Abstract
Cutaneous leishmaniasis is a neglected tropical disease caused by the parasite Leishmania and transmitted by sandflies. It has become a major health problem in many tropical and subtropical countries, especially in regions of conflict and political instability. Currently, there are only limited drug treatments and no available licensed vaccine; thus, the need for more therapeutic interventions remains urgent. Previously, a DNA vaccine encoding a 15 kDa sandfly (Phlebotomus papatasi) salivary protein (PpSP15) and recombinant nonpathogenic Leishmania tarentolae secreting PpSP15 have been shown to induce protective immunity against Leishmania major in mice, demonstrating that PpSP15 is a promising vaccine candidate. In this study, we developed a fermentation process in yeast with a yield of ~1g PpSP15/L and a scalable purification process consisting of only 2 chromatographic purification steps with high binding capacity for PpSP15, suggesting that PpSP15 can be produced economically. The biophysical/biochemical analysis of the purified PpSP15 indicated that the protein was of high purity (>97%) and conformationally stable between pH 4.4 and 9.0. More importantly, the recombinant protein had a defined structure similar to that of the related PdSP15 from Phlebotomus duboscqi, implying the suitability of the yeast expression system for producing a correctly folded PpSP15.
Keywords: chromatography, biopharmaceutical characterization, analytical biochemistry, biotechnology, vaccine(s), protein(s)
Abbreviations used: CL, cutaneous leishmaniasis; PpSP15, 15 kDa sandfly (P. papatasi) salivary protein; PdSP15, 15 kDa sandfly (P. duboscqi) salivary protein; OD, optical density; DO, dissolved oxygen; BH, bed height; ID, internal diameter; CV, column volume
Introduction
Leishmaniasis is a neglected tropical disease caused by protozoan parasites of the genus Leishmania and transmitted to humans by phlebotomine sandflies. There are approximately 2 million new cases each year1 and 70,000 annual deaths from leishmaniasis,2 and in addition, the infection accrues 3.3 million disability-adjusted life years annually.1 Alarmingly, approximately 350 million people living in endemic areas are at risk. Cutaneous Leishmaniasis (CL) is the most common manifestation, with the highest prevalence reported in Afghanistan, Iran, Syria, Algeria, Brazil, and Colombia.3 Some of the highest rates of CL now occur in areas of political unrest and conflict.4 Lesions caused by CL can leave severe, disfiguring scars often causing social stigmatization.5 Indeed, emerging evidence indicates that up to 40 million people may be affected by the chronic scarring of CL,6 of whom a significant percentage suffer comorbid depression and other psychiatric sequelae.7 Therefore, current global burden of disease estimates likely fail to capture the full range of human disease and its humanitarian impact.
To obtain blood meals, sandflies inject salivary proteins to counteract hemostasis.8 During this process, parasites, if present, are transmitted to the human host. Some of these salivary proteins are immunogenic in humans, canids, and mice.9, 10, 11, 12 Repeated exposure to sandfly salivary gland homogenate or sandfly bites has been shown the cause delayed-type hypersensitivity to salivary components, which has been shown to impede the ability of the parasite to infect its host, yielding protection against infection.13 , 14 Similar protective effects from insect salivary proteins are also seen in other vector-borne diseases; for example, animals pre-exposed to tick bites were protected against Borrelia infection15 and mice pre-exposed to mosquito bites were protected against malaria.16 In addition, animals vaccinated with the respective vector’s salivary proteins also were shown to be protected against encephalitis virus,17 Mycobacterium ulcerans infection,18 and dengue virus.19 Such findings have prompted efforts to assess host-vector interactions as a potential vaccine strategy.
Our collaborators at the National Institutes of Health, NIAID, Laboratory for Malaria and Vector Research have recently identified a 15 kDa salivary protein from Phlebotomus papatasi, PpSP15, as a promising vaccine antigen candidate against leishmaniasis.20, 21, 22 They discovered that wild-type and B-cell-deficient mice immunized with PpSP15 encoding DNA vaccine mounted an efficacious cellular immune response against Leishmania major infection20 and confirmed that this protection was due to the development of a delayed-type hypersensitivity response to PpSP15.20 , 23 In addition, as an alternative vaccine strategy, they genetically modified nonpathogenic Leishmania tarentolae parasites to secret PpSP15 and then demonstrated that L. tarentolae expressing PpSP15 boosted the desired immune response and led to protection against L. major in a mouse model.22 In collaboration with the National Institutes of Health and the Uniformed Services University of the Health Sciences, we are now developing a leishmaniasis vaccine consisting of the sandfly antigen, PpSP15, and a well-characterized Leishmania parasite antigen, LdNH36.24 , 25 Although the immunogenicity and efficacy studies of PpSP15 are ongoing, in parallel, we have been developing a scalable production process for this protein, thus allowing its rapid advancement to the clinic. During molecular cloning, the Escherichia coli expression platform was first explored since a protein homolog with close to 70% sequence similarity, PdSP15, from Phlebotomus duboscqi, had already been expressed in E. coli and then crystallized.26 However, PpSP15 formed inclusion bodies in bacteria. Thus, an expression platform using Pichia pastoris X33 was investigated.
In this study, we developed a production process to express and purify tag-free recombinant PpSP15 in yeast (P. pastoris), which included improving the fermentation yield and developing a scalable purification scheme. At the conclusion of these studies, the fermentation yield was improved from 779 mg PpSP15 per liter of fermentation supernatant (FS) to 994 mg, an improvement of 27.6%. The purification process comprised only 2 chromatography steps, ion-exchange chromatography (IEX) using SP Sepharose XL resin (SPXL) and hydrophobic interaction chromatography (HIC) with Phenyl Sepharose 6 Fast Flow high sub resin (PS6FF). The dynamic binding capacities for IEX and HIC were determined as 52 mg/mL and 15 mg/mL, respectively. This purification process increased PpSP15 purity from 64.6% to 97.2%.
Subsequently, biophysical and biochemical analyses demonstrated that the protein was not only highly pure (>97%) and monomeric, but also maintained a well-defined secondary structure (approximately 63% alpha-helix) and tertiary structures similar to that of the related P. duboscqi PdSP15, suggesting the correct folding of PpSP15. pH screening using thermal shift assay also suggested that PpSP15 was stable between pH 4.4 and pH 9.0. Collectively, the data from the process characterization and biophysical and biochemical protein characterization indicate that this process is thus ready for upscaling and reproducibility analysis. The material generated from the scaled-up process will be further assessed for long-term stability and stability under stress.
