2 polymer chains. It was found that gelation occurs above a critical target concentration of L2. Furthermore, addition of a removal strand, R1, that could hybridise completely with L2 was shown to disrupt the cross-linking and return the gel to a liquid state, demonstrating reversible transitions that do not require heating or chemical species that break covalent bonds (Fig. 2C).
Similarly, Mi and co-workers demonstrated a reversible sol–gel transition based on a thrombin-binding aptamer [102]. Acrylamide was co-polymerised with Acrydite-modified ssDNA sequences G1 and G2, separately. Strand A is designed to be partially complementary to G1 and G2 while also self-hybridising to form a tertiary structure with strong binding affinity for thrombin. The aptamer–thrombin complex acted as a cross-linker by hybridising with G1 and G2 from adjacent polymers. The cross-linking is reversible by adding another strand, D, which is fully complementary to A. When strand D completely hybridises with A, the matrix is dissolved and thrombin is released. This system demonstrated the ability to capture a protein within a hydrogel and release it with an oligonucleotide trigger.
Murakami and Maeda have developed DNA-responsive hydrogels that shrink or swell in the presence of a particular ssDNA sequence [103]. Standard radical polymerisation was used to prepare polyacrylamide hydrogels with 5′- and 3′-methacryloyl-modified ssDNA as the cross-linker. The cross-linking ssDNA then acted as a probe sequence that hybridised with a target sequence. The hydrogels exhibited different shrinking and swelling behaviours for a variety of different target sequences. The resolution was such that even a single-base mismatch produced a characteristic swelling profile, while completely non-complementary sequences did not affect the swelling at all.
3. Branched DNA Structures and Dendrimers
Branched DNA junctions, through the rational design of single-stranded oligomers, have paved the way for intricate DNA-based nanostructures that could not be realised with linear DNA alone. The expanded utility of branched DNA architectures arises from the multiple termini that are available for functionalisation by different moieties. In addition, this allows for further hybridisation between branched DNA subunits resulting in more complex two- and three-dimensional (3D) DNA assemblies.
3.1. Design of Branched DNA Motifs
Seeman and colleagues pioneered the assembly of branched DNA nanostructures that mimic four-armed Holliday junctions that exist transiently in the cell. Holliday junctions are unstable due to the internal sequence symmetry that allows for migration of the branch point during genetic recombination [10]. By designing oligonucleotides with partial complementary sequences and specific sequence-symmetry constraints, Seeman's group assembled the first synthetic four-armed DNA junction [104]. This seminal work has inspired an assortment of novel branched DNA architectures, including various DNA shapes (cubes, octahedrons, hexagons, etc [105], [106], [107].), DNA catenanes and knots [108] and multi-armed branched DNA structures [109], [110], [111]. Early branched DNA junctions, however, suffered from high conformational flexibility, preventing the formation of regular superstructures made from DNA junctions. As a solution, Seeman and co-workers developed more rigid motifs based on the concept of crossover hybridisation. Here, two or three helices of DNA were connected by cross-allocation of one or more strands, resulting in double-crossover (DX), triple-crossover DNA (TX) and paranemic crossover (PX) architectures [112], [113], [114]. Branched DNA molecules have also inspired the construction of more complex structures, such as DNA nanomachines [115].
While a thorough treatment of the potential applications of branched DNA is beyond the scope of this discussion, we focus on recent efforts to incorporate branched architectures into biological systems. Inspired by Seeman's work, we have designed branched DNA structures, namely three- and four-armed branched DNA (‘Y-DNA’ and ‘X-DNA’, respectively), and have successfully utilised them in biological and biomedical applications. In collaboration with the Baird group, trivalent Y-DNA was used as a tool for assessing structural and spatial constraints in receptor-signal initiation [116]. In this study, trivalent Y-DNA functioned as rigid spacers between 2,4-dinitrophenyl (DNP) groups that bind specifically to anti-DNP IgE on RBL mast cells. The Baird group found that cellular degranulation, tyrosine phosphorylation responses and calcium mobilisation were all affected by the length of the Y-DNA spacer used. These results are also a clear demonstration of how DNA can be useful in biology for structural, rather than genetic, purposes.
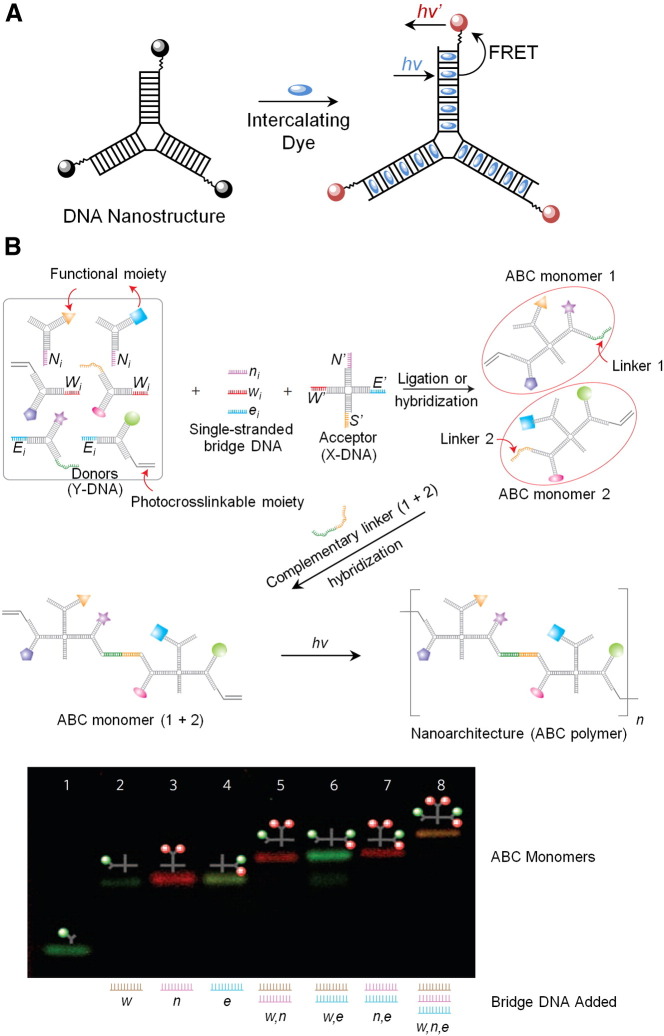
Armitage and co-workers also exploited the structural features of DNA in designing branched fluorescent DNA nanotags [117]. The structural regularity of the DNA double helix provides an ideal environment for packing a high density of fluorescent intercalating dyes with adequate separation to eliminate self-quenching effects (Fig. 3A). In a synergistic fashion, the dyes, in turn, serve to stabilise the branched DNA structures from thermal denaturation. As a result, the fluorescent DNA nanotags generated were stable and exhibited dramatically increased quantum yield from the dye molecules. These nanotags could potentially be used as biological labels in flow cytometry and fluorescence microscopy.
