Abstract
We synthesized on cellulose membranes 4942 ten-amino-acid peptides which included all of the sequences predicted for the severe acute respiratory syndrome (SARS) corona virus. We probed these membranes with four pairs of acute and convalescent sera from recovered SARS cases. We correlated positively reacting peptides with the in vitro SARS-CoV neutralizing activity of the samples. We found that convalescent sera with high neutralizing activity recognized exclusively only a limited number of peptides on the membranes. This suggests that antibodies against the epitopes represented by these peptides could be responsible for much of the SARS-CoV neutralizing activity. The findings have implications for monitoring humoral responses to SARS-CoV as well as for developing a successful SARS vaccine.
Keywords: Immune response, SARS vaccine, Peptide membranes
Introduction
A newly identified virus named SARS-CoV has been established as the etiological agent of severe acute respiratory syndrome (SARS). A total of 8422 probable cases of this highly infectious disease had been reported to WHO by August 2003 including 916 deaths (http://www.who.int/csr/sars/en/). The viral genome has been sequenced Marra et al., 2003, Rota et al., 2003. It consists of 29 751 nucleotides which contain 15 identifiable open reading frames (AY274119).
Peptides incorporating all of the sequences predicted in the open reading frames of the SARS-CoV genome were prepared on derivatized cellulose membranes using a robotic peptide synthesizer (Autospot ASP 222, Intavis Bioanalytical Instruments, Lagenfeld, Germany). The peptides were 10 amino acids long and overlapped by eight residues. Each peptide on a membrane was therefore shifted from the one previous by two amino acids towards the C-terminal end. The arrangement of the 4942 peptides on sets of four membranes covering the 15 open reading frames (Orfs) is illustrated in Fig. 1 .
Fig. 1.
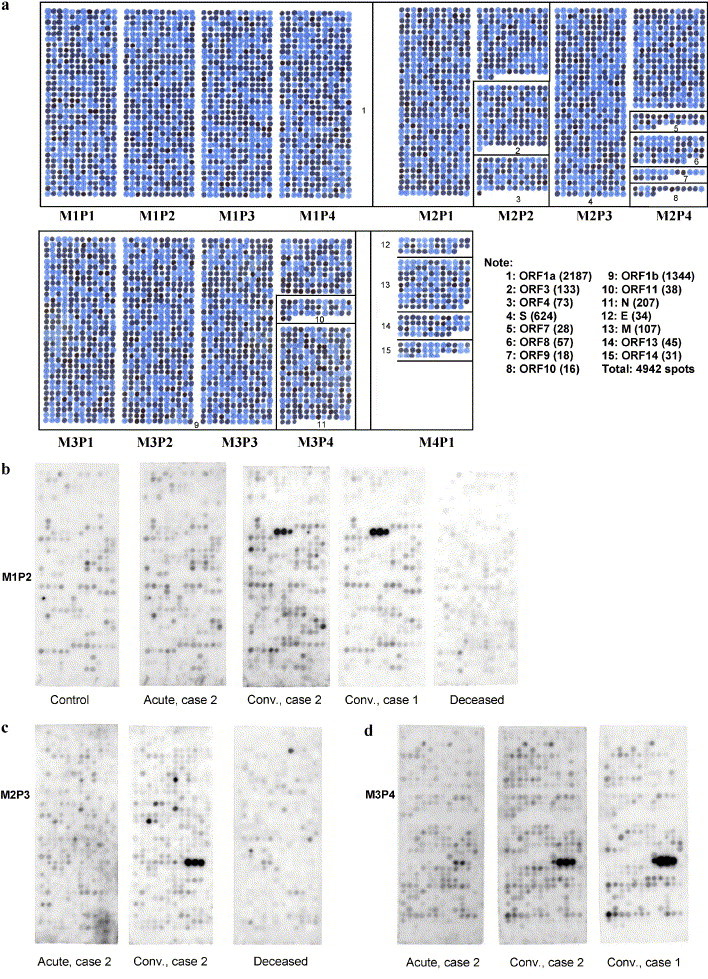
(a) Outline of the overlapping peptide set covering membranes M1, M2, M3, and M4. Panels on the membranes are designated from left to right P1, P2, P3, and P4. Numbers in the boxes designate the location of each open reading frame. The note to the right of M4P1 shows the key to the Orfs with peptide totals in brackets. This membrane was spotted with dye instead of amino acids. In actual membranes, each spot is a 10-amino-acid peptide with adjacent spots being shifted by two amino acids. Characterization of the immune response against these single case epitopes promises to provide important insights into their role in the resolution of infection. However, epitopes recognized by multiple convalescent sera may be the most important targets of neutralizing antibodies. (b–d) Examples of membrane panels probed with various serum samples and developed with peroxidase-labeled goat antihuman IgA; (b) M1P2 probed with control serum, acute as well as convalescent serum from case 2, convalescent serum from case 1, and serum from the deceased