Materials and Methods
Cloning and Expression of PpSP15
The recombinant expression vector pPICZαA containing the DNA sequence encoding PpSP15 (NCBI protein_id = “AAL11047.1”, GenBank ID: AF335487.1)20 was first purchased from GenScript (Piscataway, NJ), followed by electroporation into P. pastoris X33. Transformants were grown for 3 days at 30°C on yeast extract peptone dextrose agar plates with either 100 or 500 μg/mL Zeocin. Thirteen colonies from each transformation were selected for small-scale (10 mL) expression in BMGY/BMMY medium, with induction with 0.5% methanol per day at 30°C for 3 days. The clone with the highest expression was chosen for generating a research seed stock.
Optimization of the Fermentation Process
One milliliter of seed stock was inoculated into 1 L BMG (buffered minimal glycerol) medium and the culture was incubated overnight at 30°C with constant shaking at 250 rpm until an OD600 of 9.6 was reached. Approximately 180 mL overnight culture were inoculated into 4 L sterile basal salts medium (containing 26.7 mL 85% phosphoric acid, 0.93 g CaSO4.2H2O, 18.2 g K2SO4, 14.9 g MgSO4.7H2O, 4.13 g KOH, 30 g glycerol, 4.35 mL PTM1 Trace Elements, 4.35 mL 0.02% d-Biotin and 1 mL antifoam per liter) to reach to an initial OD600 of 0.4. The yeast cells were expanded overnight in batch mode at 30°C and pH 5 and then, on glycerol depletion, the glycerol fed-batch phase was initiated at 15 mL/L/h of 50% glycerol for 5 h. During the last hour of the glycerol fed-batch phase, the temperature was reduced from 30°C to 26°C and the pH was increased from 5.0 to 5.2. For the induction phase, methanol supply was ramped from 1 to 12 mL/L/h over an 8-h period followed by maintenance at 12 mL/L/h for 21 h. After that, methanol supply was increased from 12 to 14 mL/L/h over a 4-h period followed by maintenance at 14 mL/L/h until the end of the fermentation process. The total induction time was approximately 72 h. During the fermentation process, samples were collected at different time points. The FS was harvested by centrifugation and kept at −80°C for later processing. This optimized process was compared with the original process and the major process differences are described in Supplementary Table S1.
Development of the Purification Process
Two liters of FS were thawed and filtered through a 0.45 μm filter. The filtered FS was further concentrated to approximately 650 mL and buffer exchanged against 100 mM potassium phosphate buffer, pH 6 using a 0.11 m2 3 kDa cutoff Pellicon 3 Ultracel cassette from Millipore (Burlington, MA) until the conductivity equaled that of 100 mM potassium phosphate buffer, pH 6 (~9.01 ms/cm); the processed material at this step was denoted as the tangential flow filtration (TFF) pool.
Because the theoretical pI of PpSP15 is approximately 9.1, cation exchange chromatography using SPXL resin was chosen as the capture step. To determine the binding capacity of the SPXL resin, the TFF pool was loaded onto a 29 mL SP XL column packed in-house (bed height [BH] 14.5 cm × ID 1.6 cm) at 149.3 cm/h with a residence time of 5.8 min, until a breakthrough was observed; the column was then washed with 5 column volumes (CVs) of binding buffer (100 mM potassium phosphate buffer, pH 6). A step elution with 30% of elution buffer (100% elution buffer contains 100 mM potassium phosphate buffer, 1 M NaCl, pH 6) over 5 CVs was applied to elute the target protein, and this collected fraction was denoted as our SPXL pool. A gradient elution from 30% to 100% elution buffer over 5 CVs then removed host cell proteins (HCPs) and other impurities from the column.
An additional polishing step with Phenyl Sepharose 6 Fast Flow high sub resin (PS6FF) was used to remove the remaining impurities. Two hundred mL of SPXL pool were retrieved and mixed with ammonium sulfate until the molarity of ammonium sulfate reached 2 M. Similarly, to study the binding capacity of the PS6FF resin, the SPXL pool with 2 M ammonium sulfate was then loaded onto a 5 mL HiTrap PS6FF column (BH 2.5 cm × ID 1.6 cm; GE Healthcare) at a flow rate of 25.6 cm/h with a residence time of 5.8 min. The column was washed with 10 CVs of binding buffer (100 mM potassium phosphate buffer, 2 M ammonium sulfate, pH 6) to remove any loosely bound protein. A step elution with 50% of elution buffer (100% elution buffer contains 100 mM potassium phosphate buffer, pH 6) over 10 CVs was applied to elute the target protein, and this collected fraction was denoted the PS6FF pool. Followed by a second step elution of 100% elution buffer over 10 CVs, the remaining impurities were removed from the column. The collected PS6FF pool was further buffer-exchanged by dialysis against 1X PBS, pH 7.4, or other buffers, as needed.
Once the binding capacity had been established, the recovery of each step was determined by not overloading the column to prevent any loss of target protein in the flow-through.
Identity and Purity Assessment by SDS-PAGE after Densitometry, Western Blot, or Proteomic Mass Spectrometry
For analysis, the in-process samples and purified PpSP15 were loaded on 14% Tris-glycine or 12% Bis-tris gels, and subsequently stained with Coomassie Blue or silver, as described previously,27 or transferred to PVDF membranes and analyzed by western blot using in-house mouse anti-PpSP15 antisera (1: 2000 dilution), or anti-P. pastoris HCP antibodies (Cygnus, Cat# F145). The Coomassie Blue–stained bands observed on gels overloaded with PpSP15 were then extracted, digested by trypsin and analyzed by mass spectrometry at Yale/NHLBI Proteomics Center to confirm the identity for each band.
Size Assessment by Dynamic Light Scattering
Approximately, 40 μL of 0.5 mg/mL PpSP15 were filtered through a 0.02 μm Anotop 10 filter (Whatman, Maidstone, United Kingdom) to remove any possible environmental contamination and loaded in quadruplicate onto a 384-well clear bottom black plate (Aurora Biotechnologies, Carlsbad, CA). Sample measurements were acquired 10 times over 5 s using a Wyatt DynaPro® plate reader II and analyzed using Dynamics Software package version 7.8 (Wyatt Technology, Santa Barbara, CA).
