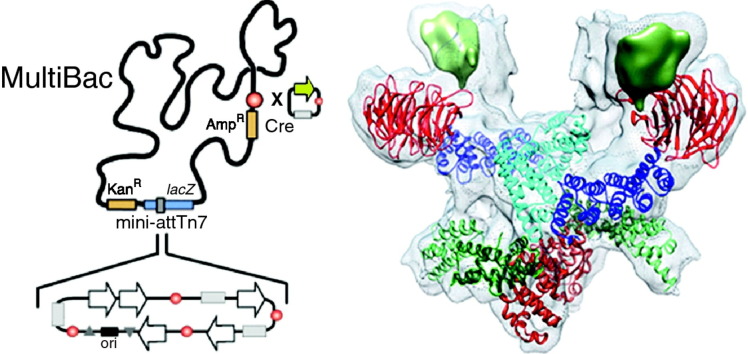
Graphical abstract
Highlights
-
•
BEVS can provide large multiprotein complexes in high quality and large quantity.
-
•
Recent improvements have rendered BEVS easy-to-use, also to non-specialist users.
-
•
Recombinant production can overcome limitations caused by sample heterogeneity.
-
•
MultiBac BEVS helped to unlock the architecture of complex multiprotein machines.
-
•
Structures of Mediator Head, APC/C, MCC, and TFIID core have been determined.
Abstract
Most essential functions in eukaryotic cells are catalyzed by complex molecular machines built of many subunits. To fully understand their biological function in health and disease, it is imperative to study these machines in their entirety. The provision of many essential multiprotein complexes of higher eukaryotes including humans, can be a considerable challenge, as low abundance and heterogeneity often rule out their extraction from native source material. The baculovirus expression vector system (BEVS), specifically tailored for multiprotein complex production, has proven itself to be uniquely suited for overcoming this impeding bottleneck. Here we highlight recent major achievements in multiprotein complex structure research that were catalyzed by this versatile recombinant complex expression tool.
Current Opinion in Structural Biology 2013, 23:357–364
This review comes from a themed issue on New constructs and expression of proteins
Edited by Imre Berger and Lorenz M Mayr
For a complete overview see the Issue and the Editorial
Available online 27th April 2013
0959-440X/$ – see front matter, © 2013 Elsevier Ltd. All rights reserved.
Introduction
Recombinant expression has had a profound impact on protein research, and heterologous expression systems are used on a daily basis in most molecular biology laboratories. The major workhorse for protein expression has been Escherichia coli, and today a very large number of expression plasmids and specialized strains, each with its own merits, exist for this prokaryotic host. The overwhelming majority of protein entries in the PBD (http://www.rcsb.org/) originate from heterologous production in E. coli. However, the increasing trend towards the study of more complex biological assemblies, sometimes with 10 or more subunits, has necessitated the development of more powerful eukaryotic expression systems that offer access to multiprotein complexes, which for a variety of reasons (large subunit size, requirement of post translational modifications and chaperones, others) cannot be made efficiently in E. coli. The baculovirus expression vector system (BEVS), introduced more than 20 years ago, has recently emerged as a particularly useful tool for producing large eukaryotic protein complexes in the quantity and quality required for detailed structural study.
MultiBac: baculovirus expression vector system for protein complex production
BEVS rely on a recombinant baculovirus carrying the gene(s) of interest to infect insect cell cultures for high-level heterologous production of the single protein or protein complex of choice. BEVS has remained for some time a relatively specialized application, but has entered mainstream in structural biology laboratories in the past several years. Engineering of the baculovirus genome towards improved protein complex production characteristics, the development of new plasmids and stream-lined methods for multigene assembly and integration into the baculovirus genome, and the establishment of standard operating procedures (SOPs) for cell culture maintenance, virus generation, infection and protein production have resulted in the MultiBac system, a BEVS that is particularly easy-to-use and sufficiently robust to be employed on a routine basis by users who do not possess specialist training [1•]. These are critically important considerations for the use of BEVS in structural biology laboratories, where the overriding objective is protein sample preparation for structure analysis, and the investment in optimizing individual expression experiments necessarily needs to stay within a reasonably short timeframe. MultiBac has been instrumental for producing a number of multiprotein complexes that could not be accessed before for structural and functional studies. Recent examples of such multiprotein complexes are provided in the following, including the Head module of the Mediator from Saccharomyces cerevisiae, the yeast anaphase promoting complex APC/C, and the human general transcription factor TFIID core complex, each of which highlighting different aspects of BEVS application that were exploited to achieve successful structure determination.