case. Notice the triad of spots recognized only in the serum of the two convalescent cases. The peptide sequences from Orf 1a are SDDYIKLNGP, DYIKLNGPLT, and IKLNGPLTVG. (c) P3M2 probed with acute and chronic serum from case 2 and serum from the deceased case. Panel 3 has peptides from S-protein. Notice the triad of spots recognized only by the convalescent serum. The peptide sequences from S-protein are FQPFQQFGRD, PFQQFGRDVS, and QQFGRDVSDF. (d) P4M3 probed with acute and convalescent serum from case 2 and convalescent serum from case 1. Notice the triad of spots recognized in the two convalescent sera. The peptide sequences from N-protein are QLPQGTTLPK, PQGTTLPKGF, and GTTLPKGFYA.
Membranes were probed with pairs of acute and convalescent sera from four cases who recovered from a SARS infection. As controls, serum from one case that failed to survive, and one from a healthy, nonexposed volunteer was utilized. Samples were diluted 100-fold in buffer and then applied to a membrane for 1 h at 37 °C. The membranes were developed with horseradish peroxidase-conjugated goat antibodies against human IgG, IgM, or IgA. Positive spots were then identified following treatment with ECL chemiluminescence reagents.
Sera were tested for the presence of antibodies against SARS-CoV in a standard virus neutralizing test. Serial 2-fold dilutions of each serum from 1/8 to 1/1024 were incubated with 100 plaque-forming units of SARS-CoV (Tor-2 isolate, Marra et al., 2003) for 2 h and then added to monolayers of Vero E6 cells. The cultures were examined after 72 h for the presence of characteristic cytopathic effects. The dilution before the one at which cytopathic effects were first noted was recorded as the antivirus titer. All sera, which showed evidence of antivirus activity, were retested following heat treatment at 56 °C for 30 min. This process confirmed the presence of neutralizing antibodies and eliminated the possibility of nonspecific, heat-sensitive factors contributing to the neutralizing activity.
Results and discussion
The results of neutralization assays for the various sera are shown in Table 1 , along with the age and sex of the respective patients. Acute sera were collected on presentation, and convalescent sera collected 1 month later in case 1, and at least 2 weeks later in cases 2, 3, and 4. The acute serum of case 3 had a neutralizing antibody titer of 1/8, which indicates the beginning of an immune response. Other acute sera as well as the acute serum of the deceased case taken 7 days after illness onset, and the healthy control, had no detectable neutralizing activity.
Table 1.
Neutralization titers of sera from recovered SARS cases plus a deceased case and a nonexposed control
| Recovered cases | Acute | Convalescent |
|---|---|---|
| Case 1 (F, 50) | <1/8 | 1/512 |
| Case 2, (F, 45) | <1/8 | 1/32 |
| Case 3, (M, 49) | 1/8 | 1/64 |
| Case 4, (F, 65) | <1/8 | 1/128 |
| Deceased case (F, 77) | <1/8 | |
| Control case (M, 76) | <1/8 |
Age and sex of the cases are shown in brackets.
