Abstract
In this study, 60 pigs with clinical signs of post-weaning multisystemic wasting syndrome (PMWS) from 20 different pig herds and 180 control pigs (without clinical signs of PMWS) were examined to get more insights into the frequencies of porcine circovirus 2 infections and the presence of co-infections in pigs with and without clinical signs of PMWS in the Netherlands.
Porcine circovirus type 2 was detected in 100% of the pigs with clinical signs of PMWS by virus isolation and/or PCR and in 50% of the pigs from PMWS-free herds. There was an association between the levels of infectious PCV2 and/or PCV2 DNA load and the severity of clinical signs as described for PMWS. A high variation in PCV2 antibody titres was found in the clinically affected pigs, and 27% of these pigs did not mount PCV2 antibody titres higher than 1:200. A concurrent infection of PCV2 and porcine reproductive and respiratory syndrome virus (PRRSV) was found in at least 83% of the pigs with clinical signs of PMWS and in 35% of the pigs from PMWS-free herds. Co-infections of European- and American-type PRRSV were detected only in PMWS herds and in one control herd with a history of PMWS clinical signs.
Abbreviations: PCV2, porcine circovirus type 2; PMWS, postweaning multisystemic wasting syndrome; PRRSV, porcine reproductive and respiratory syndrome virus
Keywords: Porcine circovirus type 2, PCV2, PMWS, PRRSV-EU and US
1. Introduction
Post-weaning multisystemic wasting syndrome (PMWS) causes considerable economic losses in pig production and the disease has been associated with porcine circovirus type 2 (PCV2) infections (Allan et al., 1999a). Under experimental conditions, the reproduction of PMWS in pigs infected with PCV2 alone was difficult to achieve, and PCV2 infected pigs developed only mild to moderate PMWS specific lesions (Balasch et al., 1999; Krakowka et al., 2000). Serological studies showed that antibodies directed against PCV2 were detected in pigs of more than 94% of the Dutch pig herds (Elbers and de Jong, 2002), but not all PCV2 infected pigs will develop PMWS. This indicated that the course of most PCV2 infections is more or less sub-clinical and that PCV2 may need co-factors to fully reproduce PMWS. Data from experimental co-infections with porcine parvovirus (PPV) (Allan et al., 1999b; Ellis et al., 2000; Krakowka et al., 2000) or PRRSV (Harms et al., 2001) supported this theory. Furthermore, the stimulation of the immune system might be a pivotal event in the production of PMWS in PCV2 infected pigs (Krakowka et al., 2001), although contradictory data have also been generated regarding this subject (Ladekjaer-Mikkelsen et al., 2002).
Field studies showed that PRRSV can be detected often in PMWS affected pigs (Sorden et al., 1998; Segalés et al., 2002), and under experimental conditions severe disease can be reproduced in pigs concurrently infected with PCV2 and PRRSV (Harms et al., 2001; Rovira et al., 2002; Stockhofe-Zurwieden et al., 2003). However, whether these findings are relevant for the field situation remains to be established as no reports have been published on the frequency of PRRSV infections in PMWS herds versus PMWS-free control herds, and there is no information on the frequency of co-infections of European- (EU) and American (US)-type PRRSV in PMWS herds and PMWS-free control herds.
A case-control study was performed to get more insights into the presence of co-infections, such as infections of EU- and US-type PRRSV, in pig herds with (cases) and without (controls) clinical signs of PMWS in the Netherlands.
2. Materials and methods
2.1. Herds and pigs
From 20 different pig herds in the Netherlands, 3 pigs each diagnosed for clinical signs of PMWS (cases) and 8 to approximately 14 weeks of age, were selected and examined postmortem (n=60). Criteria for the selection of cases were the combination of wasting, pallor, fever (>39.7 °C), respiratory distress and enlarged inguinal lymph nodes as observed by palpation. For each case, control pigs of the same age as cases but without clinical signs of PMWS were also selected and examined postmortem. Control pigs were selected: (1) housed within the same compartment as cases (n=60), (2) from a different compartment but within the same case herd (n=60), and (3) from another pig herd within the same geographical region as the case herd, and without a recent history of PMWS and porcine dermatitis and nephropathy syndrome (PDNS) (n=60).
2.2. Macroscopic lesions and sample collection
At necropsy, each pig was examined for gross pathological findings, and at least four different organs and tissues e.g. lung, mesenteric and inguinal lymph nodes, spleen and kidney were sampled for virology and bacteriological examinations. For histological examinations, parts of e.g. kidney, lung and mesenteric and inguinal lymph nodes were fixed in 10% neutral buffered formalin, in paraffin embedded, sectioned at 5 μm, and stained with haematoxylin and eosin (HE). Blood was collected from all pigs and serum was used for antibody detection (stored at −70 °C).