Aggregation Assessment by Size-Exclusion HPLC (SE-HPLC)
A Waters® HPLC system (Alliance® 2695 Separation Module) equipped with a TSKGel G2000SWxl (Tosoh bioscience, Cat # 0008540) connecting a guard column (Tosoh Bioscience, Cat # 0008543) was used to assess the aggregation of PpSP15. One hundred microliters of 0.68 mg/mL PpSP15 were injected onto the column and the protein was eluted in phosphate buffer (5.6 mM Na2HPO4, 1.1 mM KH2PO4 and 225 mM NaCl at pH 7.4) at 0.5 mL/min over 40 min. Gel filtration standard (Bio-Rad, Cat # 1511901) was used as a control. The eluted protein was monitored at 280 nm.
Structure Assessment and Thermal Stability by Circular Dichroism
Purified PpSP15 was diluted with deionized water to a final concentration of approximately 0.3 mg/mL and analyzed by circular dichroism (CD) on a Jasco-810 spectropolarimeter (Easton, MD). Far-UV CD spectra were obtained from 260 to 190 nm with a Jasco J-1500s spectrophotometer set at 100 nm/min and a response time of 1 s at 25°C. The obtained CD data were analyzed using CDPro software by comparing to the reference set SDP48 using 2 data-fitting programs (CONTIN and CDSSTR). In addition, the sample was heated from 25°C to 90°C to analyze the thermal stability of its secondary structure.
pH Screening by Thermal Shift Assay
Purified PpSP15 was diluted and buffer-exchanged to a final concentration of 0.5 mg/mL in 150 mM sodium citrate at different pH values ranging from 3.1 to 6.0, in citrate-phosphate buffer28 at pH values ranging from 7.2 to 8.0 and 160 mM sodium borate at pH 9.0. Deionized water was included as a negative control. A protein thermal shift assay was then performed by heating the sample from 25°C to 99°C and monitoring the fluorescence intensity change using the Protein Thermal Shift™ Dye kit (Thermo Fisher Scientific, Cat # 4461146) on a ViiA™ 7 Real-Time PCR System, and analyzed with ViiA™ Software v1.2.
Results
Process Characterization
Fermentation Process Optimization
The optimized fermentation process was compared to the default fermentation process and analyzed using biomass by wet cell weight measurement and expression level of PpSP15 by SDS-PAGE (Supplementary Fig. S1 and Supplementary Table S1). The major difference of the optimized process was the addition of a glycerol fed-batch phase to increase the cell mass before induction, thus potentially increasing protein yield. The default process produced 427.3 g of biomass/L of culture and 779 mg of PpSP15/L of FS, whereas the optimized process produced approximately 4% more biomass (442.2 g/L of culture) but 27.6% more PpSP15 (994 mg/L of FS).
Purification Process Characterization
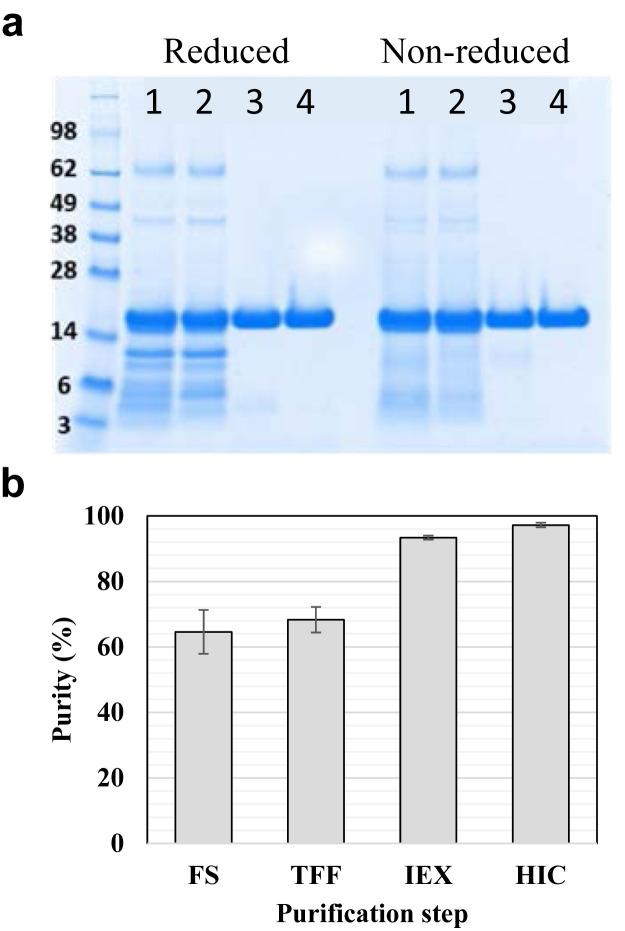
During purification, in-process samples were used to characterize protein purity (Fig. 1 and Table 1 ). The binding capacity of each chromatographic step was also determined, as this information will facilitate upscaling the process. The low purity of PpSP15 (64.6%) in the FS as determined by densitometry (Fig. 1b and Table 1) was likely due to heavy contamination with host cell impurities or degraded PpSP15 (Fig. 1a, lane 1). To purify PpSP15 more efficiently, the FS was first filtered through a 0.45 μm filter and then concentrated 3-fold by TFF (Fig. 1a, lane 2) to reduce the volume and thus processing time. Owing to the high theoretical pI of PpSP15 (pI = 9.1), cation exchange chromatography was chosen, and thus, the concentrated FS was further buffer exchanged using the same filtration device. It is noted that only 3.3 to 4 diavolumes were required to reduce the sample conductivity to the level of binding buffer (100 mM potassium phosphate buffer, pH 6), and 95% of the protein was recovered. After buffer exchange, the FS was loaded onto a cation exchange column (IEX) made from SPXL resin. The binding capacity was determined as 52 mg/mL SPXL at a flow rate of 149.3 cm/h on a column with a BH of 14.5 cm. In the IEX step, 92% of PpSP15 was recovered from the TFF step; the purity of PpSP15 in the SPXL pool increased to 93.4%, suggesting that most of the impurities, that is, host cell proteins, were removed (Fig. 1a, lane 3). The conductivity of the SPXL pool was then adjusted to ~200 ms/cm by adding ammonium sulfate to 2 M and the sample was loaded onto an HIC column containing Phenyl Sepharose 6 Fast Flow High Sub (PS6FF) to remove small impurities from 3 to 14 kDa (Fig. 1a, lane 4). It is noted that even though approximately 486 mg of PpSP15 were bound to the column during loading, it did not bind very tightly to this column and approximately 411 mg was being eluted during the wash step, leaving only 75 mg being eluted at the elution step (Supplementary Figs. S2a and S2b). When SPXL pool containing only 75 mg of PpSP15 was loaded, no or very little of PpSP15 was observed in the wash step (Supplementary Fig. S2c). Thus, binding capacity was reported as remaining PpSP15 after the wash step per unit volume of PS6FF resin, that is, 75 mg/5 mL PS6FF resin or 15 mg/mL PS6FF at a flow rate of 25.6 cm/h with a residence time of 5.8 min. After the PS6FF run, the purity of the product increased to 97.2%, and 91% of target protein was recovered from the IEX step, which led to a high overall process recovery rate of 79%.