The mediator head module from S. cerevisiae
Mediator is a large multiprotein complex that regulates most, if not all, gene transcription by RNA polymerase II (Pol II) [2]. Mediator is structurally and functionally conserved in all eukaryotes [3, 4]. In the yeast S. cerevisiae, where it was first discovered [5], Mediator comprises 21 subunits with a combined molecular mass exceeding 1 MDa [6, 7]. Mediator functions as an interface between DNA-bound protein activators of transcription and Pol II [2, 8, 9]. Of medical relevance, the mutations on several human Mediator subunits have been linked to neurological disorders and cancers [10, 11, 12, 13, 14, 15, 16]. Thus, elucidation of the molecular mechanisms of Mediator function is a major research subject in biomedical sciences. Despite its paramount importance in gene regulation, the large size, complexity, and low abundance of Mediator have made biochemical and structural studies extremely difficult, if not impossible, for a long time.
Single-particle electron microscopy (EM) provided first impressions of the complexity underlying Mediator architecture. Unlike Pol II, which adopts a compact globular structure, Mediator from yeast is composed of three distinct modules [17, 18], termed Head, Middle, and Tail (Figure 1 ). As suggested by structural, genetic and biochemical studies [19, 20, 21], the modules consist of 7–9 individual subunits each (Figure 1a). Mediator has multiple activities [5], and each module is thought to perform its own defined set of functions [22]. For example, the subunits from the Head module genetically interact with the C-terminal domain (CTD) of Pol II [23, 24], while several Middle and Tail module subunits have been shown to directly interact with transcriptional activators [9, 21, 25, 26, 27]. A reasonable strategy therefore is to address particular Mediator functions by studying the modules individually as first steps towards deciphering the overall mechanism of the whole complex. While still a formidable challenge, this approach certainly reduces the otherwise intimidating complexity of studying Mediator structure and function.
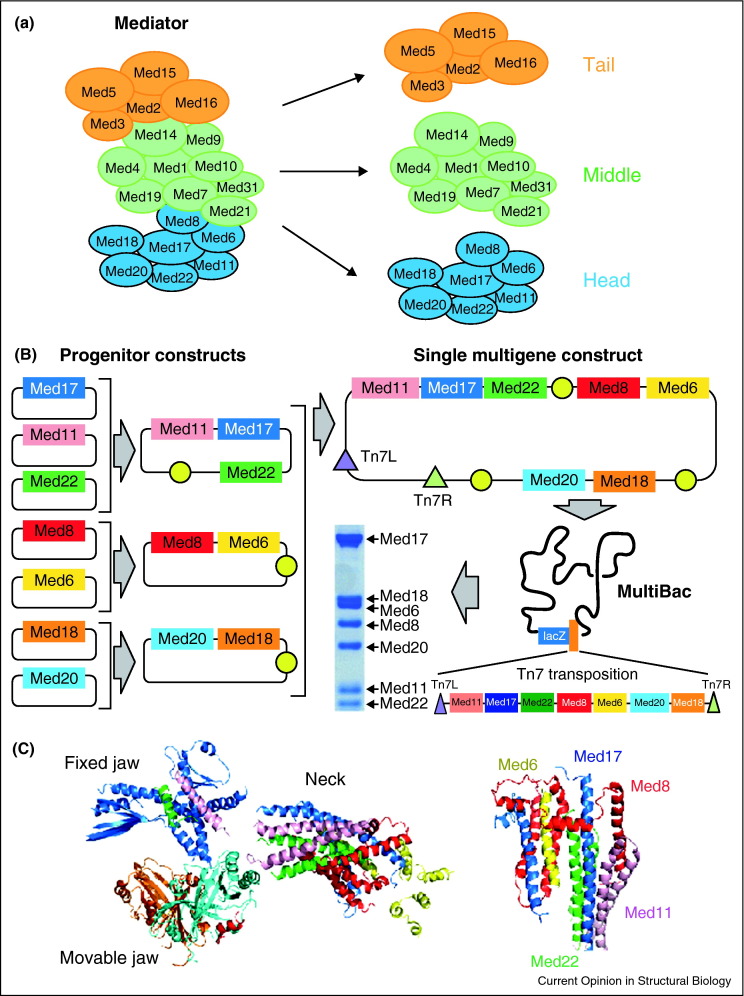
Figure 1.
S. cerevisae Mediator Complex. (a) Subunit organization of Mediator. Head module subunits are colored in blue; those of the Middle module are in green and those of the Tail module are in ochre. (b) Genes encoding the Mediator Head module were assembled from multiple DNA progenitors (left) into a single multigene expression construct (right, top) for insertion by Tn7 transposition (Tn7L, Tn7R) into the MultiBac baculovirus to produce the Head module in infected insect cells (right, bottom). (c) X-ray crystal structure of the recombinant Mediator Head module showing Med17 (blue), Med11 (purple), Med22 (green), Med6 (yellow), Med8 (red), Med18 (cyan), and Med 20 (orange). The Head consists of three distinct domains, fixed jaw, movable jaw and neck (left). The neck domain is arranged in a novel multihelical bundle (right) [30••].