Fig. 3.
Application of branched DNA polymeric topologies for generating fluorescent nanotags and for multiplexed detection. (A) Creation of a fluorescent DNA nanotag by self-assembly of fluorescent intercalating dyes within a branched DNA nanostructure. The regularity of the DNA double-helix spaces out the dyes and eliminates self-quenching effects while dramatically increasing the quantum yield due to the high-density packing of dyes. Acceptor dye molecules can be conjugated on the termini of the branched DNA structures for fluorescence resonance energy transfer (FRET) that allow for wavelength shifting. Reproduced from reference [117] with permission. (B) Assembly of anisotropic branched DNA into multifunctional polymeric structures. Upon UV illumination the monomers assemble into spherical polymeric particles, but only in the presence of a target DNA sequence. Encoding the different monomers with fluorescent labels allows for multiplexed detection. Reproduced from reference [122].
3.2. Dendrimer-Like DNA
In the classification of polymeric topologies, dendrimers are considered a subgroup of the branched topology [14]. Based on molecular recognition properties of multivalent DNA subunits, it is possible to rationally design DNA dendrimers. We first reported the controlled assembly of dendrimer-like DNA (DL-DNA) from the ligation of Y-DNA building blocks to each other [118]. We demonstrated that these stable and monodisperse DL-DNA structures could be produced through a robust and efficient assembly process for up to five dendrimer generations. To create these structures, the Y-DNA building blocks were designed to have non-palindromic cohesive ends to eliminate the occurrence of self-ligations and to ensure uni-directional ligations with each increasing generation. The multiple end termini of the DL-DNA can be attached to various ligands and molecules, and can easily be tailored to be either anisotropic for multivalent signalling or isotropic for signal amplification. These DL-DNA constructions are a clear example that novel functional nanostructures can be assembled with control and precision through the manipulation of DNA.
The properties of these nonlinear DNA structures are being investigated for nanomedicine. Similar to Mirkin's work on DNA biobarcodes (using linear DNA), we developed a version of DNA nanobarcodes based on second generation DL-DNA. By ligating a combination of fluorophores and a molecular probe to the terminal ends of the dendrimer structure, we created nanobarcodes with fluorescent signals unique to a specific probe sequence [119]. We demonstrated that these DNA nanobarcodes could be used for highly efficient multiplexed detection of pathogen DNA (e.g., anthrax, Ebola and the severe acute respiratory syndrome (SARS) viruses) with attomole detection limits in less than a minute. In addition, the terminal ends of DL-DNA can be designed to be isotropic and conjugated to a specific fluorophore to create a highly amplified signal. Single-molecule diffusion studies of DL-DNA have also been performed to investigate its behaviour as a potential drug-delivery material [120], [121].
The modularity of the assembly of DL-DNA allows for different functional moieties to be precisely placed, which enables the construction of building blocks that are both multivalent and anisotropic. This strategy was recently used to assemble anisotropic, branched, and cross-linkable monomeric units (ABC-monomers) for the purpose of sensitive pathogen detection [122]. The monomers consisted of colour combinations of QDs, photo-cross-linkable moieties and sticky ends for ligation to a target sequence. The sticky ends were designed to hybridise with pathogen DNA sequences, which resulted in the formation of dimers. Upon ultraviolet (UV) illumination, the dimers polymerised via their cross-linkable groups, forming spherical particles with unique fluorescent signals. Without the target DNA, polymerisation would not have occurred, making this a novel technique for ‘target-driven’ polymerisation (Fig. 3B).
4. DNA Networks
A network topology can be attained by cross-linking DNA, either through the interaction of the DNA backbone with other materials or by self-assembly of subunits based on sticky-end recognition. These approaches have resulted in films, hydrogels and even 2D periodic lattices, all of which have numerous prospective applications that are beyond the scope of this article. We discuss both approaches to designing DNA networks in this section. As discussed previously, we define DNA hydrogels as swollen networks in which DNA either is the primary structural component or is cross-linked through physical or chemical interactions with other materials.
4.1. Backbone-Mediated Networks
The polyanionic backbone of dsDNA can be cross-linked to form both hydrogels and insoluble films depending on the method of cross-linking. Ethylene glycol diglycidyl ether (EGDE) has been used to covalently cross-link long, highly ionic strands of salmon-sperm DNA [123], [124]. Electrostatically cross-linked DNA networks, on the other hand, rely on the interactions between the anionic phosphate groups of the DNA backbone with cationic materials, such as surfactants, metal ions, or other polymers [125], [126]. The early work of Amiya and Tanaka showed that covalent DNA gels, similarly to many other polyelectrolyte gels, undergo a discontinuous volume transition depending on the solvent conditions [123]. The implications of such studies have made DNA networks worth investigating for biomedical applications [124].