Figure 1.
(a) Purity profile analysis using SDS-PAGE with 5 μg of PpSP15 loaded at each step and stained with Coomassie Blue. (b) Estimated step purity presented as a histogram.
Table 1.
Summary of the Purification Process Improvements, Binding Capacity of Resins, Purity Profile, and Recovery After Each Purification Step.
| Step | Purification Device | Binding Capacity (mg of PpSP15/mL of Resin) | Purity of PpSP15 (%) | Step Recovery (%) | Overall Recovery (%) | Improvement |
|---|---|---|---|---|---|---|
| FS | – | – | 64.6 ± 6.7 | 100 | 100 | – |
| TFF | 3 kDa membrane | – | 68.3 ± 3.9 | 95 | 95 | The FS volume was concentrated 3-fold to reduce the processing time for diafiltration and SPXL step |
| IEX | SPXL resin | 52 mg/mL SPXL | 93.4 ± 0.6 | 92 | 87 | Step elution for target protein |
| HIC | PS6FF resin | 15 mg/mL PS6FF | 97.2 ± 0.7 | 91 | 79 | Step elution for target protein |
Biophysical and Biochemical Characterization of Purified PpSP15
Integrity and Purity by SDS-PAGE, DLS, and Western Blot
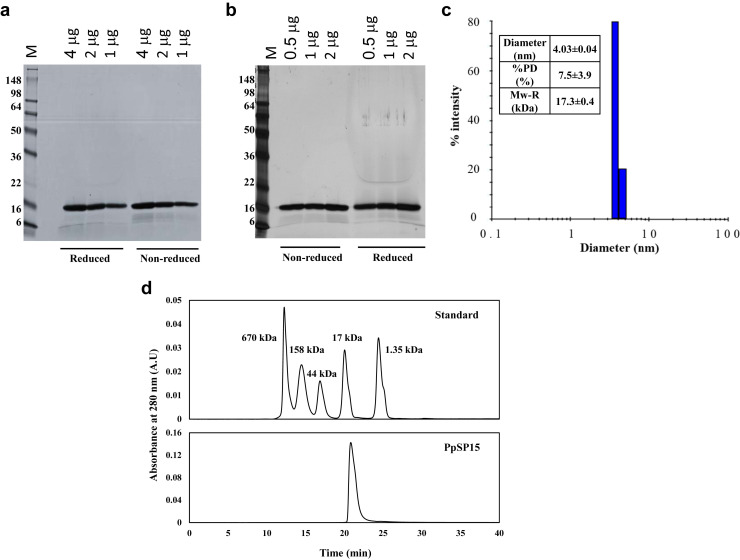
Purified PpSP15 was analyzed by SDS-PAGE to study its integrity and purity (Fig. 2 and Supplementary Fig. S3). When less than 2 μg of purified PpSP15 were loaded onto an SDS-PAGE gel and stained with Coomassie Blue (Fig. 2a and Supplementary Fig. S3), only a single band was observed at approximately 16 kDa on a Tris-glycine gel (Fig. 2a) and 17 kDa on a Bis-tris gel (Supplementary Fig. S3). However, when more than 2 μg were loaded, a faint band at approximately 4.5-5 kDa was observed under reduced conditions and a light smear at approximately 6-14 kDa under nonreduced conditions. The 2 distinct bands observed on the overloaded gel (Supplementary Fig. S3) were recovered, trypsin-digested, and analyzed by proteomic mass spectrometry. The results indicated that the 17 kDa protein band contained 99.9% PpSP15, whereas the 4.5 kDa band contained approximately 52.5% of degraded PpSP15 and 47.5% of HCPs (Table S2). On the silver-stained SDS-PAGE gel, a similar profile was obtained: when more than 1 μg of purified PpSP15 was loaded, small molecular weight impurities were observed (Fig. 2b), but no aggregation or dimer formation was observed. DLS indicated the size of PpSP15 was approximately 17 kDa, which was consistent with the data shown by SDS-PAGE. The low polydispersity (<8%) further indicated that the protein was monodispersed (Fig. 2c). To further assess the level of any potential aggregation, SE-HPLC was also used. The results (Fig. 2d) indicated that PpSP15 eluted as a single peak at 20.8 min, slightly later than the retention time of the 17 kDa protein standard (20.0 min) from a well-resolved Bio-Rad gel filtration standard, suggesting the size of PpSP15 was likely slightly less than 17 kDa. No detectable peak eluted before 20.8 min again demonstrated that PpSP15 was likely not aggregated.
Figure 2.
(a) 1-4 μg of PpSP15 were loaded on an SDS-PAGE gel and stained with Coomassie Blue (b) 0.5-2 μg of PpSP15 were loaded on an SDS-PAGE gel and silver stained. M: SeeBlue Plus2 protein standards. (c) Size analysis of PpSP15 by DLS. (d) Aggregation analysis by size exclusion HPLC; Bio-Rad gel filtration standard (top) was used as a control to compare with the size of PpSP15 (bottom).