2.3. Virus isolation
Small blocks of organs and tissues from each pig (lung, mesenteric and inguinal lymph nodes, spleen and kidney) were selected and of each organ or tissue a 10% organ suspension was prepared in Earle's minimal essential medium (EMEM). Suspensions were homogenised and subsequently clarified by centrifugation at 1500g for 10 min (4 °C). Porcine circovirus 2 isolation was performed on PK-15 cells and DULAC cells and after incubation for 3–4 days at 37 °C (5% CO2), a serum sample, collected from a specified pathogen free (SPF) pig infected with PCV2 only, was used for the detection of viral antigens in infected cell cultures. The immunoperoxidase monolayer assay (IPMA) was further performed as described earlier (Wellenberg et al., 2000), using a HRPO-labelled rabbit anti-swine-Ig conjugate (Dakopatt, Glostrup, Denmark). Non-inoculated PCV-free PK-15 cells and DULAC-cells served as negative cell control and an organ pool suspension collected from a PMWS affected pig served as positive control.
2.4. DNA and RNA extractions and PCR tests for the detection of viral genomic sequences
Total DNA and total RNA from lung, mesenteric and inguinal lymph nodes, spleen and kidney were extracted from 10% organ suspensions by column chromatography using the QIAamp Blood and Tissue kit (Qiagen, Westburg, The Netherlands) for DNA and the High Pure RNA Isolation kit for RNA (Roche Diagnostics, Germany). Cell lysis, precipitation, and elution of DNA or RNA were performed as recommended by the manufacturer. The elution products were stored at −20 °C. PCRs were performed for the detection of PCV2 (Wellenberg et al., 2000) and porcine parvovirus (PPV) DNA (Soares et al., 1999), and an rt-PCR for the detection of EU and US type PRRSV RNA. The type of PRRSV strain (EU or US) was determined by fragment-length rt-PCR (Oleksiewicz et al., 1998) and sequence analyses.
2.5. Quantitative PCV2 PCR
To quantify the amount of PCV2 DNA copies in organ samples, a real-time fluorescent-probe PCR was used. The Light-Cycler probes (LC red 640 – ATC TCA TCA TGT CCA CCG CCC AGG A) (FL fluorescein – CGT TGT ACT GTG GTA CGC TTG ACA GT) and the primers (1391; 5′-CTC CCC TGT CAC CCT GGG TG -3′) and (1577; 5′-CTC TCC CGC ACC TTC GGA TAT-3′) amplifying a 186-bp fragment from the cap gene of PCV2 were designed. The Light-Cycler PCV2 PCR was performed in a total volume of 20μl containing a 10μl aliquot of DNA preparation. Final concentrations were 1× standard buffer (containing dNTPs and Taq polymerase (Roche Diagnostics GmbH, Mannheim, Germany)), 4 mM magnesium chloride, 0.5 μM of each primer and 0.15 μM of each probe. Cycling parameters were 95 °C for 10 min and 45 cycles at 95 °C for 10 s, 58 °C for 12 s, and 72 °C for 20 s. For standard curve, serial dilutions of plasmid (PCV2 cloned in pCR21) of 1–1011/μl copies were used.
2.6. Antibody detection
For the detection of porcine circovirus type 1 (PCV1) or PCV2 antibodies in porcine sera immunoperoxidase monolayer assays (IPMA) were used (Wellenberg et al., 2000). Briefly, PCV1 or PCV2 infected PK-15 cells were formaldehyde-fixed and pre-incubated with a solution of 10% Triton X-100 and additionally with 10% horse serum in phosphate-buffered salt solution. The antibody titres were determined by preparing twofold dilutions in 10% horse serum in phosphate-buffered salt solution starting at a dilution of 1:100. Serum dilutions were incubated for 1 h at 37 °C. The IPMA was further performed as described above, using a HRPO-labelled rabbit-anti-swine-Ig conjugate (Dakopatt, Glostrup, Denmark).
ELISAs were used for the specific detection of PPV (Cedi-Diagnostics, Lelystad, the Netherlands) and PRRSV (Idexx, USA) antibodies. ELISAs were performed according to the instructions of the manufacturers. Additionally, an ELISA has been developed to discriminate between antibodies against PRRSV-EU and PRRSV-US (ID-Lelystad, the Netherlands). In addition, 20 randomly chosen serum samples from PMWS cases and controls from PMWS-free herds were screened for the presence of antibodies against other viral infections such as the coronaviruses; porcine epidemic diarrhoea virus (PEDV), transmissible gastroenteritis virus (TGEV) and porcine respiratory coronavirus (PRCV), the influenza viruses H1N1, H1N2, and H3N2. This to get an impression on the occurrence of other (endemic) virus infections in case herds and PMWS-free herds.