In one study, Nishi and co-workers demonstrated the ability of a DNA hydrogel to encapsulate and protect probiotic bacteria in the harsh conditions associated with oral delivery [127]. Lactic acid bacteria, which is present in yoghurt, was encapsulated in a complex of salmon-milt DNA, gelatin and a polysaccharide extracted from seaweed. The hydrogel was able to protect the bacteria when submerged in a simulated gastrointestinal environment, while also providing conditions that were suitable for bacterial growth. Both the biocompatibility and bioactivity of DNA-based hybrid systems demonstrate its merit as a functional biomaterial. In addition to hydrogels, insoluble films have also been reported. One such film was formed through complexation of DNA with cationic metal ions [126]. The complexes were found to be stable and the metal ions remained functional, making these films particularly interesting as antibacterial and biosensing materials. DNA-alginate films have also been reported, in which the film was used as a filter to remove toxic materials upon intercalation with the DNA backbone [128]. In addition, the films were also used in a solid-phase assay as an antigen to detect an anti-DNA antibody [129].
4.2. Networks Formed via Subunit Self-Assembly
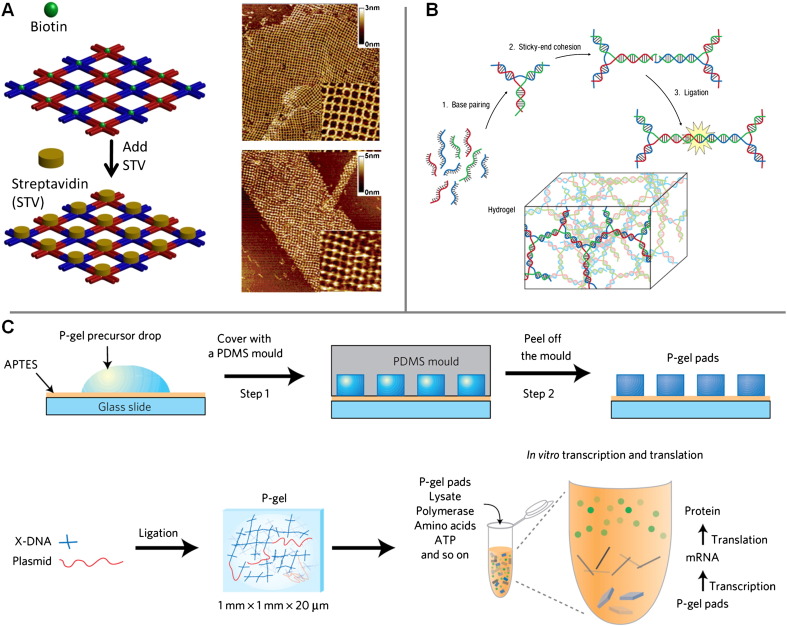
What makes DNA networks particularly fascinating is that one can engineer branched monomers that form highly ordered 2D arrays as well as 3D hydrogels – a morphological versatility that cannot be realised with conventional polymers. Self-assembled 2D crystals have been demonstrated with a variety of different branched DNA motifs, including crosses, three-point stars and triangular junctions, resulting in supramolecular structures with interesting tuneable tiling patterns [113], [130], [131], [132], [133], [134] (Fig. 4A). Two-dimensional lattices have displayed much organisational control over positioning of nanomaterials [135]. This has also been demonstrated with biomolecules, as aptamer sequences have been incorporated into rationally designed DNA architectures to produce periodically spaced proteins [136], [137]. Programmable protein arrays may allow for studies of highly controlled distance-based macromolecular interactions, which could prove useful for drug design.
Fig. 4.
Application of networked DNA polymeric topology for protein arrays and protein production. (A) Schematic showing a self-assembled DNA scaffold from branched DNA tiles containing biotin and the subsequent binding of streptavidin into an ordered protein array. AFM images corresponding to each step are shown on the right. Adapted from reference [137] with permission. (B) Assembly of a DNA hydrogel from branched DNA building blocks by ligation of sticky-ends. Reproduced from reference [139] with permission. (C) Fabrication of protein-producing DNA hydrogels (P-gel) in a micromould format. The P-gels contains multiple copies of a gene that are expressed through transcription and translation in the presence of cell lysate. Reproduced from reference [140].
In addition, amorphous hydrogels have been assembled from branched DNA subunits. We successfully demonstrated the first DNA hydrogel whose assembly is governed by sticky-end recognition [138] (Fig. 4B). Using the same rationale behind DL-DNA, branched DNA-building blocks were designed so that all arms had identical palindromic sticky ends. The sticky ends were then ligated to form a 3D hydrogel network. Without CpG repeats within the sequence of the DNA motifs, the DNA hydrogel is naturally biocompatible, biodegradable and non-immunogenic, making it a promising candidate for drug delivery and tissue engineering [139]. In particular, it was observed that two drugs, porcine insulin and Camptothecin, after encapsulation in the DNA hydrogel, could be released in a controlled manner over time. Live mammalian cells, such as Chinese Hamster Ovarian (CHO) cells, could also be encapsulated in the gel and remain alive for up to 3 days later.
Recently, the DNA hydrogel was adapted into a cell-free, protein-producing hydrogel system (P-gel) [140]. DNA subunits were once again enzymatically cross-linked, but this time in the presence of protein-encoding DNA with palindromic sticky ends. This resulted in a DNA hydrogel with genes covalently linked within the matrix. With the addition of cell lysates, P-gel was able to produce proteins with higher yield and production efficiency (up to 300 times more efficient) than conventional cell-free protein-synthesis systems. Furthermore, P-gel was utilised to produce a variety of proteins, including reporter proteins, membrane proteins, kinases, protein hormones and toxic proteins, all with comparable efficiency and yield. The substantial improvement in protein production by P-gel was attributed to factors such as enhanced transcription, a higher surface-to-volume ratio and an improved protection against degradation of the gene. For these reasons, P-gel could be potentially used in high-throughput protein engineering or as an implantable on-site protein-expression system in the future.