These data indicate that the four recovered cases developed antibodies with viral neutralizing potency between the time of acute and convalescent serum sampling. Therefore, those peptides strongly recognized on membranes probed with convalescent sera, but not with acute or control sera, should be the most immunodominant and may include SARS-CoV epitopes that are vulnerable to neutralization by antibody. This analysis is limited to linear epitopes since peptides on the membranes do not replicate conformational epitopes or those that develop from posttranslational modifications such as glycosylation.
Three examples of membranes reacted with acute and convalescent sera and developed with anti-IgA antibodies are shown in Fig. 1. Fig. 1b compares the second panel of membrane 1 probed with serum from the unexposed control, the deceased case, convalescent serum from case 1, and acute and convalescent serum from case 2. The panel has a total of 408 peptides encoded from the Orf 1a open reading frame (see Fig. 1a). No spots were strongly immunostained except for a single triad in the two convalescent sera. The peptides in the triad have the following sequences: SDDYIKLNGP, DYIKLNGPLT, and IKLNGPVG. The sequence IKLNGP is common to all three. Fig. 1c compares the third panel of membrane 2 probed with the acute and convalescent serum of case 2, and the deceased case. The panel has 408 peptides encoded from the S-protein open reading frame. One triad of spots was immunopositive, in the convalescent, but not the acute serum. The triad has the following sequences: FQPFQQFGRD, PFQQFGRDVS, and QQFGRDVSDF. The common sequence is QQFGRD. Fig. 1d compares the fourth panel of membrane 3 probed with acute and convalescent serum of case 2, and the convalescent serum of case 1. The bottom half of the membrane has peptides encoded for the complete nucleocapsid (N) protein open reading frame. One triad was immunopositive in the two convalescent sera but was not positive in the corresponding acute serum of case 2. The triad has the following sequences: QLPQGTTLPK, PQGTTLPKGF, and GTTLPKGFYA with the common sequence being GTTLPK.
Shown in Table 2 are the 24 overlapping membrane peptides that were recognized exclusively, or much more strongly, in multiple pairs of convalescent compared with the respective acute sera. None of these peptides was recognized by the serum from the deceased case or the healthy control. There were two overlapping sequences from Orf 1a, one from Orf 1b, six from N-protein, eleven from S-protein, two from M-protein, one from E protein and one from Orf 10. The remaining open reading frames were not reactive in more than one serum sample. In the table, those peptides listed with a sequence of 14 amino acids were identified as a combination of three adjacent spots on the membranes, whereas those listed with 12 were identified as a combination of two adjacent spots. Common sequences therefore include residues 5–10 of the 14-amino-acid peptides and 3–10 of the 12-amino-acid peptides.
Table 2.
Overlapping membrane peptides recognized in SARS convalescent sera by antibodies to IgG, IgM, and IgA*
| Orf1a |
| QVASDNIKDCVKCF (case 1 IgG; case 4 IgM) aa 665–678 |
| SDDYIKLNGPLTVG (cases 1, 3, 4 IgA) aa 1065–1078 |
| Orf1b NQDVNLHSSRLS (case 2 IgA, IgM; case 4 IgA) aa 4725–4736 |
| Nucleocapsid (N)-protein |
| QLPQGTTLPKGFYA (cases 1, 2 IgG; cases 1, 3, 4 IgA) aa 161–174 |
| PTVTLLPAADMDDF (case 1 IgG; cases 3, 4 IgM) aa 391–404 |
| QPLPQRQKKQPT (case 3 IgG; case 1 IgG) aa 381–392 |
| YKTFPPTEPKKD (cases 1, 3, 4 IgA) aa 361–372 |
| GGSQASSRSSSR (cases 1, 2 IgG; case 2 IgM) aa 179–190 |
| IRQGTDYKHWPQ (cases 1, 2 IgG; case 1 IgM) aa 293–304 |
| Spike (S)-protein |
| SDTLYLTQDLFLPF (cases 1, 2 IgM) aa 49–62 |
| IDKGIYQTSNFR (cases 1, 3 IgA) aa 295–306 |
| CPFGEVFNATKF (cases 2, 4 IgA) aa 323–334 |