2.7. Bacteriology
All affected organs at necropsy were investigated for the presence of pathogenic bacteria, e.g. Haemophilus spp., Mycoplasma spp. Fifteen randomly chosen faeces samples were collected for the detection of Salmonella spp. Standard procedures were used for the detection of bacteria in affected organs and tissues.
2.8. Epidemiological data
Data on the PMWS and PDNS history, and the use of vaccines against e.g. PRRSV and Mycoplasma hyopneumoniae were collected from each case and control herd.
2.9. Statistical methods
Statistical analysis (χ 2) was performed to compare; (a) the proportion of PCV2 positive pigs (by virus isolation and/or PCR) within the case group with the proportion of PCV2 positive control pigs in the 3 control groups, (b) the proportion of cases with high PCV2 DNA loads in mesenteric and inguinal lymph nodes (> 105 copies/μl) with the proportion of control pigs within the three control groups with high PCV2 DNA loads, and (c) the proportion of PRRSV infected pigs (rt-PCR and/or ELISA antibodies positive) within the case group with the proportion of PRRSV positive control pigs within the three control groups.
Fisher exact test was performed to compare the proportion of co-infections of PRRSV-EU and -US in case herds with that of the PMWS-free control herds.
3. Results
3.1. Macroscopic and histopathological lesions
Enlargement of the lymph nodes and spleen, pneumonia and kidneys with white spots were the most dominant macroscopic changes in cases (Fig. 1 ). In most of the cases, pneumonia was characterised by mild to severe interstitial oedema, firm tissue with decreased retraction and focal catarrhal or fibrous inflammation. Stomach ulcers were observed in 32% of the PMWS cases. Histopathological examinations of mesenteric and inguinal lymph nodes revealed lymphocytic depletion in follicular and parafollicular areas in most but not in all of the examined pigs with clinical signs of PMWS. Gross lesions consisted of hyperplasia, probably of the follicular dendritic cells in the follicular areas, were also found in lymph nodes from pigs with clinical signs of PMWS. Most of these lesions were characterised histologically by infiltration of mononuclear cells often in combination with the presence of giant cells. Interstitial pneumonia with bronchovasculo-interstitial infiltrations of leukocytes was a dominant histopathological finding in pigs with clinical signs of PMWS. In 76% of the kidneys from pigs with clinical signs of PMWS an interstitial nephritis was found, which varied in severity.
Fig. 1.
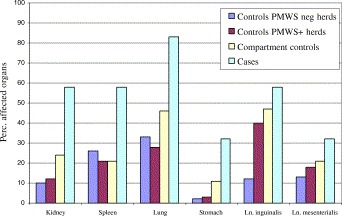
Frequencies of macroscopic lesions in organs from controls of PMWS-free herds, from controls within case herds but from another compartment as cases, controls within the same compartment as cases, and cases, respectively (n=60 per group).
3.2. Porcine circovirus type 2
In 87% of the 60 cases, moderate to high levels of infectious PCV2 were detected in lung, mesenteric and inguinal lymph nodes, spleen and kidney by virus isolation on PK-15 cells, and DULAC cells as well (Table 1 ). In the remaining 13% of the cases no infectious or low levels of infectious PCV2 were detected. However, high levels of PCV2 DNA (> 105 copies/μl organ suspension) were found in lymph nodes of these pigs by the quantitative real-time PCV2 PCR, and in sera of 5 of these 8 cases high antibody titres against PCV2 (>1:102400) were detected.
Table 1.
Detection of PCV2 in cases and in pigs within the three control groups by virus isolation (VI) and PCR (n=60 per group)
| Group | PCV2 (VI) | PCV2 (VI and PCR) |
|---|---|---|
| Cases | 87% | 100% |
| Compartment controls | 73% | 87% |
| Controls within PMWS + herd but from another compartment | 53% | 67% |
| Controls from PMWS-free herds | 30% | 50% |
Infectious PCV2 was detected by virus isolation in 30% of the control pigs from PMWS-free herds, but the number of PCV2 infected PK-15 or DULAC cells after inoculation with organ suspensions from control pigs was much lower than the number of PCV2 infected PK-15 or DULAC cells after inoculation with organ suspensions from cases (data not shown). PCV2 DNA was detected in 100% of the examined cases and in 50% of the controls from PMWS-free herds. The mean 10log PCV2 DNA copy number in mesenteric and inguinal lymph nodes was higher in cases than in the control groups, and at least 105 times higher than the mean 10log PCV2 DNA copy number in controls of PMWS-free herds (Table 2 ).
