Abstract
One of the important pursuits in science and engineering research today is to develop low-cost and user-friendly technologies to improve the health of people. Over the past decade, research efforts in microfluidics have been made to develop methods that can facilitate low-cost diagnosis of infectious diseases, especially in resource-poor settings. Here, we provide an overview of the recent advances in microfluidic devices for point-of-care (POC) diagnostics for infectious diseases and emphasis is placed on malaria, sepsis and AIDS/HIV. Other infectious diseases such as SARS, tuberculosis, and dengue are also briefly discussed. These infectious diseases are chosen as they contribute the most to disability-adjusted life-years (DALYs) lost according to the World Health Organization (WHO). The current state of research in this area is evaluated and projection toward future applications and accompanying challenges are also discussed.
Keywords: Infectious diseases, Microfluidics, Malaria, Diagnostics, HIV, Sepsis, Point-of-care
1. Introduction
Microfluidics refers to the science and technology of systems which manipulate small (10− 9 to 10− 18 L) amounts of fluids (Whitesides, 2006). It offers advantages such as low sample and reagent volumes, high throughput, and more precise control of spatial and temporal factors (Beebe et al., 2002). Due to its desirable attributes, such platforms have been employed as biosensors for health monitoring and proposed to be used as micro-total analysis systems (μTAS) where automatic transport, mixing and reacting of samples and reagents can occur in a small device to facilitate timely readouts (Reyes et al., 2002).
Although the World Health Organization (WHO) has continually refined its framework to manage diseases such as malaria (Furdui and Harrison, 2004), these infectious illnesses remain prevalent in resource-scarce regions. It is generally agreed that diagnosis is important for managing infectious disease as it facilitates early detection and access to care, delays the development of resistance and saves money on alternative treatments (Wongsrichanalai et al., 2007). The WHO recommends that diagnostics for the developing countries fulfill the following criteria: (i) affordable, (ii) sensitive, (iii) specific, (iv) user-friendly, (v) rapid and robust, (vi) equipment-free and (vii) deliverable to end-users, summarized as ASSURED (Martinez et al., 2010) which are compatible with the characteristics of microfluidic technologies (Lee et al., 2010).
In this review, emphasis is placed on the study and diagnosis of the malaria, sepsis and HIV due to their high mortality rates. Other prevailing infectious diseases such as SARS, tuberculosis, and dengue are also briefly discussed. These infectious diseases are chosen as they are most prevalent in resource-scarce communities and they contribute the most to disability-adjusted life-years (DALYs) lost according to the World Health Organization (WHO) (Mabey et al., 2004, Yager et al., 2006). This review is written in a disease-centered structure in contrast to the technique-centered approach by Jung et al. (2015), Su et al. (2015) and also Damhorst et al. (2015) . Individual diseases are described here with their current diagnostic/treatment platforms. Gaps in managing these infectious diseases are also identified and the potential of microfluidic technologies to bridge the gaps is discussed. We believe that the disease-centered approach can enhance the knowledge of readers researching in specific disease fields on the utility of microfluidics. Additionally, we hope to use this review to encourage further research in infectious diseases such as SARS which have generated low scientific interest in the microfluidics community thus far (Fig. 1 ).
Fig. 1.
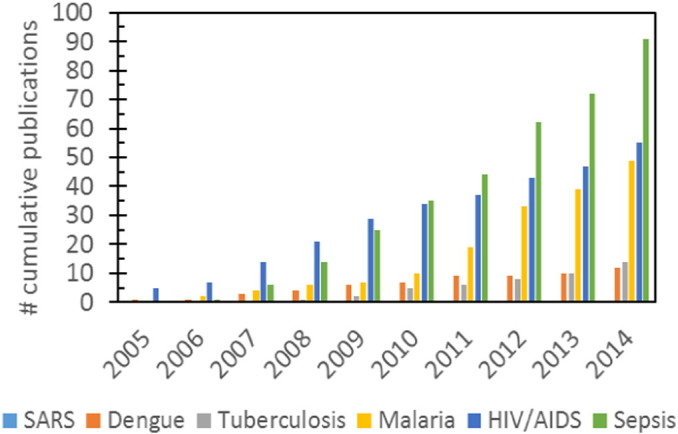
Cumulative number of publications using microfluidic for the study and diagnosis of various infectious diseases (from 2005 to 2015). There were only 3 publications on SARS around 2002–2004 where there was SARS outbreak in Asian-Pacific countries. The infectious diseases that experience a steady rise in number of publications were HIV/AIDS and sepsis which affect both developing and developed nations equally. On the other hand, the number of publications in malaria and tuberculosis is stagnating despite the continued need for better diagnostic tools for resource-scarce communities. This shows that diseases prevalent in developing nations are still very much neglected by the scientific community that is concentrated in developed nations. Regrettably, many of the publications in dengue were also from groups in Latin America and South-East Asia, once again highlighting the lack of international interest in these infectious diseases. Note: Publication numbers were calculated using PubMed and ScienceDirect.