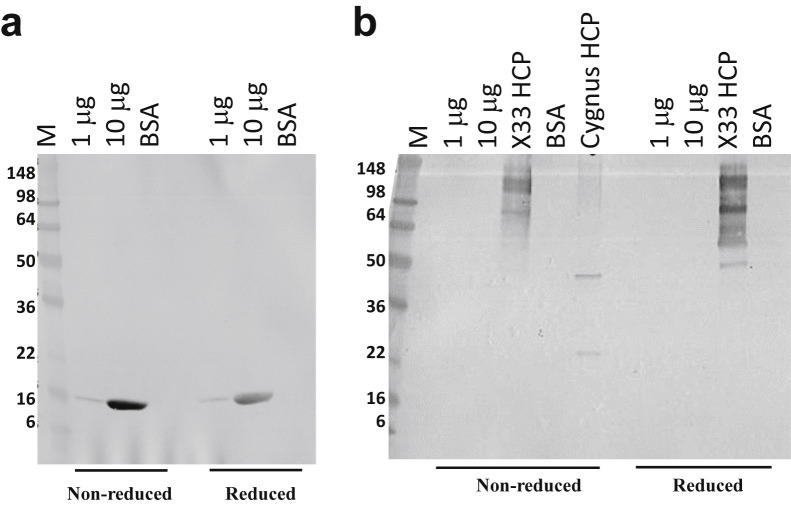
The identity and purity of the sample were also evaluated using western blotting with PpSP15-specific polyclonal antiserum (Fig. 3a ) or P. pastoris X33 HCP-specific antibodies (Fig. 3b). The PpSP15 antisera were able to recognize the protein band at approximately 16 kDa, thus confirming the identity as PpSP15, of which the theoretical molecular weight was 14.75 kDa. However, the proteomic mass spectrometric data indicated that small molecular weight impurities containing degraded PpSP15 and HCPs were present and that these were not recognized by either PpSP15-specific antisera or P. pastoris X33 HCP-specific antibodies, suggesting that the impurities could be low in quantity and required a higher concentration of detection antibodies or that they were not antigenic.
Figure 3.
Western blot (a) 1 μg, 10 μg of PpSP15 and BSA were probed with mouse anti-PpSP15 antisera (1:2000 dilution) and (b) 1 μg, 10 μg of PpSP15, 7.2 μg of in-house Pichia pastoris X33 HCPs, BSA, and Cygnus HCP were probed with goat anti-P. pastoris HCP antibody (1:1000 dilution). M: SeeBlue Plus2 protein.
Structure Assessment Using Circular Dichroism
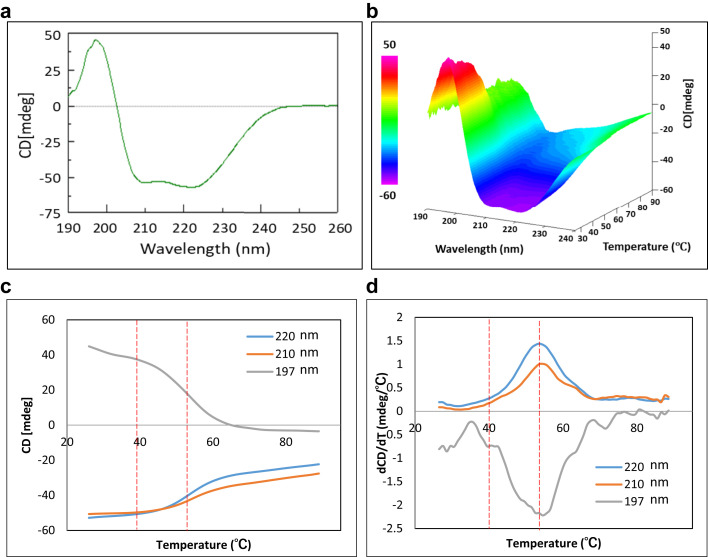
Far-UV CD was performed to investigate the secondary structure of PpSP15. The result indicates that the yeast expressed PpSP15 was rich in alpha-helices (62.6% alpha-helix, 7.5% beta-sheet, and 12.7% turn/loop) (Fig. 4a ). The thermal stability of its secondary structure was evaluated by heating the sample from 25°C to 90°C (Figs. 4b-4d) and the CD melting curves were further extracted at 197 nm, 210 nm, and 220 nm (Figs. 4c and 4d) because these wavelengths showed the best signal to noise data. Based on the derivative, the onset of structural changes begins at approximately 40°C (Ton1) and the average melting temperature (Tm1) was found to be 55°C for all 3 wavelengths (Figs. 4c and 4d).
Figure 4.
Circular dichroism for PpSP15. (a) CD spectrum obtained at 25°C to evaluate the protein’s secondary structure; (b) 3-dimensional CD spectra obtained by heating PpSP15 from 25°C to 90°C; (c) CD melting curves extracted at 197 nm, 210 nm, and 220 nm; (d) derivatives of CD melting curves at 197 nm, 210 nm, and 220 nm.