Table 2.
PCV2 DNA load in 10% organ suspensions of mesenteric and inguinal lymph nodes from cases and pigs within the three control groups
| Group | n | PCV2 DNA load ranged from: (copies/μl) | PCV2 load >105 copies/μl(%) | Mean 10log PCV2 DNA (copies/μl) |
|---|---|---|---|---|
| Cases | 43 | 103–2 × 1010 | 88 | 7.4 |
| Compartment controls | 27 | <10–2 × 109 | 48 | 5.1 |
| Controls PMWS + herds but from another compartment | 15 | <10–1 × 108 | 47 | 3.6 |
| Controls from PMWS-free herds | 35 | <10–6 × 108 | 17 | 2.1 |
In 6 (17%) of the 35 investigated lymph nodes from controls of PMWS-free herds, the PCV2 DNA loads were between 105 and 6 × 108 DNA copies/μl organ suspension. In 4 of these 6 control pigs without clinical signs of PMWS, mild to moderate macroscopic changes such as white spotted kidneys, pneumonia, enlarged spleen, and/or enlarged and haemorrhagic lymph nodes were observed.
Statistical analysis (χ 2) revealed that the proportion of PCV2 positive pigs (by virus isolation and/or PCR) within the case group was significantly higher than the proportion of PCV2 positive control pigs; (a) housed within the same compartment (p=0.003), (b) from other compartments within case herds (p=0.0000), and (c) from PMWS-free herds (p=0.0000). χ 2 Analysis also showed significant differences between the proportion of cases with high PCV2 DNA copy loads in mesenteric and inguinal lymph nodes (>105 copies/μl) and the proportion of control pigs within the three control groups with high PCV2 DNA copy loads (p<0.001).
In 27% of the cases no PCV2 antibodies could be detected by IPMA, or the PCV2 antibody titre was very low (<1:400). The PCV2 antibody titres in 61% of the cases varied between 1:400 and approximately 1:102400, while very high PCV2 antibody titres of ⩾ 1:409600 were found in the remaining cases (Fig. 2 ). In herds from these latter cases, also pigs showing signs of PDNS were observed.
Fig. 2.
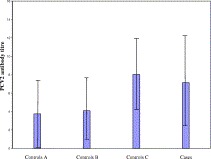
Mean PCV2 antibody titres in cases versus controls (controls from PMWS-free herds (a), controls within case herds but from another compartment as cases (b), controls within the same compartment as cases (c), and cases, respectively) (n=60 per group). SD are expressed per group as lines. PCV2 antibody titres are expressed as twofold dilution steps starting a serum dilution of 1:100 (i.e. “1”= 1:100 dilution and “12”=1:204.800).
3.3. Porcine circovirus type 1
PCV1 antibodies were detected in serum of 38% of the cases and in 22% of the controls from PMWS-free herds. The PCV1 antibody titres in cases ranged between <1:100 and 1:6400 and in sera of PMWS-free control pigs the PCV1 antibody titres ranged between <1:100 and 1:1600.
3.4. Porcine parvovirus
PPV DNA could not be detected in lung, mesenteric and inguinal lymph nodes, spleen and kidney of 38 examined cases. Porcine parvovirus antibodies were detected in 63% of the examined cases and in 67% of the PMWS-free controls. However, ELISA positive reactions might have been the results of the presence of PPV maternal antibodies.
3.5. PRRSV
In 90% of the pigs from the case herds and 45% of the pigs from control herds PRRSV RNA and/or PRRSV antibodies were detected. PRRSV RNA and/or PRRSV antibodies were detected in 83% of the cases and 35% of the control pigs from PMWS-free herds (Table 3 ), while PRRSV RNA was detected in 73% of the examined PMWS cases and in 27% of the controls from PMWS-free herds. PRRS-virus RNA was not only found in lung tissue and in mesenteric and inguinal lymph nodes, but could also be detected in spleen and in kidneys. Statistical analysis (χ 2) further revealed that the proportion of PRRSV infected pigs (rt-PCR and/or ELISA antibodies positive) within the case group was significantly higher than the proportion of PRRSV positive control pigs; (a) housed within the same compartment (p=0.02), (b) from another compartment (p=0.0000), and (c) from PMWS-free herds (p=0.0000).
Table 3.