2. Malaria
Malaria infection by Plasmodium protozoa through the intermediate host, Anopheles mosquito is currently most common (Gardner et al., 2002). The infected red blood cells (iRBCs) can disrupt microcirculation (Maier et al., 2009), manifesting into anemia and organ (Dondorp et al., 1999) failure in severe cases. Despite improvements in malaria management, 600,000 people continue to fall victim to the disease annually (World Health Organization, 2013).
The ‘gold’ standard test for malaria is the microscopic method (thin and thick Giemsa-blood smear) that allows trained technicians to detect parasitemia level up to 1 iRBC in 106 cells (~ 1000 parasites/μL) (Murray et al., 2008). Although this test is one of the most affordable means for malaria diagnosis, its specificity relies critically on the quality of the microscope and skills of the technician. There can be misinterpretation of microorganisms like bacteria and fungi as Plasmodium protozoa and difficulty in differentiating different Plasmodium strains (Erdman and Kain, 2008). Logistical challenge of transporting microscopes to remote areas may also delay diagnostic efforts (Gascoyne et al., 2004).
Another important malaria diagnostic tool is the rapid diagnostic test (RDTs) which detects for malaria antigen, usually in 5–15 μL of blood with monoclonal antibodies specific to the target parasite antigen. RDTs circumvent the challenge of poor road and electricity infrastructure in remote areas with results obtainable in 5‐20 min via simple self-testing and interpretation (Mills et al., 1999). Nonetheless, humid and warm climate can degrade chemicals adsorbed onto RDTs (Chiodini et al., 2007). Most RDTs detect only Plasmodium falciparum through targeting the histidine-rich protein 2 (HRP-2) (Bell et al., 2006) or all Plasmodium. proteins such as lactate dehydrogenase (Iqbal et al., 2001). Hence RDTs results may not be sufficiently specific to guide treatments when mixed infections occur. The ability to differentiate the strain of malaria parasite infections is crucial in the light of increasing evidence that mixed infections are more fatal (Genton et al., 2008), infection by Plasmodium vivax is more lethal in young children (Tjitra et al., 2008) and emergence of artemisinin-resistant P. falciparum (Dondorp et al., 2009). The sensitivity of RDTs also declines as parasitemia fall below 100 parasites/μL (Mills et al., 1999). The price of RDTs which stands at about US $0.55–1.50 (depending on the manufacturers and order quantities) can also be prohibitive for resource-scarce communities (Wongsrichanalai et al., 2007).
Laboratory alternatives such as polymerase chain reaction (PCR) have also been used to diagnose malaria and are capable of detecting low parasitemia levels (~ 15–25 parasites/μL) or mixed infections (Makler et al., 1998). Unfortunately, the accuracy of PCR result is subject to the suitability of primers, sample collection, storage, transport procedures and nucleic acid extraction protocols. Furthermore, the susceptibility of sample and reagent to contamination and the logistics of transporting samples to distant laboratories discourage routine use of PCR as a malaria diagnosis tool as it is incompatible for immediate patient care (Hänscheid and Grobusch, 2002).
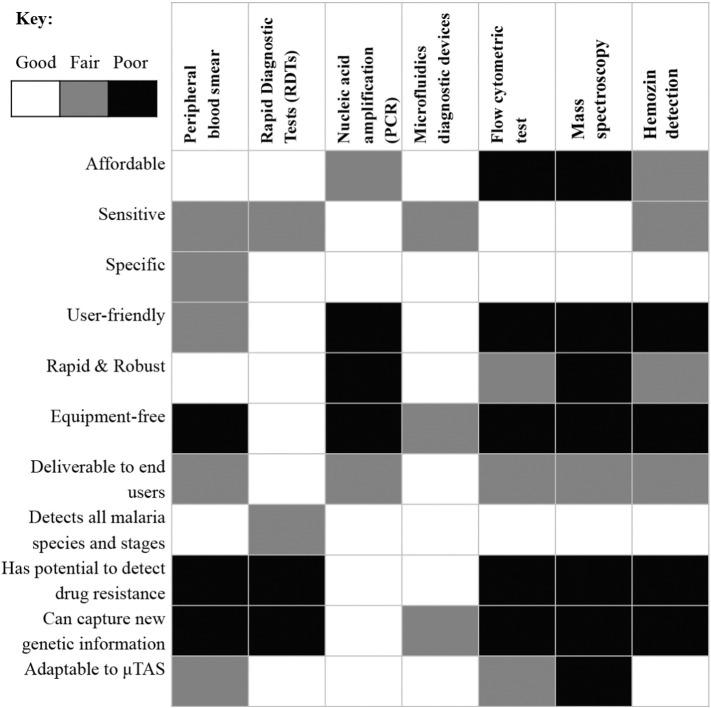
One other technique for malaria diagnosis is through hemozin detection. Hemozin is converted by the malaria parasite from the degraded product of hemoglobin, heme. Heme can be visualized by polarization and/or dark field microscopy. However, the sensitivity of heme detection depends strongly on the parasitemia level and the amount of heme present (Rebelo et al., 2012). Thus this technique may not be suitable for detection of ring and trophozoite stages of malaria. Fig. 2 summarizes the strengths and weaknesses of the various malaria diagnostic tools available in the market.