5. Future Outlook
The field of DNA nanomedicine is rapidly growing with the realisation that DNA can be as good a generic material as it is a genetic material. This paradigm shift has opened up a plethora of options for manipulating and assembling DNA into active constructions. The majority of these are individually or collectively engineered from branched and linear DNA topologies. Some of the notable developments include DNAzymes, DNA switches, DNA tweezers as well as DNA walkers [141], [142], [143], [144]. Breaker and Joyce pioneered the DNAzyme and demonstrated that DNA could be used as an enzyme to cleave RNA. Turberfield and co-workers developed the first DNA tweezers based on the hybridisation of linear DNA in a simple yet elegant process. The use of DNA hybridisation to induce changes in the state of DNA nanodevices is common in many of the other DNA molecular systems and machines that have been assembled. The design of DNA nanoactuators, DNA gears and DNA nanomotors, coupled with the use of DNA as a logical molecular computer, make the future of autonomous DNA nanomachines in nanomedicine an approaching reality [115], [145], [146], [147], [148], [149], [150], [151], [152].
There exist structural, functional and biological challenges that must be overcome before many of these DNA structures will play a practical role in nanomedicine. A fundamental requirement of nanoscale building materials is that they must be structurally durable, nontoxic and biochemically stable [153]. However, the issue of DNA immunogenicity in humans still remains controversial. Although ssDNA were reported to elicit antibodies, dsDNA, such as plasmid DNA, has been reported to be only weakly immunogenic in humans [154], [155]. From a chemical stability perspective, natural DNA is prone to degradation in serum by endonucleases and exonucleases. In the case of DNA aptamers, a short serum circulation life in vivo results from both nuclease degradation and rapid renal clearance due to small molecular size (< 40 kDa). However, appropriate chemical modifications, such as methylation and pegylation, could provide protection from degradation and increase circulation time substantially. In general, the interactions between living cells and synthetic nanomaterials have yet to be fully understood. Hence, more empirical data is needed regarding the survival of DNA motifs in a cellular environment before such nanostructures could be applied in vivo.
Directed self-assembly of DNA subunits into functional nanostructures still remains a non-trivial task. The interactions between subunits are particularly important in the formation of more complex structures. For example, a delicate balance between stress and flexibility of individual subunits is necessary for the formation of 2D lattices [156]. In contrast with 2D DNA networks, which generally require high rigidity, flexibility within DNA motifs is favourable for producing amorphous, isotropic 3D gels. However, the bulk behaviour of DNA networks that comprise of monomeric subunits is difficult to predict. Nevertheless, numerical models for the self-assembly of branched and networked DNA structures could be helpful in the design process. One such study showed that gelation is highly dependent on temperature and diffusivity [157], indicating that it may be possible to optimise the conditions for which the macroscale transitions of DNA nanoarchitectures are controllable. A greater understanding of bulk behaviour of nanoscale subunits will be necessary for the development of more complex architectures in the future.
Despite these challenges, substantial progress has been made over the last decade in developing novel DNA nanoarchitectures. In this article, we have proposed a contextual framework by which DNA architectures can be classified based on their polymeric topologies. Each of these topologies possesses functional characteristics that gear it towards a particular system, demonstrating the versatility of DNA that make it an ideal candidate for the wide range of applications in nanomedicine. We are confident that current research endeavours will continue to steer progress in DNA nanomedicine towards more real-world applications.
Footnotes
This review is part of the Advanced Drug Delivery Reviews theme issue on “From Biology to Materials: Engineering DNA and RNA for Drug Delivery and Nanomedicine”.
References
- 1.Luo D. Controlled DNA delivery systems. Pharm. Res. 1999;16:1300–1308. doi: 10.1023/a:1014870102295. [DOI] [PubMed] [Google Scholar]
- 2.Luo D., Saltzman W.M. Synthetic DNA delivery systems. Nat. Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 3.Luo D. Nanotechnology and DNA delivery. MRS Bull. 2005;30:654–658. [Google Scholar]
- 4.Putnam D. Polymers for gene delivery across length scales. Nat. Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 5.Smith L.C. Synthetic peptide-based DNA complexes for nonviral gene delivery. Adv. Drug Deliv. Rev. 1998;30:115–131. doi: 10.1016/s0169-409x(97)00111-7. [DOI] [PubMed] [Google Scholar]
- 6.Vijayanathan V., Thomas T., Thomas T.J. DNA nanoparticles and development of DNA delivery vehicles for gene therapy. Biochemistry. 2002;41:14085–14094. doi: 10.1021/bi0203987. [DOI] [PubMed] [Google Scholar]
- 7.Pedroso de Lima M.C. Cationic lipid-DNA complexes in gene delivery: from biophysics to biological applications. Adv. Drug Deliv. Rev. 2001;47:277–294. doi: 10.1016/s0169-409x(01)00110-7. [DOI] [PubMed] [Google Scholar]
- 8.Nayak S., Lyon L.A. Soft nanotechnology with soft nanoparticles. Angew. Chem. Int. Ed. 2005;44:7686–7708. doi: 10.1002/anie.200501321. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y.C. Long-term in vivo gene expression via delivery of PEI-DNA condensates from porous polymer scaffolds. Hum. Gene Ther. 2005;16:609–617. doi: 10.1089/hum.2005.16.609. [DOI] [PubMed] [Google Scholar]