PRRSV infections in cases and in pigs within the three control groups (n=60 per group and otherwise as indicated)
| Group | PRRSV (rt-PCR and ELISA)(%) | PRRSV (rt-PCR) |
|---|---|---|
| Cases | 83 | 73% (n=55) |
| Compartment controls | 65 | 59% (n=34) |
| Controls within PMWS + herds but from another compartment | 42 | 28% (n=29) |
| Controls from PMWS-free herds | 35 | 27% |
Sequence analyses on PRRSV rt-PCR products of ORF6 and ORF7 revealed the presence of EU- and US-type PRRSV in Dutch pigs. Co-infections of EU- and US-type PRRSV were detected in 60% of the case herds while in only one (5%) out of 20 PMWS-free control herds a co-infection of EU- and US-type PRRSV was recorded (Fisher exact test: p=0.0005). This control herd had clinical signs of PMWS 1–1.5 year before sampling (Table 4 ). In 8 PMWS-free control herds, PRRSV infections of only EU- or US-type could be detected in pigs.
Table 4.
EU- and US-type PRRSV infections on 20 case herds and 20 PMWS-free herds
| PRRSV infection type |
Case herds |
PMWS control herds |
|---|---|---|
| European (EU) | 6 | 2 |
| American (US) | 0 | 6 |
| EU and US | 12 | 1 |
| No PRRSV | 4 | 11 |
3.6. Vaccinations
In 14 out of 20 case herds and in none of the 20 control herds vaccines against PRRSV (modified live vaccines) were used (χ 2 analysis, p=0.0000). Twelve case herds started with the vaccination against PRRSV after the observation of PRRS-like/PMWS symptoms. In most cases, the PRRS diagnosis was based on clinical observations and not confirmed by laboratory diagnoses. Co-infections of EU- and US-type PRRSV were also detected in 3 (50%) out of 6 case herds that did not use vaccines against PRRSV.
In 8 case herds and 4 control herds, vaccines against Mycoplasma hyopneumoniae were used. No significant differences were recorded between case and control herds concerning the use of vaccines against e.g. PPV and Escherichia coli. Case and control herds used on compulsory bases vaccines against pseudorabies virus.
3.7. Other viral co-infections
No infections of TGEV or PEDV could be detected in cases or in control pigs from PMWS-free herds by serological methods. Comparable frequencies of antibodies against H1N1, H3N2 and PRCV could be detected in cases and controls as well (data not shown). In 5% of the cases, antibodies against H1N2 were detected and in none of the examined control pigs from PMWS-free herds.
3.8. Bacteriology
Bacteriological examination of organs at necropsy revealed by occasion the detection of pathogenic bacteria in cases, e.g. Streptococcus suis type 2 (1×), Actinobacillus pleuropneumoniae (3×), Mycoplasma spp. (3×), Streptococcus intermedius (1×), and Haemophilus. (1×), but in control pigs of PMWS-free herds also Actinobacillus pleuropneumoniae (1×), Mycoplasma (1×), and Haemophilus (1×) were detected including Aerococcus viridans (1×). Salmonella spp., namely S. typhimurium, S. reading, and S. london, were detected in 3 out of 15 faeces samples collected from cases. No Salmonella was found in faeces samples from controls of PMWS-free herds.
4. Discussion
This case-control study indicates an association between the amount of infectious PCV2 (and PCV2 DNA copy load) and clinical signs as documented for PMWS (Ellis et al., 1999; Quintana et al., 2001). The mean PCV2 DNA copy number in mesenteric and inguinal lymph nodes of pigs with clinical signs of PMWS was more than 105 times higher than in control pigs from PMWS-free herds, and 102 times higher than in control pigs within the same compartment. However, within this latter group, mild to moderate macroscopic lesions were frequently observed, which might be an indication of pré-clinical PMWS stages. Infectious PCV2 or PCV2 DNA was detected in 50% of the control pigs from PMWS-free herds, but the amount of infectious PCV2 and the PCV2 DNA copy load remained low in most of these pigs. In these pigs the course of the PCV2 infections was mild and sub-clinical. Above-mentioned data on PCV2 loads indicate that for the development of clinical signs of PMWS a threshold level of PCV2 is required. This hypothesis is supported by the data recently published by Rovira et al. (2002).
The PCV2 IPMA, in combination with the PCV2 virus isolation method and the quantitative real-time PCV2 PCR, showed that in 27% of the cases the PCV2 antibody titres were low or even not detectable while high PCV2 virus and DNA copy loads were detected. This indicates that in PMWS the immune system may be suppressed. Above-mentioned data are in agreement with those found in our experimental infection study, in which PCV2 infected specified pathogen free (SPF) pigs developed clinical signs and macroscopic and histopathological lesions comparable to those described for PMWS. In 2 out of the 4 PCV2-infected pigs, the PCV2 DNA copy loads in e.g. mesenteric and inguinal lymph nodes were also very high (>108 copies/μl organ suspension) while the PCV2 antibody titres were very low (<1:100 and 1:100) (Stockhofe-Zurwieden et al., 2003).