Fig. 2.
A framework to evaluate the suitability of different malaria diagnostic tools in the market. The first 7 criteria are adapted from WHO (ASSURED) (Martinez et al., 2010) while the rest are adapted from Gascoyne et al. (2004). μTAS: micro-total analysis system. From the evaluation, it can be seen that microfluidic platforms can perform on par or even superior to many existing malaria diagnostics. Note: affordability is calculated without considering the fixed cost of machine.
The predominant method for malaria diagnosis is symptom-based which is arguably ineffective as malaria shares many clinical symptoms such as fever with other poverty-associated febrile illnesses (Bell et al., 2006). Bell and colleagues proposed the adoption of parasite-based diagnosis for early diagnosis and treatment. This could allow formal health services at village level to reduce morbidity and mortality in a more cost-effective manner (Ghebreyesus et al., 2000). Microfluidic diagnostic platforms are especially useful as they can facilitate parasite-based preventive diagnostic in an affordable and manner, potentially bridging this healthcare gap to improve malaria management.
In addition, in early stage malaria, parasitemia level may be as low as 5–20 parasites/μL (Hänscheid, 1999) that is beyond the accurate detection limit of Giemsa smears, RDTs and most PCR. Therefore, microfluidic techniques can be very useful for direct diagnostic or as a processing step to remove interfering blood cells and to concentrate the malaria parasites to enhance the sensitivity of the test.
2.1. Cell deformability
Erythrocytes with the normal average diameters of 7.5 μm must deform considerably as they traverse through capillaries which have significantly smaller diameters of 3–7 μm (Alizadehrad et al., 2012). iRBCs become progressively less deformable as the parasites mature (Cranston et al., 1984). The marked decrease in membrane flexibility can be attributed to two main factors. Firstly, as the relatively non-deformable parasites mature into the trophozite and schizont stages, it causes the internal viscosity of RBCs to increase sharply (Quinn et al., 2011). Secondly, membrane rigidity can be attributed to the export of neoantigens to the surfaces of RBCs and the parasites exerting oxidative stress on the host RBCs (Becker et al., 2004). The increase in deformability can precipitate important pathophysiological outcomes such as reduction in flow velocity (McWhirter et al., 2011), increased adherence to vasculature, channel clogging and thickness of cell-free layer (Yin et al., 2013).
Based on this knowledge, Hou et al. investigated the use of deformability as a biomarker for on-site iRBC enrichment using inertial microfluidics (Fig. 3A) (Hou et al., 2010). The team exploited the margination phenomenon to isolate iRBCs. It was found that with the increase of hematocrit to 40%, more than 80% of the iRBCs (in trophozoite/schizont stages) were displaced to the side channels, demonstrating the importance of cell–cell interactions to observe margination. However, the maximum flow rate was only 5 μL/min and this platform could not differentiate different species of the Plasmodium protozoas. Furthermore, as RBC deformability is also a characteristic of other diseases such as sickle cell anemia, improvements in specificity is warranted. Geislinger and colleagues also enriched less deformable ring-stage iRBCs with an average enrichment factor of 4.3 and throughput of 12,000 cells/h based on a separation process driven by the non-inertial lift effect (Geislinger et al., 2014). Warkiani et al. made use of the negative depletion approach to remove leukocytes from lysed blood before also using inertial focusing (Warkiani et al., 2015). Approximately 70% of the parasites released from iRBCs could be captured as identified using quantitative (q) PCR and 99.9% of the leukocytes was depleted. The device also boosts a sensitivity of 2–10 parasites/μL, better than current sensitivity of RDTs. This device can be coupled to other diagnostic methods to form an integrated platform as it can enrich the malaria parasites to facilitate sensitive detection.