Producing the Mediator Head module in insect cells
The Mediator Head module is composed of 7 subunits (Med17, Med6, Med8, Med11, Med22, Med18, Med20) with a molecular mass of 223 kDa. Originally, a conventional co-infection approach with seven different baculoviruses, each encoding for one subunit, was pursued [28]. Highly expressing baculoviruses were carefully isolated and optimized for maximum expression, and then used in combination to co-infect insect cell cultures. This procedure was exceedingly lengthy (each experiment taking two months or more) and complicated logistics as seven viruses had to be maintained at high titers simultaneously in parallel. X-ray crystallography, however, requires rapid turnover of expression experiments. Often, the production of a large number of variants of the protein or complex studied is necessary, to identify the sample which produces diffraction quality crystals, and this must be within a timeframe that is as short as feasible.
The solution to this problem came by expressing all Mediator Head subunits simultaneously from a single multigene baculovirus using the MultiBac system, thereby resolving a fundamental impediment. MultiBac utilizes a procedure termed tandem recombineering (TR) which considerably facilitates the iterative assembly of multigene cassettes from small DNA progenitors [1•]. All genes encoding for the Mediator Head subunits were thus combined into a single construct by TR (Figure 1b) [29, 30••], significantly reducing experimental complexity. The multigene construct was next integrated into the engineered MultiBac baculovirus genome that is characterized by reduced proteolysis and delayed host cell lysis, thus improving the quality of the sample produced [1•]. The recombinant virus was used to infect insect cell cultures to produce the Mediator Head (Figure 1b).
Crystal structure of recombinant Mediator Head module
The simplified recombinant production propelled structure determination of the Head module by X-ray crystallography. Modification of MED17 and MED18 by eliminating flexible regions was critical to obtain well-ordered single crystals suitable for diffraction data collection (Figure 1c). Incorporation of seleno-methionine in the recombinantly produced sample was instrumental for structure elucidation. The crystal structure of the Mediator Head reveals how this essential complex is built from its components combining stability as well as flexibility for transcription regulation, providing a platform for other transcription factors [30••]. New features in the structure provide first impressions about architectural design principles of such large multiprotein machines. Notably, a portion of the Head named ‘neck domain’ confers stability and integrity of the complex by forming a novel multihelical bundle, engaging five of the seven subunits of the Head simultaneously in one compact structural unit (Figure 1c).
The anaphase promoting complex (APC/C)
The APC/C is an unusually large multisubunit RING E3 ubiquitin ligase that regulates cell cycle progression through the proteasome-dependent proteolysis of cell cycle regulatory proteins [31, 32, 33]. The core APC/C, formed from at least 13 different proteins, is activated on association of a regulatory coactivator subunit. The APC/C is highly conserved across eukaryotes, and 12 of the 13 S. cerevisiae APC/C subunits are conserved in Saccharomyces pombe and human. Owing to the presence of two copies of many APC/C proteins, the holo-APC/C is comprised between 18 and 19 subunits with an overall molecular mass ranging from 1 to 1.2 MDa [34••].
Producing recombinant APC/C by BEVS: co-infection with two viruses
Early research on the APC/C was restricted to the use of native systems. Because most APC/C subunits are essential, genetic manipulations were intrinsically difficult, and the low natural abundance of APC/C limited structural and biochemical studies. The recent development of overexpression systems for S. cerevisiae and human APC/C [34••, 35], based on the MultiBac BEVS that reconstitutes all APC/C proteins, now enables a range of structural, biochemical and biophysical investigations. The expression and reconstitution of recombinant S. cerevisiae APC/C [34••] used the first generation MultiBac cloning system for insect cell-baculovirus expression [36, 37, 38, 39, 40, 41]. To express S. cerevisiae APC/C, two viruses encoding five and eight subunits were combined for co-expression. The resultant recombinant co-expression system yielded ∼200-fold more intact APC/C than the endogenous system (0.5 mg/L insect cell culture). The reconstituted S. cerevisiae APC/C was correctly assembled, as judged by its structural correspondence to native APC/C (Figure 2 a and b), and its capacity to ubiquitinate mitotic cyclin in the presence of coactivator in a D box and KEN box dependent manner [34••]. The ability to recapitulate the endogenous APC/C catalytic and regulatory activity using recombinant reconstituted APC/C provided strong evidence that the molecular composition of S. cerevisiae APC/C had been completely defined, a crucial prerequisite for understanding the complete system.