In this case-control study, organs from cases had high PCV2 DNA copy loads and showed severe macroscopic lesions. However, in controls the course of the PCV2 infection was more or less sub-clinical, or resulted in only mild to moderate macroscopic lesions. Not all PCV2 infected pigs will develop severe clinical signs as described for PMWS. This suggests that certain co-factors modulate the severity of PCV2 infections. Our case-control study, which was also performed to get more insights in which co-factors might play a role in development of clinical signs as described for PMWS, showed that, except for PRRSV, no clear differences were found in the frequencies of PCV1, PPV, TGEV, PEDV, PRCV, H1N1, H1N2 and H3N2 infections between cases and controls from PMWS-free herds. Also others reported the occurrence of concurrent infections of PCV2 and PRRSV in PMWS affected pigs (Sorden et al., 1998; Segalés et al., 2002). Rovira et al. (2002) showed that a concurrent infection of PCV2 and PRRSV could result in increased PCV2 viral copy numbers, and that PRRSV infections enhanced PCV2 replication. The PCV2 load in serum was higher in pigs PRRSV inoculated one week before PCV2 challenge, than in pigs inoculated with PCV2 only. Studies from Rovira et al. (2002), this case-control study and the PCV2-PRRSV experimental infection study (Stockhofe-Zurwieden et al., 2003) all support the hypothesis that PRRSV can be an important predisposing factor to the development of PMWS under experimental and field conditions.
Remarkably, restriction-length PCR and sequence analyses showed that PCV2 and concurrent co-infections of PRRSV-EU and -US occurred only in case herds, and in one control herd with a PMWS history 1.5 years before sampling. A high frequency of co-infections with different types of PRRSV in herds with clinical signs of PMWS has not been reported before, and therefore it might be of interest in the further research for co-factors involved in the development of PMWS. In most case herds, vaccines against PRRSV were used. But according to our opinion, the use of vaccines in case herds had no influences on our data concerning the reported frequencies of PRRSV infections and on the frequencies of concurrent co-infections of EU- and US-type PRRSV as found in cases and controls. This conclusion is based on: (a) the sequences within regions of ORF-6 and ORF-7 from EU- and US-type PRRSV strains found in cases and control pigs were not similar to the (modified)-live virus strains used in PRRSV-vaccines (data not shown), (b) the detection of PRRSV ELISA antibodies in pigs older than 8–9 weeks indicates an active immune response to PRRSV (Nodelijk et al., 1997), and (c) also the detection of PRRSV RNA in pigs at this age indicates an acute PRRSV infection. In addition, also in 50% of the case herds that did not use vaccines against PRRSV, concurrent co-infections of EU- and US-type PRRSV were recorded.
We do not have any clear evidence whether the introduction of new PRRSV strains, or the appearance of PRRSV mutants, has played a role in the increase of clinical PMWS cases in the Netherlands within the last 5 years. However, in 8 case herds with severe clinical cases of PMWS only the US-type PRRSV in combination with EU- and/or US-type PRRSV antibodies were detected. As far as we know we had no PRRSV-US strains in the Netherlands at the beginning of the '90. Also new PRRSV-EU variants with increased pathogenicity might have been responsible for new epizootics within the Dutch pig populations and might have contributed to an increase of reported PMWS cases. Mutations in strains of EU- or US-type PRRSV can result in co-infections of new PRRSV strains with different phenotypes within herds, or even within the same pig. The appearance of quasi-species, and emergence of a PRRSV sub-population during infection in pigs has been reported (Rowland et al., 1999). Subsequently, mutations within the PRRSV genome can be responsible for a reduced but also for an increased virulence (Allende et al., 2000; Nielsen et al., 2001). Semen isolates from another Arterivirus, equine arteritis virus (EAV), seems generally of low pathogenicity but variants with increased pathogenicity might arise during secondary spread of the virus and are perhaps responsible for new epizootics (Timoney and McCollum, 1993). Harms et al. (2001) reported, besides PCV2, the detection of PRRSV strains with increased pathogenicity in swine with atypical PRRS in many Midwestern swine herds in the United States. Beginning in 1996, outbreaks of atypical PRRS often occurred in sow herds that had received multiple doses of modified live PRRSV vaccines (Harms et al., 2001). Whether the use of vaccines against PPRSV has any influence on the chance for the appearance of new PRRSV mutants is unknown.
Infections of pathogenic bacteria, e.g. Streptococcus suis type 2, Actinobacillus pleuropneumoniae, Mycoplasma spp., Haemophilus spp., and Salmonella spp. were found in cases, but the frequency was low or the pathogenic bacteria could also be detected in control pigs from PMWS-free herds, as found for Actinobacillus pleuropneumoniae, Mycoplasma, and Haemophilus. The increased frequency of (secondary) endemic bacterial infections in our cases might be due to accompanying PCV2 and/or other viral infections such as PRRSV.