| CTPPALNCYWPLND (case 1 IgG, IgM; case 2 IgA) aa 467–480 |
| FQPFQQFGRDVSDF (cases 3, 4 IgA) aa 545–558 |
| RDVSDFTDSVRD (case 4 IgG, IgM) aa 553–564 |
| PIGAGICASYHT (case 2 IgG; cases 1, 2, 4 IgA; cases 1, 3, 4 IgM) aa 651–662 |
| VSLLRSTSQKSI (cases 2, 4 IgA; cases 2, 3 IgM) aa 663–674 |
| AIPTNFSISITTEV (case 4 IgG; cases 1, 2, 4 IgA; cases 1, 3, 4 IgM) aa 695–708 |
| QYGSFCTQLNRA (case 3 IgG; case 1 IgM) aa 737–748 |
| PFAMQMAYRFNG (cases 2, 3, IgM) aa 879–890 |
| Membrane (M)-protein |
| KEITVATSRTLS (case 1 IgG; cases 1, 2, 3, 4 IgA; case 1 IgM) aa 165–176 |
| GTITVEELKQLL (cases 3, 4 IgG; case 2 IgA; cases 1, 2, 3 IgM) aa 5–16 |
| E-protein |
| TVYVYSRVKNLNSSEG (cases 1, 2, 3, 4 IgG; cases 1, 2 IgA; case 2 IgM) aa 55–70 |
| Orf10 |
| HVLEDPCKVQH (cases 1, 2 IgA) aa 28–39 |
Sequences with 14 amino acids represent three adjacent peptides with a common sequence from residues 5 to 10. Sequences with 12 amino acids represent two adjacent peptides with a common sequence from residues 3 to 10. The amino acid positions (aa) in SARS-CoV open reading frames are taken from BLAST at NCBI: http://www.ncbi.nlm.nih.gov/blast/Blast.cgi.
Additional peptides to those shown in Table 2 were recognized only by convalescent serum from one of the four cases. Among these single case examples were peptides from Orf 3, Orf 4, Orf 9, Orf 13, and Orf 14, indicating that the virus expresses these proteins and that antibodies against certain of their epitopes are being produced. Characterization of the immune response against these single case epitopes promises to provide important insights into their role in the resolution of infection. However, epitopes recognized by multiple convalescent sera may be the most important for developing antibodies that will protect against the virus.
Our knowledge of the functions of the SARS-CoV proteins is mostly derived from analogies with those of other well-characterized corona viruses. Over 21 kb of the total genome is taken up by Orf 1a and Orf 1b, which are presumed to encode for proteins such as proteases and polymerases that are associated with virus replication Marra et al., 2003, Ruan et al., 2003. The genome is sufficiently unique to be proposed as belonging to a separate group designated as Group IV Marra et al., 2003, Ruan et al., 2003, but this view is not consensual and it has been noted that SARS-CoV is most closely related to group 2 coronaviruses (Snijder et al., 2003). Nevertheless, the structural proteins S, M, N, and E have been clearly identified in virus grown in infected cells Marra et al., 2003, Ruan et al., 2003. The spike, or S-protein, protrudes from the outer surface and is a strong candidate for facilitating viral entry into host cells.
Li et al. (2003) have reported that the SARS-CoV S-protein efficiently binds to angiotensin converting enzyme 2 of SARS-CoV permissive Vero E6 cells, suggesting that this binding is a key step in SARS-CoV infectivity. Interaction between the S-protein of a corona virus and specific mammalian receptors can determine species sensitivity. For example, when the S-protein ectodomain of mouse corona hepatitis virus was replaced with that of the feline infectious peritonitis virus, the viral particles were no longer infective for mouse cells, but became infective for feline cells (Kuo et al., 2000). SARS-CoV has been reported to infect domestic cats and ferrets (Martina et al., 2003) so these relationships may have relevance to this broader species susceptibility. A further example of interaction between the S-protein and mammalian receptors involves CD13 or aminopeptidase N. It is the main mammalian receptor for the S-protein of many coronaviruses. Substitution on human cells of the human CD13 N-glycosylation site with the porcine N-glycosylation site resulted in a failure to bind human corona virus-229E (Wentworth and Holmes, 2001).