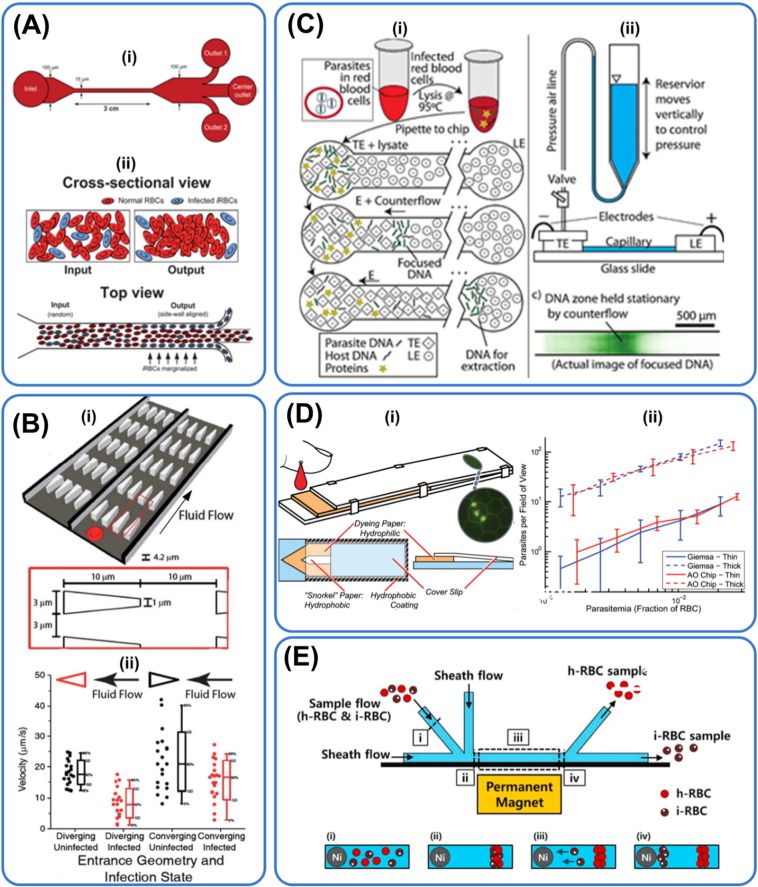
Fig. 3.
Microfluidic technologies used in malaria diagnosis. Principles for diagnosis: (A & B) deformability, (C) PCR, (D) optical and (E) magnetic. (A) Schematic illustration of working principle of a microfluidic device working with the concept of margination. iRBCs which are less deformable than healthy RBCs are displaced to the peripheral walls where they are collected. Making use of the margination phenomenon, 75% of early stage iRBCs and 90% of late stage iRBCs can be recovered, reprinted with permission from Hou et al. (2010). (B) Schematic illustration of a microfabricated deformability-based flow cytometer (i) that measures dynamic mechanical responses of RBCs. Experimental results (ii) demonstrating measured velocities of RBCs as a function of infection state for RBCs infected with late ring stage parasites at a pressure gradient of 0.24 Pa μm− 1, reprinted with permission from Bow et al. (2011). (C) Schematic showing a microfluidic technique for purification of nucleic acids from iRBCs using isotachophoresis (ITP). LE, TE: leading, trailing electrolytes. Isotachophoresis was used to extract DNA from malaria parasites. Nucleic acid yield was maximized using counterflow that increased focusing time. A limit of detection of 0.5 parasites/nL was achieved, reprinted with permission from Marshall et al. (2011). (D) Schematic showing working mechanism of a paper based microfluidic device for automated staining of malaria parasites with an embedded microscopy window. Paper cartridge consisting of both thin (single cell layer) and thick (multiple cell layer) smears where blood is stained with acridine orange dye. The cartridge is then optically examined for iRBCs, reprinted with permission from Horning et al. (2014). (E) Schematic of a label-free microfluidic device for separation of iRBCs based on their paramagnetic characteristics. Presence of paramagnetic hemozin in iRBCs is used to separate iRBCs from healthy RBCs based on their differential lateral migration in a magnetic field gradient. Collection of ring-stage iRBCs (with low hemozin concentration) was made possible with the use of steep magnetic gradient, reprinted with permission from Nam et al. (2013).
Bow et al. coupled iRBCs' deformability to fluorescence measurements to relate the mechanical properties of iRBCs to their biochemical properties (Bow et al., 2011). The team discovered that the geometry of constriction affected RBC transit time as geometries with sharper corners better differentiate RBCs with different deformability for a given pressure difference. Their microfluidic device also demonstrated the likelihood that the rigidities of ring stage P. falciparum iRBCs overlapped with older, healthy RBCs which is a valid concern in using RBC deformability as a biomarker (Fig. 3B).
Guo and colleagues capitalized on the differences in pressure threshold (attributed to deformability differences) between healthy RBCs and iRBCs as they passed through progressively narrower constrictions (5 to 1 μm) to study the malaria disease (Guo et al., 2012). Previously, Tomaiuolo and co-workers have also provided a description of RBC viscoelastic properties such as shape as a function of the applied pressure drop using microfluidic devices (Tomaiuolo et al., 2011). Guo et al. found that iRBCs in the schizont stages were typically blocked by the 3 μm constriction and this blockage significantly elevated the deformation pressure. Similar results have been found by Shelby et al. who found that trophozoite staged iRBCs experienced difficulties passing through 4 and 2 μm constriction and schizont stage iRBCs blocked these small constrictions (Shelby et al., 2003). Using their microfluidic device, Guo et al. then showed that the stiffness of ring, trophozoite and schizont stages iRBCs to be 1.5, 7 and 200 times more than uninfected RBCs. These results differed from the measurement using optical tweezers where the values are 3, 4 and 10 respectively (Hosseini and Feng, 2012). The discrepancy in membrane stiffness highlights the importance of establishing standards for tools that measure cellular membrane properties. This microfluidic device may also be suitably adapted to study phenomena such as pitting and RBC recovery after traversing through narrow constriction. It has been shown that under flow conditions, healthy RBCs do not become part of a vascular occlusion but rather wind their ways through the center of the iRBCs to exit from the constriction (Shelby et al., 2003). The usefulness of microfluidic devices is hence not limited to malaria diagnosis as it can mimic channel blockage at different bifurcations (Huang et al., 2013) to test the clinical usefulness of therapies such as transfusion (Riddle et al., 2002) and also drugs.