In summary, this case-control study indicates an association between the amount of infectious PCV2, the PCV2 DNA copy load, and clinical signs as documented for PMWS. Although, most of the pigs within the clinically affected PMWS group showed histopathological lesions characteristic for PMWS, in lymph nodes from some of the pigs within this group, lesions consisted of hyperplasia often in combination with the presence of giant cells. As lymphoid hyperplasia is not a common feature of PMWS, some of the pigs within this group probably cannot be fully characterised as PMWS-affected, despite the presence of: (a) clinical signs as documented for PMWS, (b) histopathological lesions in e.g. lung and kidneys, and (c) the detection (high levels) of PCV2 in affected organs. Furthermore, concurrent PCV2 and PRRSV infections were recorded significantly more often in Dutch pigs with clinical signs than in healthy controls, and co-infections with both EU- and US-type PRRSV were detected very frequently in Dutch case herds.
Acknowledgments
The authors thank Marieke van Es, Renate Hakze-van der Honing and Judith Maneschijn-Bonsing for excellent technical assistance. This research was financed by the Dutch Product Boards for Livestock, Meat and Eggs and the Dutch Ministry of Agriculture, Environment and Fisheries.
References
- Allan G.M, McNeilly F, Kennedy S, Daft B, Clarke E.G, Ellis J.A, Haines D.M, Meehan B.M, Adair B.M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the Usa and Europe. Journal of Veterinary Diagnostic Investigation. 1999;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- Allan G.M, Kennedy S, McFeilly F, Foster J.C, Ellis J.A, Krakowka S.J, Meehan B.M, Adair B.M. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. Journal of Comparative Pathology. 1999;121:1–11. doi: 10.1053/jcpa.1998.0295. [DOI] [PubMed] [Google Scholar]
- Allende R, Kutish G, Laegreid W, Lu Z, Lewis T, Rock D, Friesen J, Galeota J, Doster A, Osorio F. Mutations in the genome of porcine reproductive and respiratory syndrome virus responsible for the attenuation phenotype. Archives of Virology. 2000;145:1149–1161. doi: 10.1007/s007050070115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasch M, Segalés J, Rosell C, Domingo M, Mankertz A, Urniza A, Plana-Duran J. Experimental inoculation of conventional pigs with homogenates from pigs with post-weaning multisystemic wasting syndrome. Journal of Comparative Pathology. 1999;121:139–148. doi: 10.1053/jcpa.1999.0310. [DOI] [PubMed] [Google Scholar]
- Elbers A.R.W, de Jong M.F. Porcine circovirus type 2 widely spread in the Netherlands. Gd Varken. 2002;26:11. [Google Scholar]
- Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan B, Martin K, Harding J, Kennedy S, McNeilly F. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. Journal of Veterinary Diagnostic Investigation. 1999;11:3–14. doi: 10.1177/104063879901100101. [DOI] [PubMed] [Google Scholar]
- Ellis J.A, Bratanich A, Clark E.G, Allan G, Meehan B, Haines D.M, Harding J, West K.H, Krakowka S, Konoby C, Hassard L, Martin K, McNeilly F. Coinfection by porcine circovirus and porcine parvovirus in pigs with naturally acquired post-weaning multisystemic wasting syndrome. Journal of Veterinary Diagnostic Investigation. 2000;12:21–27. doi: 10.1177/104063870001200104. [DOI] [PubMed] [Google Scholar]
- Harms P.A, Sorden S.D, Halbur P.G, Bolin S, Lager K, Morosow I, Paul P.S. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and PRRSV. Veterinary Pathology. 2001;38:528–539. doi: 10.1354/vp.38-5-528. [DOI] [PubMed] [Google Scholar]
- Krakowka S, Ellis J.A, Meehan B, Kennedy S, McNeilly F, Allan G. Viral wasting syndrome of swine: experimental reproduction of post-weaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Veterinary Pathology. 2000;37:254–263. doi: 10.1354/vp.37-3-254. [DOI] [PubMed] [Google Scholar]
- Krakowka S, Ellis J.A, McNeilly F, Ringler S, Rings D.M, Allan G. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2) Veterinary Pathology. 2001;38:31–42. doi: 10.1354/vp.38-1-31. [DOI] [PubMed] [Google Scholar]