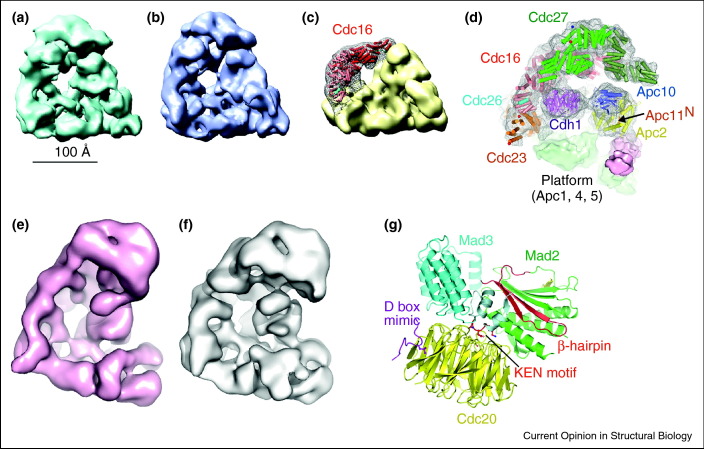
Figure 2.
Structures of the APC/C and MCC. 3-D EM reconstructions of (a) recombinant S. cerevisiae APC/C and (b) endogenous S. cerevisiae APC/C. (c) Localisation of the Cdc16 dimer in Cdc16-assigned density (mesh). (d) Pseudo atomic structure of S. cerevisiae APC/C, adapted from [34••]. (e) Endogenous human APC/C [51•]. (f) Recombinant human APC/C [35]. (g) Structure of the S. pombe mitotic checkpoint complex [50•].
Dissecting recombinant yeast APC/C
Reconstitution of the holo-APC/C and APC/C subcomplexes allowed an accurate determination of the APC/C mass. An APC/C subcomplex of eight subunits was analyzed using native mass spectrometry and measured as 698.8 kDa, in good agreement with that predicted for a complex containing all subunits in unit stoichiometry plus an additional copy of Cdc23 [34••]. Generation of APC/C subcomplexes, guided by an architectural map of the APC/C [42], and subsequent determination of their EM structures allowed the assignment of the three-dimensional molecular boundaries of individual subunits within the APC/C molecular envelope. As an example, comparing two APC/C subcomplexes differing by Cdc16–Cdc26 indicated difference density that corresponded closely to the crystal structure of the Cdc16–Cdc26 heterotetramer (Figure 2c) [43]. This subunit deletion approach allowed a systematic assignment of the majority of large APC/C subunits and by integrating crystal structures, and homology models of individual APC/C subunits, with a cryo-EM reconstruction of an APC/CCdh1•D-box ternary complex at 10 Å resolution [44], a high-resolution description of the subunit organization and pseudo-atomic model of the APC/C was determined (Figure 2d) [34••, 44]. The approach of subunit deletion employed by [34••] accurately defines the molecular boundaries of specific APC/C subunits, enabling more reliable docking of subunit atomic models compared with that possible from the approximate locations of subunits defined by labeling N-termini and C-termini.
BEVS expression of complete human APC/C
More recently the overexpression and reconstitution of human APC/C in the insect/baculovirus system were reported by two groups [35, 45]. One group generated multigene vectors modified from the MultiBac plasmids pFBDM and pUCDM vectors [36], using USER ligation-independent cloning methods [46, 47] and incorporated all 14 genes (45 kb) into two baculoviruses [35], whereas the second group generated 14 individual viruses, each expressing a single APC/C subunit, which were combined for co-expression in insect cells for biochemical studies [45]. Interestingly Apc15 and Apc16 were only recently discovered as human APC/C subunits. Efforts to generate fully assembled recombinant human APC/C before the identification of Apc16 were unsuccessful [35]. This indicated that Apc16, located within the TPR subcomplex [48], is necessary for the optimal assembly of recombinant human APC/C in insect cells. Thus, the successful assembly and reconstitution of human APC/C using the insect cell/baculovirus expression system indicated that all human APC/C subunits necessary for a functional and catalytically active APC/C had finally been identified (Figure 2e and f).
X-ray structure of the mitotic checkpoint complex (MCC), a regulator of APC/C
To ensure correct chromosome segregation in mitosis, the spindle assembly checkpoint imposed by the MCC inhibits the APC/C until all chromosomes have achieved correct bipolar attachment to the mitotic spindle [49]. The MCC comprises the APC/C coactivator Cdc20 associated with the checkpoint proteins Mad2, Mad3/BubR1 and Bub3 and blocks D box and KEN box recognition by the APC/C. The crystal structure of S. pombe MCC (without Bub3) expressed using the MultiBac system revealed that Cdc20, Mad2 and Mad3 assemble into a triangular-shaped heterotrimer (Figure 2g) [50•]. Mad3 coordinates the overall organization of the complex by forming numerous inter-subunit interactions with Mad2 and Cdc20. A helix-loop-helix motif at the N-terminus of Mad3 binds simultaneously to Mad2 and Cdc20, orienting the KEN box that acts as a pseudo-substrate inhibitor towards its receptor on Cdc20.