Microfluidic platforms using membrane deformability as a biomarker for malaria may hold great promises but its suitability is arguably challenged in the face of emerging knowledge on malaria. It has been reported that the deformability of uninfected RBCs can be reduced in cases of severe malaria (Dondorp et al., 1999). The exact mechanisms are unknown but it has been suggested that in vitro cultures of P. falciparum can produce heat-labile exoantigen that decrease RBC deformability (Naumann et al., 1991). The damaging effect of lipid peroxidation on RBC membrane flexibility has also been proposed to explain the aforementioned phenomenon (Mohan et al., 1995). The use of microfluidic has also provided valuable insights in the differences between P. falciparum and P. vivax infected RBCs. Handayani et al. constructed a microfluidic device with a narrow constriction of 2 μm and observed that in contrast to P. falciparum infected RBCs, P. vivax infected RBCs at all developmental stages were able to transverse the gap smoothly (Handayani et al., 2009). They postulated that the increase in internal viscosity of P. vivax infected RBCs could be offset by the increase in membrane deformability. These findings highlight the importance of enhancing our knowledge about the malaria disease and the interactions of malaria parasites and RBC hosts. With greater understanding of the malaria biology, deformability-based diagnostic tests can be more robustly designed. Other groups have utilized concepts relating to electrical impedance (Zheng et al., 2013) and transit time (Rosenbluth et al., 2008) to differentiate blood and cancer cells. These concepts may be extended for malaria detection provided that RBCs and iRBCs are sufficiently distinct in these key parameters.
2.2. Electrical signature
Dielectrophoresis which refers to spatially non-uniform electric fields either due to direct current or alternating current has also been utilized for malaria-related study and diagnosis. Dielectrophoretic (DEP) forces are generated when dielectric particles i.e. polarizable electrical insulators such as cells are subject to direct or alternating electric field. When the cells move in the direction of increasing electric field gradient or intensity, the behavior is called positive DEP and the converse is termed negative DEP (Pethig and Markx, 1997). Readers can refer to Pethig for mathematical equations governing the generation of DEP forces on spherical and non-spherical particles (Pethig, 2010). As the DEP responses of cells depend on the composition (permeability, capacitance, conductivity) and conformation of their cell membranes, cells of different types, sizes and physiological states can be differentially isolated (Gascoyne et al., 2002) using different media and frequency of the electric field. Gascoyne et al. demonstrated the possibility of using DEP to isolate iRBCs (Gascoyne et al., 2004) as the electrical conductivity of iRBC is significantly higher than uninfected RBCs (Aceti et al., 1990) attributing to membrane permeation pathways induced by malaria parasites. In their DEP-field-flow fractionation (FFF) microfluidic device, they observed that nucleated blood cells remained in the DEP-FFF chamber and emerged only after the electric field was turn off, preventing leukocytes contamination. Next, by applying a certain frequency of electrical signal (40–250 kHz), iRBCs were levitated more strongly than uninfected RBCs and emerge more rapidly from the DEP-FFF chambers. Consequently, the initial cell isolate contained only Plasimodium protozoa. Du and coworkers also utilized DEP stretching to characterize the differences in biophysical properties of healthy and infected RBCs (Du et al., 2014). The latter was found to have expectedly lower stretching ratio due to rigid membranes.
Nonetheless, it should be noted that one of the critical challenge of DEP is the strong size dependence as most cells exhibit variations up to 10% which may interfere with cell-type specific differences in DEP particle separation. The conductivity of the media or extracellular solution could also lead to Joule heating near electrodes, bubble generation and heat-related cell death (Bhagat et al., 2010). This device is also incompatible for diagnosis in geographical remote areas with no access to stable electricity.
2.3. Molecular analysis
PCR is an extremely useful molecular biological tool to replicate DNA. Through the steps of denaturation, annealing and extension each at specific temperatures, millions of copies of DNA can be generated from minute quantities of DNA fragments. While conventional PCR may not be as useful for malaria diagnosis (Hänscheid and Grobusch, 2002) due to time lag in sample transportation, sample processing and result dissemination (Srinivasan et al., 2000), microfluidic PCR may prove otherwise. Microfluidic PCR can have higher sensitivity than conventional PCR (Rubio et al., 1999). Furthermore, the faster speed, adaptability to μTAS and portability of microfluidic PCR greatly boosts its usefulness in resource-scarce regions where malaria is endemic (Wang et al., 2009). In addition, microfluidic devices which only require small sample volume and have high surface area to volume ratio offer higher temperature transition speed, more uniform heat distribution and reaction efficiency.