- Ladekjaer-Mikkelsen A.S, Nielsen J, Stadejek T, Storgaard T, Krakowka S, Ellis J, McNeilly F, Allan G, Botner A. Reproduction of post-weaning multisystemic wasting syndrome (PMWS) in immunostimulated and non-immunostimulated 3-week-old piglets experimentally infected with porcine circovirus type 2 (PCV2) Veterinary Microbiology. 2002;89:97–114. doi: 10.1016/S0378-1135(02)00174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Oleksiewicz M, Forsberg R, Stadejek T. Reversion of a live porcine reproductive and respiratory syndrome virus vaccine investigated by parallel mutations. Journal of General Virology. 2001;82:1263–1272. doi: 10.1099/0022-1317-82-6-1263. [DOI] [PubMed] [Google Scholar]
- Nodelijk G, Leengoed Van L.A.M.G, Schoevers E.J, Kroese A.H, De Jong M.C.M, Wensvoort G, Verheijden J.H.M. Seroprevalence of porcine reproductive and respiratory syndrome virus in Dutch weaning pigs. Veterinary Microbiology. 1997;56:21–32. doi: 10.1016/S0378-1135(96)01349-1. [DOI] [PubMed] [Google Scholar]
- Oleksiewicz M.B, Botner A, Madsen K.G, Storgaard T. Sensitive detection and typing of porcine reproductive and respiratory syndrome virus by RT-PCR amplification of whole viral genes. Veterinary Microbiology. 1998;64:7–22. doi: 10.1016/S0378-1135(98)00254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana J, Segalés J, Rosell C, Calsamiglia M, Rodriquez-Arrioja G.M, Chianini F, Folch J.M, Maldonado J, Canal M, Plana-Duran J, Domingo M. Clinical and pathological observations on pigs with postweaning multisystemic wasting syndrome. Veterinary Record. 2001;149:357–361. doi: 10.1136/vr.149.12.357. [DOI] [PubMed] [Google Scholar]
- Rovira A, Balasch M, Segalés J, Garcia L, Plana-Durán J, Rosell C, Ellerbrok H, Mankertz A, Domingo M. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. Journal of Virology. 2002;76:3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R, Steffen M, Ackerman T, Benfield D. The evolution of porcine reproductive and respiratory syndrome virus: Quasi-species and emergence of a virus subpopulation during infection of pigs with VR-2332. Virology. 1999;259:262–266. doi: 10.1006/viro.1999.9789. [DOI] [PubMed] [Google Scholar]
- Segalés J, Calsamiglia M, Rosell C, Soler M, Maldonado J, Martin M, Domingo M. Porcine respiratory and reproductive syndrome virus (PRRSV) status in pigs naturally affected with post-weaning multisystemic wasting syndrome (PMWS) in Spain. Veterinary Microbiology. 2002;85:23–30. doi: 10.1016/s0378-1135(01)00474-6. [DOI] [PubMed] [Google Scholar]
- Soares R.M, Durigon E.L, Bersano J.G, Richtzenhain L.J. Detection of porcine parvovirus DNA by polymerase chain reaction assay using primers to the highly conserved non-structural protein gene, NS-1. Journal of Virological Methods. 1999;78:191–198. doi: 10.1016/s0166-0934(98)00177-3. [DOI] [PubMed] [Google Scholar]
- Sorden S, Harms P, Sirinarumitr T, Morozov I, Halbur P, Yoon K, Paul P. American Association of Veterinary Laboratory Diagnosticians; Minneapolis, USA: 1998. Porcine circovirus and PRRSV co-infection in pigs with chronic bronchointerstitial pneumonia and lymphoid depletion: an emerging syndrome in Midwestern swine; p. 75. (Proceedings of the Annual Meetings of the American Association of Veterinary Laboratory Diagnosticians). [Google Scholar]
- Stockhofe-Zurwieden, N., Wellenberg, G.J., Schuurman, G., Zwart, R., 2003. Experimental inoculation of specified pathogen free pigs with porcine circovirus 2 only or in combination with other porcine viruses or immunostimulation. In: Proceedings 4th International Symposium on Emerging and Re-emerging Pig Diseases, Rome, June 29th–July 2nd. pp. 164–165
- Timoney P, McCollum W. Equine viral arteritis. Veterinary Clinics of North America: Equine Practice. 1993;9:295–309. doi: 10.1016/S0749-0739(17)30397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenberg G.J, Pesch S, Berndse F.W, Steverink P.J, Hunneman W, Van Der Vorst T.J, Peperkamp N.H, Ohlinger V.F, Schippers R, Van Oirschot J.T, De Jong M.F. Isolation and characterisation of porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in The Netherlands. Veterinary Quarterly. 2000;22:167–172. doi: 10.1080/01652176.2000.9695049. [DOI] [PubMed] [Google Scholar]