Human general transcription factor TFIID
Initiation of transcription by RNA polymerase II (Pol II) requires the controlled step-wise assembly of the preinitiation complex (PIC), comprising a large ensemble of proteins and protein complexes including Pol II and the general transcription factors GTFs (TFIIA, B, D, E, F, H) [52, 53, 54]. The promoter recognition complex, TFIID, is thought to be the cornerstone of PIC assembly. In addition, TFIID functions as a coactivator, mediating signals from sequence-specific activators to other components of the transcription machinery. In humans, TFIID comprises 14 protein subunits, TBP and the TBP associated factors (TAFs), ranging in size from 250 kDa to less than 20 kDa, with a multitude of functions [52, 53]. TAFs and TBP assemble into a large multisubunit complex with around 20 subunits and an overall molecular weight of ∼1.6 MDa [54].
Our understanding of TFIID and its crucial role in transcription regulation is considerably hampered by a lack of detailed knowledge of the molecular architecture of this essential factor, its assembly in the cell, and its interactions with chromatin and other factors in the context of activated transcription. The overall shape of human and yeast TFIID was shown by EM, revealing an asymmetric tri-lobed structure resembling a molecular clamp [54]. The paucity and heterogeneity of the endogenous material used in these studies have limited structural insight to moderate resolution (∼30–40 Å for human TFIID), prohibiting molecular level interpretation of TFIID architecture notwithstanding considerable effort.
The current consensus regarding the copy-number of subunits within TFIID is that a subset of TAFs exists in two copies, while TBP and the remaining TAFs are present in single copy [52]. Owing to evolutionary conservation and overall similarity in shape of the yeast and human TFIID complexes, this subunit composition is probably to be present in TFIID from most species. The concept emerged in which TAFs present in duplicate form a putative, 2-fold symmetric scaffold, around which the remaining TAFs and TBP organize as peripheral subunits [52], suggesting that a transition from symmetry to asymmetry may occur in the TFIID assembly pathway. Compelling functional support for this bipartite architectural design came from studies in Drosophila cells [55]. By means of RNAi to knockdown specific TAFs, a stable and functional core-TFIID complex, composed of TAF4, 5, 6, 9 and 12, was revealed in vivo. Further support stems from cryo-EM studies of TFIID preparations from yeast, in which a quasi-symmetric smaller shape was also found [56]. Together, these results point to the existence of a symmetric core-TFIID module of pivotal importance for the integrity and assembly of holo-TFIID.
Expressing human TFIID core complex: polyproteins adjust stoichiometries
The architecture of the human TFIID core complex, consisting of 10 subunits (two copies each of TAF4, 5, 6, 9 and 12), was recently elucidated at nanometer resolution by an integrated approach combining recombinant overproduction by MultiBac, cryo-EM, data from X-ray crystallography and homology models [57••] (Figure 3 ). The structure revealed a symmetric arrangement of the subunits, validating previous experiments. Initially, the TFIID core complex was produced by using a single baculovirus containing the encoding genes arranged as a multigene construct made up of individual expression cassettes [36], similar to the strategy adopted for the successful study of the Mediator Head module from S. cerevisiae [30••]. In the case of core-TFIID, however, it turned out that the expression levels of the individual subunits varied substantially, thus reducing overall yield of the recombinant complex dramatically. Large amounts of heterologous protein was produced in excess while the subunit characterized by the lowest expression level relative to the others dictated the final amount of properly assembled complex containing all subunits. A rescue strategy involving tagging of this weakly expressing subunit with an affinity tag did result in the purification of complete core-TFIID, however, EM analysis revealed the presence of smaller subassemblies lacking subunits that were contaminating the sample, impeding structure determination.
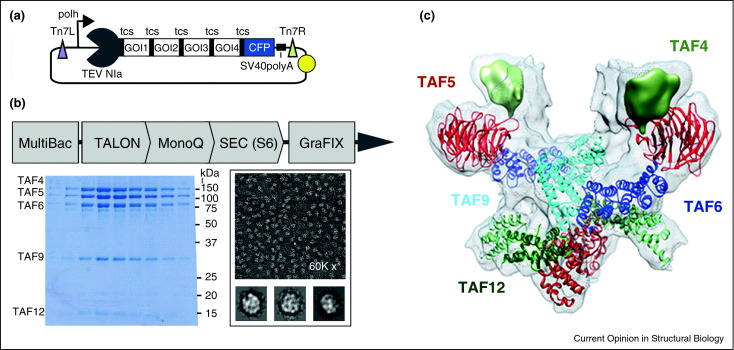
Figure 3.
Human general transcription factor TFIID core complex. (a) Generally applicable polyprotein strategy for balancing the production levels of multiprotein complex subunits. Genes of interest (GOI), a protease encoding gene (TEV NIa) and a fluorescent marker (CFP) are present in a single ORF under control of a strong baculoviral promoter (polh) and flanked by a poly A signal (black square) on a MultiBac plasmid (cf. Figure 1B), spaced apart by the specific cleavage sequence of TEV NIa (tcs) [1•, 58]. (b) Polyprotein expression and purification of human core-TFIID. Negative stain EM and 2-D classification (bottom, right) were used to optimize purification until high-quality sample was obtained as demonstrated by SDS-PAGE (bottom, left). (c) TFIID core complex structure at nanometer resolution, determined by hybrid methods [57••].