There are two main approaches to perform microfluidic PCR i.e. stationary and continuous flow (Park et al., 2011). In the former, the PCR mixture is housed in one unit which experiences temperature change during the PCR cycle. This method requires lower sample volume but runs the risks of longer reaction time and non-uniform heating and cooling. In the latter method, there are pre-fixed temperature zones and the PCR mixture flows from one unit to another. This approach, however, only operates with a fixed number of PCR cycles and may be more complex to fabricate, resulting in bulky devices. Currently, droplet microfluidic PCR is also being tested (Tewhey et al., 2009). Microfluidic PCR readout has been integrated with electrochemical, fluorescence gel electrophoresis that minimizes loss due to transferring of amplified nucleic acids and contamination (Park et al., 2011). Zhang et al. and Park et al. have done an excellent review on microfluidic PCR and readers are encouraged to refer these remarkable references for better technical information on this subject (Zhang et al., 2006, Park et al., 2011). Given the specificity of the primers in the PCR process, this approach can be applied to diagnose for most infectious diseases as long as sufficient genomic material can be extracted. The technique described in this section is therefore also applicable for other infectious diseases.
Generally, the innovations in microfluidic PCR can be categorized into i) pre-PCR sample processing, ii) PCR-reaction and iii) post-PCR analysis. Pre-PCR steps include cell separation, cell concentration, cell lysis and nucleic acid extraction. These steps are necessary to overcome PCR inhibitors present in samples and to reduce background due to large amount of hematologic cells in blood sample (Lim et al., 2005). There are also several ways that cell lyses have been performed in microfluidic devices such as using mini-sonication, nanofabricated shearing structures (Di Carlo et al., 2003), stored chemicals like detergents (Kim et al., 2004), enzymes (Sethu et al., 2004) and externally applied electric field (Lee and Tai, 1999).
Although there are many microfluidic PCR technologies out there, there are not many that specifically target malaria diagnosis. Marshall and colleagues developed a microfluidic platform (Fig. 3C) that purify DNA released from malaria parasites residing in iRBCs and the isolated DNA was compatible to quantitative PCR as it was able to offer sensitivity of 500 parasites/μL (Marshall et al., 2011). This innovation can be useful when coupled to downstream microfluidic platform to create a μTAS for malaria diagnosis which was also suggested by Gascoyne et al. (2004). Although there has been much discussion on a μTAS that incorporates concepts such as deformability and PCR for more comprehensive diagnosis of malaria, to the best of our knowledge, such system is probably still at the developmental phase. It can also be beneficial if loop meditated thermal amplification (LAMP) PCR, one of the most sensitive and specific PCR method (Hopkins et al., 2013), can be integrated with a portable microfluidic device. We also stress the need to combine innovations created for different PCR components to create a truly functional prototype. This is because various improvements have been made for instance, the creation of microfluidic digital recombinase polymerase amplification (RPA) SlipChip for quantitative detection of DNA by Shen et al. (2011), but amplification is only one step of PCR. Technologies facilitating better mixing of reagents or concentrating samples should be integrated to build the most effective μTAS for diagnosis.
2.4. Optical
Optical means have been used traditionally for the diagnosis of diseases, including malaria (Giemsa smears). The ubiquity of optical instruments and their wide range of resolution and lenses angles make them valuable tools for the study and monitoring disease progression (Psaltis et al., 2006). Nonetheless, conventional optical microscopy is bulky and expensive. Recently, however, this field has evolved with the emergence of cheaper optic alternatives enhanced with add-ons such as wave guides to create portable diagnostics (Hunt and Wilkinson, 2008). The charge-coupled device (CCD)-based platforms (Ozcan and Demirci, 2008) have been developed to monitor cells and particles movement through shadow imaging without fluorescence imaging. Other detection modes include absorbance, chemiluminescence and interferometric (Myers and Lee, 2008). For instance, Juul et al. made use of combined DNA cleavage-ligation event with droplet microfluidic for malaria parasite enzyme activity detection. This microfluidic platform can operate with a detection limit of less than 1 parasite/μL of unprocessed blood (Juul et al., 2012). Mobile phones, which can also function as a imaging platform, have also been employed in imaging thick and thin Giemsa-stained smears of P. falciparum iRBCs in brightfield with a mounted light microscope (Breslauer et al., 2009). Recently, Horning et al. described a paper cartridge coupled with an automated image processing software to detect for malaria parasites (Horning et al., 2014). The cartridge consisted of both thin and thick smear regions for species identification and limit of detection (~ 100 parasites/μL) enhancement respectively (Fig. 3D).
There are two main ways in which advances in optical study can be tied in with microfluidic for study and detection of malaria. Firstly, it is the emergence of cheaper and portable optical instrument that can be used to observe cellular interactions in microfluidic. Optical microscopy can be combined with microfluidic to enhance on-chip detection of RBC deformability or changes that are specific for diseases. In this regard, Kim et al. created a mini-microscope for in-situ monitoring of cells by modifying off-the-shelf components of a commercial webcam (Kim et al., 2012). Comparison of images of cells taken with the in-house microscope and a conventional microscope revealed that the magnification of the former was comparable to a 40 × objective lens (approximately 400 × magnification). Affordable and yet powerful optical tools can too facilitate the emergence of cheap optical detection such as by secondary speckle sensing (S3) microscopy (Cojoc et al., 2012).