- 10.Seeman N.C. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 11.Hagerman P.J. Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 12.Luo D. The road from biology to materials. Mater. Today. 2003;6:38–43. [Google Scholar]
- 13.Roberts R.J. REBASE - restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu L.Y., Bae Y.H. Polymer architecture and drug delivery. Pharm. Res. 2006;23:1–30. doi: 10.1007/s11095-005-9046-2. [DOI] [PubMed] [Google Scholar]
- 15.Freitas R.A., Jr What is nanomedicine? Nanomedicine Nanotechnol. Biol. Med. 2005;1:2–9. [Google Scholar]
- 16.Freitas R.A., Jr Current status of nanomedicine and medical nanorobotics. J. Comput. Theor. Nanosci. 2005;2:1–25. [Google Scholar]
- 17.Seeman N.C. DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct. 1998;27:225–248. doi: 10.1146/annurev.biophys.27.1.225. [DOI] [PubMed] [Google Scholar]
- 18.Seeman N.C. At the crossroads of chemistry, biology, and materials: structural DNA nanotechnology. Chem. Biol. 2003;10:1151–1159. doi: 10.1016/j.chembiol.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Seeman N.C. From genes to machines: DNA nanomechanical devices. Trends Biochem. Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldkamp U., Niemeyer C.M. Rational design of DNA nanoarchitectures. Angew. Chem. Int. Ed. 2006;45:1856–1876. doi: 10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]
- 21.Liu X.D. Functional Materials Derived from DNA. Adv. Polym. Sci. 2007;209:149–178. [Google Scholar]
- 22.LaBean T.H., Li H.Y. Constructing novel materials with DNA. Nano Today. 2007;2:26–35. [Google Scholar]
- 23.Liedl T., Sobey T.L., Simmel F.C. DNA-based nanodevices. Nano Today. 2007;2:36–41. [Google Scholar]
- 24.Bath J., Turberfield A.J. DNA nanomachines. Nat. Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 25.Seeman N.C. An overview of structural DNA Nanotechnology. Mol. Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elghanian R. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 27.Rosi N.L. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 28.Stoeva S.I. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew. Chem. Int. Ed. 2006;45:3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 29.Minard-Basquin C. Gold-nanoparticle-assisted oligonucleotide immobilisation for improved DNA detection. IEE Proc.-Nanobiotechnol. 2005;152:97–103. doi: 10.1049/ip-nbt:20055019. [DOI] [PubMed] [Google Scholar]
- 30.Wang L.X. Progress in the delivery of therapeutic oligonucleotides: organ/cellular distribution and targeted delivery of oligonucleotides in vivo. Antisense Nucleic Acid Drug Dev. 2003;13:169–189. doi: 10.1089/108729003768247637. [DOI] [PubMed] [Google Scholar]
- 31.Giljohann D.A. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirkin C.A. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 33.Thaxton C.S., Georganopoulou D.G., Mirkin C.A. Gold nanoparticle probes for the detection of nucleic acid targets. Clin. Chim. Acta. 2006;363:120–126. doi: 10.1016/j.cccn.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 34.Storhoff J.J. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 1998;120:1959–1964. [Google Scholar]
- 35.Lee J.S., Han M.S., Mirkin C.A. Colorimetric detection of mercuric ion (Hg2+) in aqueous media using DNA-functionalized gold nanoparticles. Angew. Chem. Int. Ed. 2007;46:4093–4096. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- 36.Lee J.S. A DNA-Gold nanoparticle-based colorimetric competition assay for the detection of cysteine. Nano Lett. 2008;8:529–533. doi: 10.1021/nl0727563. [DOI] [PubMed] [Google Scholar]
- 37.Sato K., Hosokawa K., Maeda M. Colorimetric biosensors based on DNA-nanoparticle conjugates. Anal. Sci. 2007;23:17–20. doi: 10.2116/analsci.23.17. [DOI] [PubMed] [Google Scholar]
- 38.Taton T.A., Mirkin C.A., Letsinger R.L. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 39.Taton T.A., Lu G., Mirkin C.A. Two-color labeling of oligonucleotide arrays via size-selective scattering of nanoparticle probes. J. Am. Chem. Soc. 2001;123:5164–5165. doi: 10.1021/ja0102639. [DOI] [PubMed] [Google Scholar]
- 40.Cao Y.C. Raman dye-labeled nanoparticle probes for proteins. J. Am. Chem. Soc. 2003;125:14676–14677. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- 41.Cao Y.W.C., Jin R.C., Mirkin C.A. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 42.Park S.J., Taton T.A., Mirkin C.A. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 43.Bailey R.C. Real-time multicolor DNA detection with chemoresponsive diffraction gratings and nanoparticle probes. J. Am. Chem. Soc. 2003;125:13541–13547. doi: 10.1021/ja035479k. [DOI] [PubMed] [Google Scholar]
- 44.Nam J.M., Park S.J., Mirkin C.A. Bio-barcodes based on oligonucleotide-modified nanoparticles. J. Am. Chem. Soc. 2002;124:3820–3821. doi: 10.1021/ja0178766. [DOI] [PubMed] [Google Scholar]
- 45.Nam J.M., Thaxton C.S., Mirkin C.A. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 46.Stoeva S.I. Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. J. Am. Chem. Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 47.Hill H.D., Mirkin C.A. The bio-barcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange. Nat. Protoc. 2006;1:324–336. doi: 10.1038/nprot.2006.51. [DOI] [PubMed] [Google Scholar]
- 48.Goluch E.D. A bio-barcode assay for on-chip attomolar-sensitivity protein detection. Lab Chip. 2006;6:1293–1299. doi: 10.1039/b606294f. [DOI] [PubMed] [Google Scholar]
- 49.Thaxton C.S. A bio-bar-code assay based upon dithiothreitol-induced oligonucleotide release. Anal. Chem. 2005;77:8174–8178. doi: 10.1021/ac0514265. [DOI] [PubMed] [Google Scholar]
- 50.Tang S.X. Nanoparticle-based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. J. Acquir. Immune Defic. Syndr. 2007;46:231–237. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- 51.Liu G.L. A nanoplasmonic molecular ruler for measuring nuclease activity and DNA footprinting. Nat. Nanotechnol. 2006;1:47–52. doi: 10.1038/nnano.2006.51. [DOI] [PubMed] [Google Scholar]