Buffer Screening Using Thermal Shift Assay
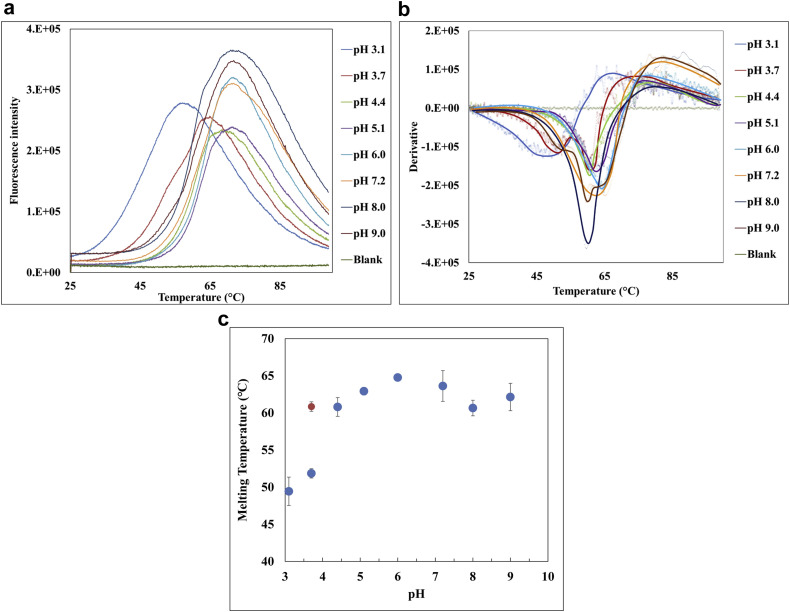
pH screening (ranging from pH 3.1 to pH 9.0) for PpSP15 was performed by evaluating the thermal stability of its tertiary structure using thermal shift assays (Fig. 5 ). Based on the derivative (Fig. 5b), the onset temperature Ton for PpSP15 at pH 3.1 could not be determined as it was lower than 25°C and the melting temperature Tm was approximately 49°C, suggesting PpSP15 may denature at room temperature at this pH. When pH increased to between 4.4 and 9.0, Ton increased to approximately 45°C. Within this pH range, a consistent Tm between 61°C and 65°C was also observed (Figs. 5b and 5c). Interestingly, at pH 3.7, 2 melting temperatures were detected: with Tm1, 52°C, close to Tm at pH 3.1, whereas Tm2, 61°C, close to Tm at higher pH (Figs. 5b and 5c), suggesting that PpSP15 might either be in a metastable state at pH 3.7 or that it had partially lost its thermal stability; hence, closer monitoring of the protein’s tertiary stability overtime at pH 3.7 needs to be considered. Nevertheless, the data suggested that to maintain the tertiary structure stability of PpSP15, a buffer system with a pH between 4.4 and 9.0 should be used.
Figure 5.
pH screening for PpSP15 using thermal shift assays. (a) Fluorescence intensity, (b) derivative fluorescence, and (c) a plot of melting temperature of PpSP15 at different pH values. Three to 4 replicates were prepared for PpSP15 at each pH and a representative curve from one sample for each pH is shown in (a and b). The detailed plots are provided in Supplementary Figure S4.
Discussions
In previous preclinical studies, a DNA vector encoding PpSP15 and a nonpathogenic L. tarentolae secreting PpSP15 were used to demonstrate protection against L. major infection in mice,20, 21, 22 suggesting PpSP15 as a promising vaccine candidate against leishmaniasis. In this study, we have generated a recombinant P. pastoris X33 construct for recombinant PpSP15 protein production, developed and optimized an upstream process at the 10 L scale, and developed a scalable downstream process consisting of IEX and HIC chromatographic steps. The purified PpSP15 was characterized by various biochemical and biophysical assays, and a more suitable pH range to stabilize PpSP15 was determined. Currently, this yeast-expressed recombinant PpSP15 protein is being evaluated for immunogenicity and efficacy against Leishmania infection.
During molecular cloning, the E. coli expression platform was first investigated, yet unable to produce soluble PpSP15. Thus, a recombinant yeast construct for the development and optimization of the upstream fermentation process was generated. The optimized process added a glycerol fed-batch step after the initial depletion of the glycerol in the basal salts medium, increasing cell density before induction. This additional step increased overall protein yield by 27.6%.
With respect to the downstream process development, owing to a high protein pI (9.1), cation exchange chromatography was logically selected as the capture step. First, the conductivity of FS was lowered by diafiltration. To reduce processing time, a 10 kDa cutoff Pellicon® 3 cassette was being tested; however, PpSP15 was found to permeate through the cassette, and thus, a 3 kDa cutoff Pellicon® 3 cassette was selected. However, instead of direct diafiltration, the FS was first concentrated 3-fold to (1) reduce the processing time required for diafiltration and cation exchange chromatography and (2) lower the volume of buffer required for diafiltration. PpSP15 in this concentrated and buffer-exchanged FS was further purified with a 14.5 cm BH IEX column at a flow rate of 149.3 cm/h using a step elution at 30% elution buffer. An additional elution step with a linear gradient from 30% to 100% elution buffer was applied to remove the bound host cell protein in the column after the target protein had been recovered. However, because gradient elution is undesirable in large-scale production, we plan to replace this step with a step elution with 100% elution buffer in future scale-up experiments. After the IEX step, it was noticed that approximately 7% of low-molecular-weight impurities were still present, and these impurities were further removed by hydrophobic HIC. During the first few trials of HIC, Butyl Sepharose High-Performance resin was tested, but it did not show enough binding affinity to PpSP15. Hence, a stronger HIC resin, Phenyl Sepharose 6 Fast Flow high sub (PS6FF), was chosen. By using PS6FF resin, impurities were reduced by an additional 3% and the purity of PpSP15 reached >97%. It was noted that typically the BH should be maintained while upscaling; however, the PS6FF column used during this development phase had a BH of 2.5 cm, and hence scaling up was not practical and required optimization. For future studies to scale up the process, we intend to pack a column with a BH between 10 and 20 cm and maintain the current residence time of 5.8 min to keep the binding capacity similar. Based on the binding capacity of PpSP15 to SPXL resin (52 mg/mL) and PS6FF resin (15 mg/mL), the minimal CVs of both resins to purify this antigen and the estimated amount of purified antigen were calculated (Supplementary Table S3). For a typical 10 L fermentation process, approximately 4.5 L of FS is obtained. With the initial yield of 994 mg of PpSP15/L of FS, it will require an SPXL column with a CV of more than 86 mL and a PS6FF column with a CV of more than 260 mL to capture all the PpSP15 during purification. With 79% of high overall recovery, we expect to collect approximately 785 mg of purified PpSP15 per litter of FS. When comparing with the purification process for several other reported recombinant protein vaccine antigens at similar fermentation scale,24 , 27 , 29, 30, 31, 32 one can find that the process for PpSP15 requires much less volume of resins; in addition, size exclusion chromatography, a process not ideal for large-scale production, had been included in the purification process for some of these recombinant protein vaccine antigens to increase their purity.24 , 27 , 30 , 31 However, the current purification scheme for PpSP15 does not require an size exclusion chromatography step to improve the purity to >95%, and all the chosen purification steps are scalable, except that dialysis was used to exchange PpSP15 into an appropriate buffer due to the small volumes at this development phase. For future upscaling, we plan to replace the dialysis step with diafiltration using 3 kDa Pellicon® cassettes.