Secondly, optics can also be combined with microfluidic to investigate cellular biophysical properties and differentiate cells of different types or states (Adamo et al., 2012, Gossett et al., 2012). Lee et al. designed a subpixel resolving optofluidic microscope (SROFM) that made use of Red–Green–Blue (RGB) LEDs to capture separate color images of iRBCs which are subsequently combined to obtain a fully colored high resolution image (Lee et al., 2011). The team stained iRBCs with 2% Toluidine blue and with its SRFOM, achieved an image resolution of 0.66 μm at the highest acuity. This method circumvents sample preparation such as fixation unlike the conventional microscopic detection method. However, SRFOM shares similar limitations as the microscopic technique as it is unable to identify ring stage iRBCs, limiting its usefulness for early malaria detection (Erdman and Kain, 2008).
Other potential optofluidics techniques which have been used for detection of other diseases but may be applicable for malaria also deserve to be highlighted. Ji et al. constructed a microfluidic cytometer and found that the transmission intensity for different cell types differ (Ji et al., 2013). Lee and colleagues has also introduced the idea of optical pressure to detect for cancer cells (Lee et al., 2007). These concepts may be adapted for malaria diagnosis.
2.5. Magnetic
The potential of using magnetic microfluidic malaria diagnosis should also be discussed although there has been no extensive literature reported. Paul et al. reported that iRBCs behave like paramagnetic particles in a magnetic field (Paul et al., 1981). This has motivated research in using magnetic means to identify iRBCs (Bhakdi et al., 2010). Zimmerman and colleagues proposed the idea of using magnets to concentrate iRBCs to enhance the sensitivity of microscopic method for detection of malaria (Zimmerman et al., 2006). Nam et al. designed a microfluidic device that capitalized on the magnetic properties of accumulated hemozin in iRBCs to separate iRBCs from malaria culture as Fe3 + in hemozin has a stronger paramagnetic effect than Fe2 + in hemoglobin (Nam et al., 2013). The team incorporated a nickel wire in the microfluidic device to create a steep magnetic gradient to achieve > 90% in ring stage iRBC recovery rate (Fig. 3E). Nonetheless, this device works below the flow rate of 1.6 μL/min and the team also has yet to show the creation of a homogenous magnetic field. In addition, the concentration of hemozin may be skewed by the number of parasites i.e. singlet, doublet and triplet (Malleret et al., 2011) in iRBCs which may decrease the specificity of the microfluidic device in informing the users about the severity and stages of malaria. As aforementioned, the amount of hemozin is also lower in ring stage iRBCs and for low parasitemia levels, posing yet another challenge for early malaria detection. Peng et al. more recently made use of magnetic resonance relaxometry to detect for hemozin in iRBCs (Peng et al., 2014). This new approach can detect for < 10 parasites/μL of whole blood and may be adapted into a microfluidic device by using portable magnets and radio-frequency detection probe.
3. Sepsis
Sepsis is a serious and life-threatening complication due to an infection. It is characterized by whole-body inflammatory state (called systemic inflammatory response syndrome or SIRS) that occurs due to chemicals secreted to fight the infection (Levy et al., 2003). The inflammation can overwhelm the blood cleansing capacity of organs and protective functions of immune cells (Hotchkiss and Karl, 2003). There are several studies published in the literature and readers who are interested on sepsis, its causes, diagnosis and prognosis are strongly suggested to read these articles (Hotchkiss and Karl, 2003, Lever and Mackenzie, 2007). Sepsis can lead to multiple organ failure, shock and death (for instance mortality rises by 7.6% every hour of delay in administering antibiotics in septic shock) (Kumar et al., 2006). Therefore, early, fast, and accurate diagnosis and treatment of sepsis is of paramount significance.
Current diagnostic and therapeutic approaches such as physical examination, analysis of blood, urine and molecular diagnostic techniques, unless integrated, can be suboptimal. Besides, due to drawbacks like high sample volume required, and being time and labor intensive, these methods are not compliant with the ASSURED criteria (Ritzi-Lehnert, 2012). Su et al. have extensively reviewed the state-of-the-art for bacteria detection by looking at how specific molecular techniques can be applied for sepsis/infection management (Su et al., 2015). Innovations that facilitate sepsis management are categorized in this review into two broad category of i) diagnosis (i.e., detection of pathogen and monitoring biochemical composition of patients' bodily fluids) and ii) treatment.