- 52.Storhoff J.J. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat. Biotechnol. 2004;22:883–887. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin W.J., Yung L.Y.L. Nanoparticle-based detection and quantification of DNA with single nucleotide polymorphism (SNP) discrimination selectivity. Nucleic Acids Res. 2007;35:e111. doi: 10.1093/nar/gkm602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niemeyer C.M., Ceyhan B. DNA-directed functionalization of colloidal gold with proteins. Angew. Chem. Int. Ed. 2001;40:3685–3688. doi: 10.1002/1521-3773(20011001)40:19<3685::aid-anie3685>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 55.Hazarika P., Ceyhan B., Niemeyer C.M. Sensitive detection of proteins using difunctional DNA-gold nanoparticles. Small. 2005;1:844–848. doi: 10.1002/smll.200500063. [DOI] [PubMed] [Google Scholar]
- 56.Hazarika P., Kukolka F., Niemeyer C.M. Reversible binding of fluorescent proteins at DNA-gold nanoparticles. Angew. Chem. Int. Ed. 2006;45:6827–6830. doi: 10.1002/anie.200602049. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C.Y. Single-quantum-dot-based DNA nanosensor. Nat. Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 58.Gerion D. Room-temperature single-nucleotide polymorphism and multiallele DNA detection using fluorescent nanocrystals and microarrays. Anal. Chem. 2003;75:4766–4772. doi: 10.1021/ac034482j. [DOI] [PubMed] [Google Scholar]
- 59.Fowler C.C., Li Y.F. Aptamers and small molecules play tug of war. Chem. Biol. 2007;14:736–738. doi: 10.1016/j.chembiol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 60.Fichou Y., Ferec C. The potential of oligonucleotides for therapeutic applications. Trends Biotechnol. 2006;24:563–570. doi: 10.1016/j.tibtech.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Ulrich H. DNA and RNA aptamers: From tools for basic research towards therapeutic applications. Comb. Chem. High Throughput Screen. 2006;9:619–632. doi: 10.2174/138620706778249695. [DOI] [PubMed] [Google Scholar]
- 62.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment - RNA ligands to bacteriophage-T4 DNA-Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 63.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 64.Jayasena S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clinical Chemistry. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 65.Bunka D.H.J., Stockley P.G. Aptamers come of age - at last. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 66.Famulok M., Hartig J.S., Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 67.Drolet D.W. A high throughput platform for systematic evolution of ligands by exponential enrichment. SELEXComb. Chem. High Throughput Screen. 1999;2:271–278. [PubMed] [Google Scholar]
- 68.Drolet D.W. Pharmacokinetics and safety of an anti-vascular endothelial growth factor aptamer (NX1838) following injection into the vitreous humor of rhesus monkeys. Pharm. Res. 2000;17:1503–1510. doi: 10.1023/a:1007657109012. [DOI] [PubMed] [Google Scholar]
- 69.Hicke B.J., Stephens A.W. Escort aptamers: a delivery service for diagnosis and therapy. J. Clin. Investig. 2000;106:923–928. doi: 10.1172/JCI11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Proske D. Aptamers - basic research, drug development, and clinical applications. Appl. Microbiol. Biotechnol. 2005;69:367–374. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- 71.Nimjee S.M., Rusconi C.P., Sullenger B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 72.Cerchia L. Nucleic acid aptamers in cancer medicine. FEBS Lett. 2002;528:12–16. doi: 10.1016/s0014-5793(02)03275-1. [DOI] [PubMed] [Google Scholar]
- 73.DiPaolo J.A., Alvarez-Salas L.M. Advances in the development of therapeutic nucleic acids against cervical cancer. Expert Opin. Biol. Ther. 2004;4:1251–1264. doi: 10.1517/14712598.4.8.1251. [DOI] [PubMed] [Google Scholar]
- 74.Tang Z.W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 75.Farokhzad O.C. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Nat. Acad. Sci. USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bagalkot V. Quantum dot-Aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on Bi-fluorescence resonance energy transfer. Nano Lett. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 77.Ireson C.R., Kelland L.R. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 78.Osborne S.E., Matsumura I., Ellington A.D. Aptamers as therapeutic and diagnostic reagents: problems and prospects. Curr. Opin. Chem. Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 79.Tombelli S., Minunni M., Mascini M. Aptamers-based assays for diagnostics, environmental and food analysis. Biomol. Eng. 2007;24:191–200. doi: 10.1016/j.bioeng.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 80.Drolet D.W., Moon-McDermott L., Romig T.S. An enzyme-linked oligonucleotide assay. Nat. Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- 81.German I., Buchanan D.D., Kennedy R.T. Aptamers as ligands in affinity probe capillary electrophoresis. Anal. Chem. 1998;70:4540–4545. doi: 10.1021/ac980638h. [DOI] [PubMed] [Google Scholar]