This challenge was resolved by adopting a strategy that certain viruses such as Coronavirus use to realize their proteome [41] (Figure 3A). These viruses express long open-reading frames (ORFs) that are translated into large polyproteins. These polyproteins are then processed into individual proteins by a highly specific protease that cuts proteolytic sites in between the functional polypeptide entities. Typically, the protease is also part of the polyprotein, and liberates itself as well by means of proteolytic cleavage. This strategy was implemented in the MultiBac system by creating long single ORFs composed of the TAF genes. The gene encoding for NIa protease from Tobacco etch virus (TEV) was added at the 5′, and corresponding recognition and cleavage sequences were inserted [1•, 41]. To follow and quantify polyprotein production during expression, the gene encoding for a fluorescent protein was appended to the 3′ end of the ORF (Figure 3a). The TEV NIa protease proved to be sufficiently specific to process the polyproteins expressed properly and to completion. This resulted in a fully balanced level of the TAF proteins resulting from polyprotein processing that assembled completely into the TFIID core complex. Core-TFIID was purified to homogeneity, enabling high-resolution structure determination (Figure 3b and c). Expression of two further TAFs, TAF8 and TAF10, likewise from a polyprotein construction, and incorporation into the core-TFIID complex followed by cryo-EM structure determination, allowed to propose a molecular mechanisms of TFIID rearrangement during assembly when the symmetric core accretes further TAFs and TBP, giving rise to the complete holo-complex that is asymmetric [57••].
Conclusions
Methodologies such as protein crystallography require large quantities of highly concentrated homogeneous sample to obtain high-resolution structural information. EM and single particle analysis methods do not require such high concentrations or total amounts of sample, but do still require high sample homogeneity, reasonable particle density on EM micrographs and optimal EM grid preparations. Native mass spectrometry that allows determination of subunit stoichiometry and insights into multimeric complex assembly processes is also dependent on highly concentrated, high quality sample in defined buffer conditions [58]. Highly homogenous specimens are likewise required for functional studies and for pharmaceutical applications. Recombinant expression enables to exactly specify the subunit composition of multiprotein complexes, thus increasing homogeneity. Further, engineering and mutating specific subunits offer the potential to test biological hypotheses, including examining the roles and locations of specific subunits within the context of the whole complex, which is a vital prerequisite for understanding biological function.
Most essential processes in cells are carried out by large multiprotein machines, and many of these are critically dependent on recombinant expression for analyzing their structure and function in detail. Eukaryotic complexes will in many cases require eukaryotic expression systems for their provision in the quality and quantity required. Remarkable advances in eukaryotic expression technologies that can provide these specimens are being made, with new and powerful systems being put at the disposal of the community. We anticipate that new expression tools including those described here will prove to be instrumental for the detailed study of many important protein complexes that have remained elusive to date.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We thank all members of our laboratories for their assistance and helpful discussions. The research contributions described were supported by Cancer Research, UK (to DB); the US National Science Foundation (NSF) grant MCB 1157688 and the Showalter Trust Fund (to YT); the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National pour la Recherche Scientifique (CNRS), the Association pour la Recherche sur le Cancer (ARC) and the Fondation pour la Recherche Médicale (FRM) (to PS); and the Agence Nationale de Recherche (ANR) and the European Commission (EC) Framework Programme 7 (FP7) projects PCUBE, BioSTRUCT-X, ComplexINC and 4DCellFate (to IB).
References
- 1•.Bieniossek C., Imasaki T., Takagi Y., Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem Sci. 2012;37:49–57. doi: 10.1016/j.tibs.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Overview description of the MultiBac BEVS for multiprotein expression, and its applications ranging from structural biology to vaccinology and gene therapy.