Antibodies directed against the 11 epitopes of S-protein identified in Table 2 are therefore potential candidates for neutralizing activity. Antibodies against M-protein and E-protein epitopes could also be important although they are smaller and are believed to be less exposed. The protein encoded by Orf 10 may be displayed on the viral surface and an immune response to it may contribute to the resolution of disease.
Antibodies against Orf 1a, Orf 1b, and N-Protein probably are unlikely to have contributed to the in vitro neutralization results. N-protein is internal and is probably inaccessible in the brief exposure time of the neutralization test, and the products of Orf 1a and Orf 1b are not believed to be structural proteins. Nevertheless, they may be very important to in vivo protection.
Given the complexity of antibodies produced in each convalescent serum, it would be difficult to predict what the relative contribution of each to the overall neutralizing activity might be, particularly when antibodies against conformational or carbohydrate-dependent epitopes might be present. Nevertheless, high titers of those consistently represented in the various convalescent sera might be sufficient to confer immunity.
Numerous strategies have been suggested for producing a SARS vaccine (DeGroot, 2003). One possibility is to use a combination of peptides, such as those listed in Table 2, as a polyvalent antigen. Peptide-based vaccines have heretofore had only limited success but this can be attributed to a lack of knowledge as to which peptides to use. Such uncertainty is reduced by analyzing the antibody pattern in sera from cases that have successfully resolved the infection. Peptides making up the antigen can be screened in advance for homologies to mammalian proteins, reducing the possibility of self-attack. They can also be easily modified to cope with viral mutations that could escape traditional vaccines. Significant mutations have already been identified in SARS-CoV Ruan et al., 2003, Martina et al., 2003, indicating the necessity of developing adaptable vaccines.
Polypeptide vaccines should be safe, highly stable, and easy to administer. Antibodies cloned against particularly sensitive peptides could be used as therapeutic agents. The type of analysis described here may have general utility in designing peptide vaccines and therapeutic antibodies against other infectious agents such as HIV.
Methods
Cellulose-bound overlapping peptides (10 mers) derived from all proteins encoded by the 15 Orfs of the SARS genome were synthesized using an AutoSpot robot provided by Intavis Bioanalytical Instruments. The software used for designing the arrays was from DIGEN, Jerini Biotools GmbH, Berlin, Germany. Membranes derivatized with a polyethylene (PEG) linker and a free amino terminal, as well as 9-fluorenylmethyloxycarbonyl (fmoc) amino acids were obtained from Intavis Bioanalytical Instruments. The fmoc amino acids (0.25 M) were dissolved in dimethylformamide (DMF, Sigma, Oakville, Ont. Canada) and then activated with 1-hydroxybenzotriazole (HOBT, Sigma) and N,N′-disopropylcarbodiimide (DIPC, Sigma) for at least 15 min. They were then delivered to the membrane in 60-nl aliquots per spot by the robotic synthesizer. Fifteen minutes after completion of each cycle, the membranes were removed from the apparatus and treated with 2% acetic anhydride in DMF to acetylate any free remaining amino groups. They were then washed and further treated with 20% piperidine in DMF to remove the fmoc protecting group. This was followed by washing with DMF and methanol, and finally by air drying. The membrane was then precisely repositioned on the robotic apparatus to initiate the next coupling cycle. After the final cycle, the side chain protecting groups were removed by treatment with a solution of the following composition: 5 ml of 50% trifluoracetic acid; 5 ml dichloromethane (DCM); 300 μl triisopropyl silane (Sigma); and 200 μl water. Membranes were then washed twice with DCM, twice with DMF, and twice with methanol. After air drying, membranes were stored in a sealed bag at 4 °C until used.