With respect to protein characterization, several biophysical and biochemical assays were performed to study the characteristics of this protein. It is well known that the recombinant proteins expressed in yeast can potentially be heavily N-glycosylated,27 , 33 an N-glycosylation assessment screening for the presence of the consensus sequence for N-glycosylation, AsnXxxSer/Thr/Cys (where Xxx can be any amino acid except proline) was conducted.34 In the PpSP15 sequence (Supplementary Fig. S5), there are 9 Asn residues, but only Asn22 is followed by Ser24. However, the amino acid in between Asn22 and Ser24 is Pro23, thus Asn22 is most likely not glycosylated. SDS-PAGE gels and the mass spectrometry also indicated the absence of N-glycans. Other more sensitive and specific assays were also applied to study the purity/impurity, including silver-stained SDS-PAGE and anti-PpSP15 western blot and anti-P. pastoris X33 HCP western blot. The results suggested that only trace amounts of HCPs and product-derived impurities, degraded PpSP15, were present in the purified PpSP15 sample. Moreover, no detectable aggregation was found, which was also supported by DLS and SE-HPLC. Indeed, its low binding affinity to the HIC column explaining this protein is likely highly hydrophilic. In addition, when comparing the PpSP15 sequence to PdSP15 and its crystal structure,26 after excluding the signal peptide, it was predicted that all 6 cysteine residues in PpSP15 form disulfide bridges, and thus, no free cysteine residue should be present to form intermolecular disulfide bridges. These findings suggest a low tendency for PpSP15 to aggregate, a desirable property for long-term stability. The secondary structure analyzed by far-UV CD indicated PpSP15 as a highly alpha-helical protein (62.6% alpha-helix and 7.5% beta-sheet). This result was consistent with the secondary structure of its homolog, PdSP15 (PDB ID 4OZD), (66% helix), which further validated the assay. Finally, pH screening using a thermal shift assay was conducted to determine the proper pH system to maintain PpSP15 stability. It is noted that the onset temperature and melting temperature remained constant between pH 4.4 and 9.0; however, a lower melting temperature was observed at both pH 3.1 and 3.7, suggesting instability of PpSP15, which further indicates buffer formulations at a pH lower than 4.4 should be avoided.
In conclusion, the production process for recombinant PpSP15 as a leishmaniasis vaccine antigen candidate has been developed. The upstream production process was optimized and generated approximately 1 g PpSP15 per liter of FS. The tag-free recombinant PpSP15 protein was able to be purified by a 2-chromatographic-step purification process, which involved an ion-exchange column and a hydrophobic interaction column, and the binding capacities for both chromatographic steps were determined. Even though some minor adjustments will be required, the process is suitable for further upscaling and studies of process recovery and process reproducibility. Characterization of PpSP15 showed that the protein was highly pure with a well-defined structure. Future studies should be focused on understanding long-term stability and stability under different stresses as these pieces of information along with the ongoing preclinical study will inform whether there is a path forward for manufacturing an economical vaccine for this neglected tropical disease.
Acknowledgments
The authors are grateful to the laboratories of Dr. Jesus Valenzuela at the NIAID and Dr. Naomi Aronson at USUHS for helpful discussions on the PpSP15 process development.
This study was supported by a grant from the Department of Defense, United States (Grant number: W81XWH-17-2-0050).
Footnotes
This article contains supplementary material available from the authors by request or via the Internet at https://doi.org/10.1016/j.xphs.2020.02.004.
Contributor Information
Wen-Hsiang Chen, Email: Wen-Hsiang.Chen@bcm.edu.
Maria Elena Bottazzi, Email: bottazzi@bcm.edu.
Appendix A. Supplementary data
References
- 1.Torres-Guerrero E., Quintanilla-Cedillo M.R., Ruiz-Esmenjaud J., Arenas R. Leishmaniasis: a review. F1000Res. 2017;6:750. doi: 10.12688/f1000research.11120.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martins-Melo F.R., Lima Mda S., Ramos A.N., Jr., Alencar C.H., Heukelbach J. Mortality and case fatality due to visceral leishmaniasis in Brazil: a nationwide analysis of epidemiology, trends and spatial patterns. PLoS One. 2014;9(4):e93770. doi: 10.1371/journal.pone.0093770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reithinger R., Dujardin J.C., Louzir H., Pirmez C., Alexander B., Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7(9):581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- 4.Hotez P.J. The rise of leishmaniasis in the twenty-first century. Trans R Soc Trop Med Hyg. 2018;112(9):421–422. doi: 10.1093/trstmh/try075. [DOI] [PubMed] [Google Scholar]
- 5.Bennis I., Thys S., Filali H., De Brouwere V., Sahibi H., Boelaert M. Psychosocial impact of scars due to cutaneous leishmaniasis on high school students in Errachidia Province, Morocco. Infect Dis Poverty. 2017;6(1):46. doi: 10.1186/s40249-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey F., Mondragon-Shem K., Hotez P. A new perspective on cutaneous leishmaniasis-implications for global prevalence and burden of disease estimates. PLoS Negl Trop Dis. 2017;11(8):e0005739. doi: 10.1371/journal.pntd.0005739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey F., Mondragon-Shem K., Haines L.R. Cutaneous leishmaniasis and co-morbid major depressive disorder: a systematic review with burden estimates. PLoS Negl Trop Dis. 2019;13(2):e0007092. doi: 10.1371/journal.pntd.0007092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribeiro J.M. Role of saliva in blood-feeding by arthropods. Annu Rev Entomol. 1987;32:463–478. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- 9.Bahia D., Gontijo N.F., Leon I.R. Antibodies from dogs with canine visceral leishmaniasis recognise two proteins from the saliva of Lutzomyia longipalpis. Parasitol Res. 2007;100(3):449–454. doi: 10.1007/s00436-006-0307-8. [DOI] [PubMed] [Google Scholar]
- 10.Barral A., Honda E., Caldas A. Human immune response to sand fly salivary gland antigens: a useful epidemiological marker? Am J Trop Med Hyg. 2000;62(6):740–745. doi: 10.4269/ajtmh.2000.62.740. [DOI] [PubMed] [Google Scholar]