- 2.Kornberg R.D. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Boube M., Joulia L., Cribbs D.L., Bourbon H.M. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 4.Bourbon H.M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y.J., Bjorklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 6.Bjorklund S., Gustafsson C.M. The yeast Mediator complex and its regulation. Trends Biochem Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Guglielmi B., van Berkum N.L., Klapholz B., Bijma T., Boube M., Boschiero C., Bourbon H.M., Holstege F.C., Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conaway R.C., Sato S., Tomomori-Sato C., Yao T., Conaway J.W. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Malik S., Roeder R.G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firestein R., Bass A.J., Kim S.Y., Dunn I.F., Silver S.J., Guney I., Freed E., Ligon A.H., Vena N., Ogino S. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rump P., Niessen R.C., Verbruggen K.T., Brouwer O.F., de Raad M., Hordijk R. A novel mutation in MED12 causes FG syndrome (Opitz–Kaveggia syndrome) Clin Genet. 2011;79:183–188. doi: 10.1111/j.1399-0004.2010.01449.x. [DOI] [PubMed] [Google Scholar]
- 12.Graham J.M., Jr., Visootsak J., Dykens E., Huddleston L., Clark R.D., Jones K.L., Moeschler J.B., Opitz J.M., Morford J., Simensen R. Behavior of 10 patients with FG syndrome (Opitz–Kaveggia syndrome) and the p.R961W mutation in the MED12 gene. Am J Med Genet A. 2008;146A:3011–3017. doi: 10.1002/ajmg.a.32553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makinen N., Mehine M., Tolvanen J., Kaasinen E., Li Y., Lehtonen H.J., Gentile M., Yan J., Enge M., Taipale M. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 14.Berti L., Mittler G., Przemeck G.K., Stelzer G., Gunzler B., Amati F., Conti E., Dallapiccola B., Hrabe de Angelis M., Novelli G. Isolation and characterization of a novel gene from the DiGeorge chromosomal region that encodes for a mediator subunit. Genomics. 2001;74:320–332. doi: 10.1006/geno.2001.6566. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann R., Straussberg R., Mandel H., Fattal-Valevski A., Ben-Zeev B., Naamati A., Shaag A., Zenvirt S., Konen O., Mimouni-Bloch A. Infantile cerebral and cerebellar atrophy is associated with a mutation in the MED17 subunit of the transcription preinitiation mediator complex. Am J Hum Genet. 2010;87:667–670. doi: 10.1016/j.ajhg.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto S., Boissel S., Zarhrate M., Rio M., Munnich A., Egly J.M., Colleaux L. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science. 2011;333:1161–1163. doi: 10.1126/science.1206638. [DOI] [PubMed] [Google Scholar]
- 17.Cai G., Imasaki T., Takagi Y., Asturias F.J. Mediator structural conservation and implications for the regulation mechanism. Structure. 2009;17:559–567. doi: 10.1016/j.str.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis J.A., Takagi Y., Kornberg R.D., Asturias F.A. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Bjorklund S., Jiang Y.W., Kim Y.J., Lane W.S., Stillman D.J., Kornberg R.D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci U S A. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y.C., Kim Y.J. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol Cell Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y.C., Park J.M., Min S., Han S.J., Kim Y.J. An activator binding module of yeast RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19:2967–2976. doi: 10.1128/mcb.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conaway R.C., Conaway J.W. Origins and activity of the Mediator complex. Semin Cell Dev Biol. 2011;22:729–734. doi: 10.1016/j.semcdb.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson C.M., Koleske A.J., Chao D.M., Young R.A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 24.Hengartner C.J., Thompson C.M., Zhang J., Chao D.M., Liao S.M., Koleske A.J., Okamura S., Young R.A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 25.Taatjes D.J. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balamotis M.A., Pennella M.A., Stevens J.L., Wasylyk B., Belmont A.S., Berk A.J. Complexity in transcription control at the activation domain-mediator interface. Sci Signal. 2009;2:ra20. doi: 10.1126/scisignal.1164302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh S.S., Ansari A.Z., Ptashne M., Young R.A. An activator target in the RNA polymerase II holoenzyme. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 28.Takagi Y., Calero G., Komori H., Brown J.A., Ehrensberger A.H., Hudmon A., Asturias F., Kornberg R.D. Head module control of mediator interactions. Mol Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Cai G., Imasaki T., Yamada K., Cardelli F., Takagi Y., Asturias F.J. Mediator head module structure and functional interactions. Nat Struct Mol Biol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Imasaki T., Calero G., Cai G., Tsai K.L., Yamada K., Cardelli F., Erdjument-Bromage H., Tempst P., Berger I., Kornberg G.L. Architecture of the Mediator head module. Nature. 2011;475:240–243. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]; First crystal structure of the Mediator Head module from S. cerevisiae. The complex was expressed using a single baculovirus containing seven individual expression cassettes. The structure shows the subunit architecture at 4 Å resolution, revealing novel interaction motifs that stabilize the Mediator Head conformation.
- 31.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan M., Morgan D.O. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 33.Barford D Structural insights into anaphase-promoting complex function and mechanism. Philos Trans R Soc Lond B Biol Sci. 2011;366:3605–3624. doi: 10.1098/rstb.2011.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Schreiber A., Stengel F., Zhang Z., Enchev R.I., Kong E.H., Morris E.P., Robinson C.V., da Fonseca P.C., Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]; Structure analysis of complete APC/C from yeast. Recombinant production using two baculoviruses is described, one virus containing eight subunits, the other five subunits. Co-infection of insect cell cultures with the two viruses results in highly homogenous and active APC/C with a full complement of subunits. The structure is determined by hybrid methods including EM, use of crystal coordinates and native MS.