Membranes were rehydrated by treating first with methanol, then 50% methanol followed by washing three times in 50 mm Tris-buffered saline/0.2% Tween 20 (TBS-T). They were then blocked with 5% skim milk in TBS-T overnight at 4 °C. After washing three times with TBS-T, the membranes were incubated at 37 °C for 1 h with 1/100 dilutions of serum samples of patients and controls. After washing, membranes were treated with 1/5000 dilutions (0.2 μg/ml) of goat antihuman IgA, IgG and IgM antibodies conjugated to horseradish peroxidase (Sigma). After washing in TBS-T, membranes were developed with an ECL chemiluminescence kit provided by Amersham Pharmacia Biotech (Buckinghamshire, England). Four milliliters of the two ECL Western blotting detection reagents were mixed before being applied on the membranes. The membranes were scanned using a Bio-Rad Fluorescent Imager (Hercules, CA) and saved for later analysis. The membranes were regenerated by incubation in 8 M urea/1% SDS overnight followed by 60 min in 50% ethanol/10% acetic acid. After this step, membranes were washed three times in methanol, and after air drying stored at 4 °C in a sealed bag until reuse.
In initial development of the SARS-CoV serum neutralization test, comparisons were made between the microneutralization test and a 50% plaque reduction assay. Testing of several sera consistently produced assay results where the 50% plaque reduction titer was 2-fold greater than in the microneutralization test. Following this validation, the microneutralization test system was adopted for greater convenience despite the 2-fold greater sensitivity of the plaque reduction assay. Typical results of this assay are illustrated in Fig. 2 .
Fig. 2.
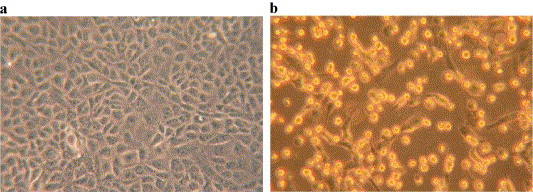
(a) Vero E6 cells were imaged before infection and (b) 32 h after infection with SARS-CoV at a multiplicity of 1 pfu per cell. Notice the extensive cell damage in b.
Acknowledgements
The authors are grateful to Drs. A McGeer and D. Skowronski for help in obtaining specimens and to Dr. K Karunakarum for initial assistance in processing the membranes. We are also grateful to Dr. Mitoshi Kunimatsu of Nagoya City University for help in programming the robotic synthesizer. This research was supported by a CIHR grant on which M.P. is a co-investigator, a grant to P.L.M. from the Province of British Columbia via the SARS Accelerated Vaccine Initiative (SAVI), and support from individual British Columbians to the Kinsmen Laboratory of Neurological Research.
References
- DeGroot A.S. How the SARS vaccine effort can learn from HIV-speeding towards the future, learning from the past. Vaccine. 2003;21:4095–4104. doi: 10.1016/S0264-410X(03)00489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasllleva N., Sul J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzurlaga K., Greenough T.C., Choe N., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Martina B.E.E., Hagmans B.L., Kuiken T., Fouchier R.A.M., Rimmelzwaan G.F., Amerongen G.V., Peiris J.S.M., Lim W., Osterhaus A.D.M.E. SARS virus infection of cats and ferrets. Nature. 2003;425:815. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Ruan Y.J., Wei C.L., Ee A.L., Vega V.B., Thoreau H., Su S.T., Chia J.M., Ng P., Chiu K.P., Lim L., Zhang T., Peng C.K., Lin E.O., Lee N.M., Yee S.L., Ng L.F., Chee R.E., Stanton L.W., Long P.M., Liu E.T. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Brendenbeek P.J., Dobbe J.C., Theil V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth D.E., Holmes K.V. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J. Virol. 2001;75:9741–9752. doi: 10.1128/JVI.75.20.9741-9752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