- 82.Shangguan D. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Nat. Acad. Sci. U.S.A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shangguan D. Optimization and modifications of aptamers selected from live cancer cell lines. ChemBioChem. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- 84.Shangguan D.H. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin. Chem. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 85.Blank M., Blind M. Aptamers as tools for target validation. Curr. Opin. Chem. Biol. 2005;9:336–342. doi: 10.1016/j.cbpa.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 86.Chen C.K. Complex SELEX against target mixture: Stochastic computer model, simulation, and analysis. Comput. Meth. Programs Biomed. 2007;87:189–200. doi: 10.1016/j.cmpb.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 87.Hoffman A.S. Hydrogels for biomedical applications. Adv. Drug Deliv. Rev. 2002;54:3–12. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- 88.Peppas N.A. Physicochemical foundations and structural design of hydrogels in medicine and biology. Annu. Rev. Biomed. Eng. 2000;2:9–29. doi: 10.1146/annurev.bioeng.2.1.9. [DOI] [PubMed] [Google Scholar]
- 89.Langer R., Peppas N.A. Advances in biomaterials, drug delivery, and bionanotechnology. AIChE J. 2003;49:2990–3006. [Google Scholar]
- 90.Peppas N.A. Hydrogels in biology and medicine: From molecular principles to bionanotechnology. Adv. Mater. 2006;18:1345–1360. [Google Scholar]
- 91.Qiu Y., Park K. Environment-sensitive hydrogels for drug delivery. Adv. Drug Deliv. Rev. 2001;53:321–339. doi: 10.1016/s0169-409x(01)00203-4. [DOI] [PubMed] [Google Scholar]
- 92.Kopeček J. Hydrogel biomaterials: A smart future? Biomaterials. 2007;28:5185–5192. doi: 10.1016/j.biomaterials.2007.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holtz J.H., Asher S.A. Polymerized colloidal crystal hydrogel films as intelligent chemical sensing materials. Nature. 1997;389:829–832. doi: 10.1038/39834. [DOI] [PubMed] [Google Scholar]
- 94.Beebe D.J. Functional hydrogel structures for autonomous flow control inside microfluidic channels. Nature. 2000;404:588–590. doi: 10.1038/35007047. [DOI] [PubMed] [Google Scholar]
- 95.Kim J., Nayak S., Lyon L.A. Bioresponsive hydrogel microlenses. J. Am. Chem. Soc. 2005;127:9588–9592. doi: 10.1021/ja0519076. [DOI] [PubMed] [Google Scholar]
- 96.Dong L. Adaptive liquid microlenses activated by stimuli-responsive hydrogels. Nature. 2006;442:551–554. doi: 10.1038/nature05024. [DOI] [PubMed] [Google Scholar]
- 97.Kwon I.C., Bae Y.H., Kim S.W. Electrically erodible polymer gel for controlled release of drugs. Nature. 1991;354:291–293. doi: 10.1038/354291a0. [DOI] [PubMed] [Google Scholar]
- 98.Lowman A.M. Oral delivery of insulin using pH-responsive complexation gels. J. Pharm. Sci. 1999;88:933–937. doi: 10.1021/js980337n. [DOI] [PubMed] [Google Scholar]
- 99.Ichikawa H., Peppas N.A. Novel complexation hydrogels for oral peptide delivery: In vitro evaluation of their cytocompatibility and insulin-transport enhancing effects using Caco-2 cell monolayers. J. Biomed. Mater. Res. A. 2003;67A:609–617. doi: 10.1002/jbm.a.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin D.C., Yurke B., Langrana N.A. Mechanical properties of a reversible. DNA-crosslinked polyacrylamide hydrogel. J. Biomech. Eng. 2004;126:104–110. doi: 10.1115/1.1645529. [DOI] [PubMed] [Google Scholar]
- 101.Lin D.C., Yurke B., Langrana N.A. Inducing reversible stiffness changes in DNA-crosslinked gels. J. Mater. Res. 2005;20:1456–1464. [Google Scholar]
- 102.Wei B. Capture and Release of Protein by a Reversible DNA-Induced Sol-Gel Transition System. Angew. Chem. Int. Ed. 2007;47:331–333. doi: 10.1002/anie.200704143. [DOI] [PubMed] [Google Scholar]
- 103.Murakami Y., Maeda M. DNA-responsive hydrogels that can shrink or swell. Biomacromolecules. 2005;6:2927–2929. doi: 10.1021/bm0504330. [DOI] [PubMed] [Google Scholar]
- 104.Kallenbach N.R., Ma R.I., Seeman N.C. An immobile nucleic acid junction constructed from oligonucleotides. Nature. 1983;305:829–831. [Google Scholar]
- 105.Chen J.H., Seeman N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 106.Zhang Y.W., Seeman N.C. Construction of a DNA-truncated octahedron. J. Am. Chem. Soc. 1994;116:1661–1669. [Google Scholar]
- 107.Aldaye F.A., Sleiman H.F. Sequential self-assembly of a DNA hexagon as a template for the organization of gold nanoparticles. Angew. Chem. Int. Ed. 2006;45:2204–2209. doi: 10.1002/anie.200502481. [DOI] [PubMed] [Google Scholar]
- 108.Seeman N.C. Synthetic DNA knots and catenanes. New J. Chem. 1993;17:739–755. [Google Scholar]
- 109.Ma R.I. 3-Arm nucleic acid junctions are flexible. Nucleic Acids Res. 1986;14:9745–9753. doi: 10.1093/nar/14.24.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y.L. Assembly and characterization of 5-arm and 6-arm DNA branched junctions. Biochemistry. 1991;30:5667–5674. doi: 10.1021/bi00237a005. [DOI] [PubMed] [Google Scholar]
- 111.Wang X., Seeman N.C. Assembly and characterization of 8-arm and 12-arm DNA branched junctions. J. Am. Chem. Soc. 2007;129:8169–8176. doi: 10.1021/ja0693441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fu T.J., Seeman N.C. DNA double-crossover molecules. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 113.LaBean T.H. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc. 2000;122:1848–1860. [Google Scholar]
- 114.Shen Z.Y. Paranemic crossover DNA: A generalized Holliday structure with applications in nanotechnology. J. Am. Chem. Soc. 2004;126:1666–1674. doi: 10.1021/ja038381e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu H., Liu D.S. DNA nanomachines and their functional evolution. Chem. Commun. 2009:2625–2636. doi: 10.1039/b822719e. [DOI] [PubMed] [Google Scholar]
- 116.Sil D. Trivalent ligands with rigid DNA spacers reveal structural requirements for IgE receptor signaling in RBL mast cells. ACS Chem. Biol. 2007;2:674–684. doi: 10.1021/cb7001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Benvin A. Fluorescent DNA nanotags: Supramolecular fluorescent labels based on intercalating dye arrays assembled on nanostructured DNA templates. J. Am. Chem. Soc. 2007;129:2025–2034. doi: 10.1021/ja066354t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Y. Controlled assembly of dendrimer-like DNA. Nat. Mater. 2004;3:38–42. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]