- 35.Zhang Z., Yang J., Kong E.H., Chao W.C., Morris E.P., da Fonseca P.C., Barford D. Recombinant expression, reconstitution and structure of human Anaphase Promoting Complex (APC/C) Biochem J. 2013;449:365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 36.Berger I., Fitzgerald D.J., Richmond T.J. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 37.Bieniossek C., Richmond T.J., Berger I. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr Protoc Protein Sci. 2008;5:20. doi: 10.1002/0471140864.ps0520s51. (chapter 5) [DOI] [PubMed] [Google Scholar]
- 38.Fitzgerald D.J., Berger P., Schaffitzel C., Yamada K., Richmond T.J., Berger I. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 39.Fitzgerald D.J., Schaffitzel C., Berger P., Wellinger R., Bieniossek C., Richmond T.J., Berger I. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure. 2007;15:275–279. doi: 10.1016/j.str.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Trowitzsch S., Bieniossek C., Nie Y., Garzoni F., Berger I. New baculovirus expression tools for recombinant protein complex production. J Struct Biol. 2010;172:45–54. doi: 10.1016/j.jsb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Vijayachandran L.S., Viola C., Garzoni F., Trowitzsch S., Bieniossek C., Chaillet M., Schaffitzel C., Busso D., Romier C., Poterszman A. Robots, pipelines, polyproteins: enabling multiprotein expression in prokaryotic and eukaryotic cells. J Struct Biol. 2011;175:198–208. doi: 10.1016/j.jsb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thornton B.R., Ng T.M., Matyskiela M.E., Carroll C.W., Morgan D.O., Toczyski D.P. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z., Kulkarni K., Hanrahan S.J., Thompson A.J., Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.da Fonseca P.C., Kong E.H., Zhang Z., Schreiber A., Williams M.A., Morris E.P., Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uzunova K., Dye B.T., Schutz H., Ladurner R., Petzold G., Toyoda Y., Jarvis M.A., Brown N.G., Poser I., Novatchkova M. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19:1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bitinaite J., Rubino M., Varma K.H., Schildkraut I., Vaisvila R., Vaiskunaite R. USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 2007;35:1992–2002. doi: 10.1093/nar/gkm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geu-Flores F., Nour-Eldin H.H., Nielsen M.T., Halkier B.A. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 2007;35:e55. doi: 10.1093/nar/gkm106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutchins J.R., Toyoda Y., Hegemann B., Poser I., Heriche J.K., Sykora M.M., Augsburg M., Hudecz O., Buschhorn B.A., Bulkescher J. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musacchio A., Salmon E.D. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 50•.Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]; Crystal structure of the mitotic checkpoint complex. Crystal structure of the fission yeast mitotic checkpoint complex. This structure explained the basis by which Mad2 and Mad3 act synergistically to inhibit Cdc20. The KEN degron binding site of Cdc20 is occupied by a KEN box mimic of Mad3, located in a helix-turn-helix motif that is stabilized and held in position on Cdc20 by Mad2. APC/C is further inhibited by disruption of the D box co-receptor (see Refs. [44, 51•]).
- 51•.Herzog F., Primorac I., Dube P., Lenart P., Sander B., Mechtler K., Stark H., Peters J.M. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper described the first EM structure of the APC/C with the mitotic checkpoint complex (MCC), showing how the MCC binds to the platform region of the APC/C. Together with Refs. [44, 50•], this explains how the MCC shifts Cdc20 away from Apc10, disrupting the D-box co-receptor.
- 52.Cler E., Papai G., Schultz P., Davidson I. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci. 2009;66:2123–2134. doi: 10.1007/s00018-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Müller F., Zaucker A., Tora L. Developmental regulation of transcription initiation: more than just changing the actors. Curr Opin Genet Dev. 2010;20:533–540. doi: 10.1016/j.gde.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Papai G., Weil P.A., Schultz P. New insights into the function of transcription factor TFIID from recent structural studies. Curr Opin Genet Dev. 2011;21:219–224. doi: 10.1016/j.gde.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright K.J., Marr MT2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci U S A. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papai G., Tripathi M.K., Ruhlmann C., Werten S., Crucifix C., Weil P.A., Schultz P. Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure. 2009;17:363–373. doi: 10.1016/j.str.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.Bieniossek C., Papai G., Schaffitzel C., Garzoni F., Chaillet M., Scheer E., Papadopoulos P., Tora L., Schultz P., Berger I. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]; Structure determination of the symmetric physiological core of human TFIID by hybrid methods. The complex was produced in insect cells by using a polyprotein strategy. Crystal structures and homology models were positioned into a high-resolution cryo-EM map. A structure of a complex containing two further subunits reveals subunit rearrangements leading to asymmetric holo-TFIID.
- 58.Sharon M., Robinson C.V. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu Rev Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]