- 11.Gomes R.B., Brodskyn C., de Oliveira C.I. Seroconversion against Lutzomyia longipalpis saliva concurrent with the development of anti-Leishmania chagasi delayed-type hypersensitivity. J Infect Dis. 2002;186(10):1530–1534. doi: 10.1086/344733. [DOI] [PubMed] [Google Scholar]
- 12.Gomes R.B., Mendonca I.L., Silva V.C. Antibodies against Lutzomyia longipalpis saliva in the fox Cerdocyon thous and the sylvatic cycle of Leishmania chagasi. Trans R Soc Trop Med Hyg. 2007;101(2):127–133. doi: 10.1016/j.trstmh.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Belkaid Y., Kamhawi S., Modi G. Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J Exp Med. 1998;188(10):1941–1953. doi: 10.1084/jem.188.10.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamhawi S., Belkaid Y., Modi G., Rowton E., Sacks D. Protection against cutaneous leishmaniasis resulting from bites of uninfected sand flies. Science. 2000;290(5495):1351–1354. doi: 10.1126/science.290.5495.1351. [DOI] [PubMed] [Google Scholar]
- 15.Wikel S.K., Ramachandra R.N., Bergman D.K., Burkot T.R., Piesman J. Infestation with pathogen-free nymphs of the tick Ixodes scapularis induces host resistance to transmission of Borrelia burgdorferi by ticks. Infect Immun. 1997;65(1):335–338. doi: 10.1128/iai.65.1.335-338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donovan M.J., Messmore A.S., Scrafford D.A., Sacks D.L., Kamhawi S., McDowell M.A. Uninfected mosquito bites confer protection against infection with malaria parasites. Infect Immun. 2007;75(5):2523–2530. doi: 10.1128/IAI.01928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labuda M., Trimnell A.R., Lickova M. An antivector vaccine protects against a lethal vector-borne pathogen. PLoS Pathog. 2006;2(4):e27. doi: 10.1371/journal.ppat.0020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsollier L., Deniaux E., Brodin P. Protection against Mycobacterium ulcerans lesion development by exposure to aquatic insect saliva. PLoS Med. 2007;4(2):e64. doi: 10.1371/journal.pmed.0040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conway M.J., Londono-Renteria B., Troupin A. Aedes aegypti D7 saliva protein Inhibits dengue virus infection. PLoS Negl Trop Dis. 2016;10(9):e0004941. doi: 10.1371/journal.pntd.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valenzuela J.G., Belkaid Y., Garfield M.K. Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J Exp Med. 2001;194(3):331–342. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliveira F., Lawyer P.G., Kamhawi S., Valenzuela J.G. Immunity to distinct sand fly salivary proteins primes the anti-Leishmania immune response towards protection or exacerbation of disease. PLoS Negl Trop Dis. 2008;2(4):e226. doi: 10.1371/journal.pntd.0000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katebi A., Gholami E., Taheri T. Leishmania tarentolae secreting the sand fly salivary antigen PpSP15 confers protection against Leishmania major infection in a susceptible BALB/c mice model. Mol Immunol. 2015;67(2 Pt B):501–511. doi: 10.1016/j.molimm.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira F., Kamhawi S., Seitz A.E. From transcriptome to immunome: identification of DTH inducing proteins from a Phlebotomus ariasi salivary gland cDNA library. Vaccine. 2006;24(3):374–390. doi: 10.1016/j.vaccine.2005.07.085. [DOI] [PubMed] [Google Scholar]
- 24.Hudspeth E.M., Wang Q., Seid C.A. Expression and purification of an engineered, yeast-expressed Leishmania donovani nucleoside hydrolase with immunogenic properties. Hum Vaccin Immunother. 2016;12(7):1707–1720. doi: 10.1080/21645515.2016.1139254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAtee C.P., Seid C.A., Hammond M. Expression, purification, immunogenicity and protective efficacy of a recombinant nucleoside hydrolase from Leishmania donovani, a vaccine candidate for preventing cutaneous leishmaniasis. Protein Expr Purif. 2017;130:129–136. doi: 10.1016/j.pep.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Oliveira F., Rowton E., Aslan H. A sand fly salivary protein vaccine shows efficacy against vector-transmitted cutaneous leishmaniasis in nonhuman primates. Sci Transl Med. 2015;7(290) doi: 10.1126/scitranslmed.aaa3043. 290ra290. [DOI] [PubMed] [Google Scholar]
- 27.Chen W.H., Chag S.M., Poongavanam M.V. Optimization of the production process and characterization of the yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1), a SARS vaccine candidate. J Pharm Sci. 2017;106(8):1961–1970. doi: 10.1016/j.xphs.2017.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIlvaine T.C. A buffer solution for colorimetric comparison. J Biol Chem. 1921;49:4. [Google Scholar]
- 29.Seid C.A., Curti E., Jones R.M. Expression, purification, and characterization of the Necator americanus aspartic protease-1 (Na-APR-1 (M74)) antigen, a component of the bivalent human hookworm vaccine. Hum Vaccin Immunother. 2015;11(6):1474–1488. doi: 10.1080/21645515.2015.1036207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curti E., Seid C.A., Hudspeth E. Optimization and revision of the production process of the Necator americanus glutathione S-transferase 1 (Na-GST-1), the lead hookworm vaccine recombinant protein candidate. Hum Vaccin Immunother. 2014;10(7):1914–1925. doi: 10.4161/hv.28872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biter A.B., Weltje S., Hudspeth E.M. Characterization and stability of Trypanosoma cruzi 24-C4 (Tc24-C4), a candidate antigen for a therapeutic vaccine against Chagas disease. J Pharm Sci. 2018;107(5):1468–1473. doi: 10.1016/j.xphs.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Curti E., Kwityn C., Zhan B. Expression at a 20L scale and purification of the extracellular domain of the Schistosoma mansoni TSP-2 recombinant protein: a vaccine candidate for human intestinal schistosomiasis. Hum Vaccin Immunother. 2013;9(11):2342–2350. doi: 10.4161/hv.25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vervecken W., Kaigorodov V., Callewaert N., Geysens S., De Vusser K., Contreras R. In vivo synthesis of mammalian-like, hybrid-type N-glycans in Pichia pastoris. Appl Environ Microbiol. 2004;70(5):2639–2646. doi: 10.1128/AEM.70.5.2639-2646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medzihradszky K.F. Characterization of site-specific N-glycosylation. Methods Mol Biol. 2008;446:293–316. doi: 10.1007/978-1-60327-084-7_21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.