- 119.Li Y.G., Cu Y.T.H., Luo D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat. Biotechnol. 2005;23:885–889. doi: 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Luo D. A dendrimer-like DNA-based vector for DNA delivery: a viral and nonviral hybrid approach. Meth. Mol. Med. 2006;127:115–125. doi: 10.1385/1-59745-168-1:115. [DOI] [PubMed] [Google Scholar]
- 121.Freedman K.O. Diffusion of single star-branched dendrimer-like DNA. J. Phys. Chem. B. 2005;109:9839–9842. doi: 10.1021/jp0444924. [DOI] [PubMed] [Google Scholar]
- 122.Lee J.B. Multifunctional nanoarchitectures from DNA-based ABC monomers. Nat. Nanotechnol. 2009;4:430–436. doi: 10.1038/nnano.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Amiya T., Tanaka T. Phase-Transitions in Cross-Linked Gels of Natural Polymers. Macromolecules. 1987;20:1162–1164. [Google Scholar]
- 124.Costa D., Miguel M.G., Lindman B. Effect of additives on swelling of covalent DNA gels. J. Phys. Chem. B. 2007;111:8444–8452. doi: 10.1021/jp067917q. [DOI] [PubMed] [Google Scholar]
- 125.Costa D. Interaction between covalent DNA gels and a cationic surfactant. Biomacromolecules. 2006;7:1090–1095. doi: 10.1021/bm0508919. [DOI] [PubMed] [Google Scholar]
- 126.Yamada M. Preparation of novel bio-matrix by the complexation of DNA and metal ions. Polymer. 2005;46:10102–10112. [Google Scholar]
- 127.Jonganurakkun B. Survival of lactic acid bacteria in simulated gastrointestinal juice protected by a DNA-based complex gel. J. Biomater. Sci. Polym. Ed. 2003;14:1269–1281. doi: 10.1163/156856203322553482. [DOI] [PubMed] [Google Scholar]
- 128.Iwata K. Utilization of DNA as functional materials: Preparation of filters containing DNA insolubilized with alginic acid gel. Int. J. Biol. Macromol. 1996;18:149–150. doi: 10.1016/0141-8130(95)01073-4. [DOI] [PubMed] [Google Scholar]
- 129.Kitamura H. DNA-alginate complex recognized by autoantibodies against DNA. Int. J. Biol. Macromol. 1997;20:75–77. doi: 10.1016/s0141-8130(97)01146-x. [DOI] [PubMed] [Google Scholar]
- 130.Qi J. Ligation of triangles built from bulged 3-arm DNA branched junctions. J. Am. Chem. Soc. 1996;118:6121–6130. [Google Scholar]
- 131.Winfree E. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 132.Mao C.D., Sun W.Q., Seeman N.C. Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy. J. Am. Chem. Soc. 1999;121:5437–5443. [Google Scholar]
- 133.Liu D. Tensegrity: Construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am. Chem. Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 134.Chelyapov N. DNA triangles and self-assembled hexagonal tilings. J. Am. Chem. Soc. 2004;126:13924–13925. doi: 10.1021/ja0458120. [DOI] [PubMed] [Google Scholar]
- 135.Zheng J.W. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu Y. Protein nanoarrays - Aptamer-directed self-assembly of protein arrays on a DNA nanostructure. Angew. Chem. Int. Ed. 2005;44:4333–4338. doi: 10.1002/anie.200501089. [DOI] [PubMed] [Google Scholar]
- 137.Park S.H. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 2005;5:729–733. doi: 10.1021/nl050175c. [DOI] [PubMed] [Google Scholar]
- 138.Um S.H. Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 2006;5:797–801. doi: 10.1038/nmat1741. [DOI] [PubMed] [Google Scholar]
- 139.LaBean T. Hydrogels: DNA bulks up. Nat. Mater. 2006;5:767–768. doi: 10.1038/nmat1745. [DOI] [PubMed] [Google Scholar]
- 140.Park N. A cell-free protein-producing gel. Nat. Mater. 2009;8:432–437. doi: 10.1038/nmat2419. [DOI] [PubMed] [Google Scholar]
- 141.Breaker R.R., Joyce G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 142.Mao C.D. A nanomechanical device based on the B-Z transition of DNA. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 143.Yurke B. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 144.Sherman W.B., Seeman N.C. A precisely controlled DNA biped walking device. Nano Lett. 2004;4:1203–1207. [Google Scholar]
- 145.Simmel F.C., Yurke B. Using DNA to construct and power a nanoactuator. Phys. Rev. E. 2001;63:041913. doi: 10.1103/PhysRevE.63.041913. [DOI] [PubMed] [Google Scholar]
- 146.Li J.W.J., Tan W.H. A single DNA molecule nanomotor. Nano Lett. 2002;2:315–318. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Feng L.P. A two-state DNA lattice switched by DNA nanoactuator. Angew. Chem. Int. Ed. 2003;42:4342–4346. doi: 10.1002/anie.200351818. [DOI] [PubMed] [Google Scholar]
- 148.Liu D.S., Balasubramanian S. A proton-fuelled DNA nanomachine. Angew. Chem. Int. Ed. 2003;42:5734–5736. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 149.Turberfield A.J. DNA fuel for free-running nanomachines. Phys. Rev. Lett. 2003;90:118101. doi: 10.1103/PhysRevLett.90.118102. [DOI] [PubMed] [Google Scholar]
- 150.Tian Y., Mao C.D. Molecular gears: A pair of DNA circles continuously rolls against each other. J. Am. Chem. Soc. 2004;126:11410–11411. doi: 10.1021/ja046507h. [DOI] [PubMed] [Google Scholar]
- 151.Seelig G., Yurke B., Winfree E. Catalyzed relaxation of a metastable DNA fuel. J. Am. Chem. Soc. 2006;128:12211–12220. doi: 10.1021/ja0635635. [DOI] [PubMed] [Google Scholar]
- 152.Benenson Y. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 153.Mustelin T. Challenges and optimism for nanoengineering. Nanomedicine. 2006;1:383–385. doi: 10.2217/17435889.1.4.383. [DOI] [PubMed] [Google Scholar]
- 154.Donnelly J.J. DNA vaccines. Annu. Rev. Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 155.Donnelly J.J., Wahren B., Liu M.A. DNA vaccines: Progress and challenges. J. Immunol. 2005;175:633–639. doi: 10.4049/jimmunol.175.2.633. [DOI] [PubMed] [Google Scholar]
- 156.He Y., Mao C.D. Balancing flexibility and stress in DNA nanostructures. Chem. Commun. 2006:968–969. doi: 10.1039/b513962g. [DOI] [PubMed] [Google Scholar]
- 157.Largo J., Starr F.W., Sciortino F. Self-assembling DNA dendrimers: a numerical study. Langmuir. 2007;23:5896–5905. doi: 10.1021/la063036z. [DOI] [PubMed] [Google Scholar]