3.1. Sepsis diagnosis
Sepsis syndrome identification is difficult as the clinical signs of septicemia can be very similar to those of other lethal diseases, including high body temperature, difficulty in breathing, abdominal pain and abnormal heart pumping function (Edmond and Zaidi, 2010). Escherichia (E.) coli is the most common cause of early-onset sepsis in neonates (Camacho-Gonzalez et al., 2013). Recently, different approaches have been presented by researchers for detection of E. coli. For instant, methods such as magnetic separation, fluorescence staining, electrical detection and microfluidic immunoassay were used to capture and identify E. coli from blood (Yang and Li, 2006, Wang et al., 2012b) (Fig. 4A). Golden et al. developed a multi wavelength microflow cytometer using grooved microfluidic channel. E. coli assays were performed to demonstrate the capability of the device in discriminating particles (Golden et al., 2009). Bisceglia et al. also utilized DEP microfluidic device to isolate E. coli (Bisceglia et al., 2015). These chips can reduce diagnosis time by bypassing bacteria culture. However, further work is desired to reduce the processing time (~ few hours to few min) and the use of highly diluted blood. Kang and colleagues integrated expertise in DNAzyme-based sensor, droplet encapsulation and particle counting system to create a comprehensive droplet digital detection (IC 3D) system (Fig. 4B) (Kang et al., 2014a). This platform can detect as low as 1 E. coli bacterium in diluted blood sample in a culture/amplification-free process. Schwartz and Bercovici leveraged on isotachophoresis and fluorescently labeled antimicrobial peptides to continuously remove E. coli that are suspended in water (Schwartz and Bercovici, 2014). This technology may be potentially applied to detect bacteria in bodily fluids especially in early sepsis onset or sepsis suspect.
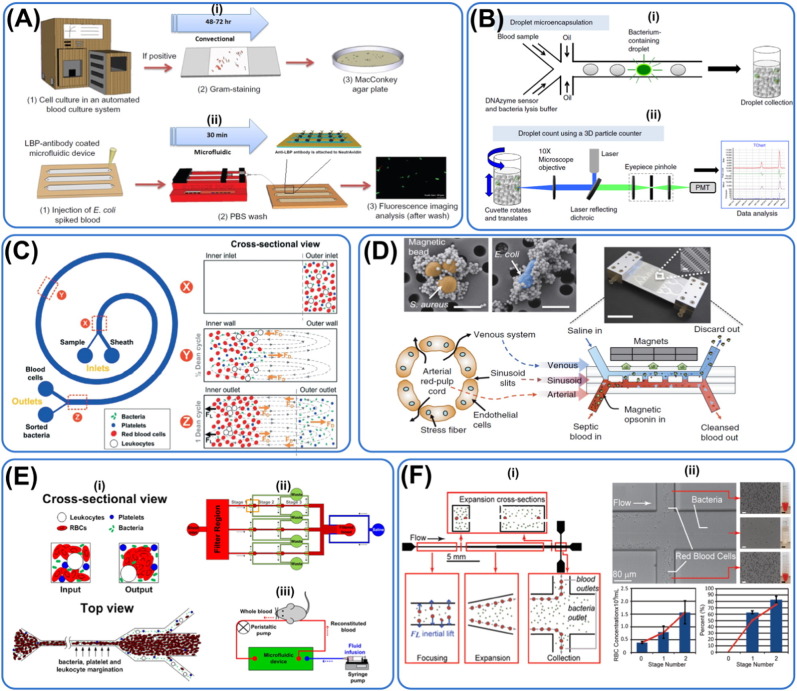
Fig. 4.
Microfluidic techniques contributing to sepsis management. Diagnosis: (A) Immunoaffinity, (B) droplet microfluidic, (C) spiral channel inertial microfluidic. Treatment: (D) straight channel inertial microfluidic, (E) margination, (F) treatment through filtration. (A) Immuno-affinity method to capture bacteria coupled with fluorescence imaging. The new technique takes just 30 min to complete compared to traditional bacteria culture that takes 2–3 days, greatly reducing diagnosis time. Reprinted with permission from Wang et al. (2012b). (B) IC 3D system where there is enrichment of bacteria and subsequent detection by fluorescence intensity. Bacteria are encapsulated in single droplets together with bacteria-specific DNAzyme. The platform is coupled with optical imaging and different species of bacteria can be differentiated by their fluorescence intensity, hence guiding therapeutic intervention. Reprinted with permission from Kang et al. (2014a). (C) Spiral microfluidic device that makes use of Dean drag forces to focus bacteria and platelets at the outer wall while RBCs and leukocytes which experience more substantial inertial lift forces focus near the inner wall. The filtration device takes 20 min to process 1 min of whole blood with 65% recovery of pathogens that could be used for downstream RNA analyses. Reprinted with permission from Hou et al. (2015). (D) Blood cleansing device to remove microorganisms from the body. The artificial biospleen mimicks the structure and role of spleen. Contaminated blood containing magnetic opsonin is passed into the microfluidic channel at high flow rate and the external magnets are used to remove pathogens bounded to the magnetic elements and discarded. Cleansed blood is then returned to the subject (rats). This process did not activate complement cascade and coagulation while reducing the amount of inflammatory cytokines in the system. Reprinted with permission from Kang et al. (2014b). (E) Microfluidic device making use of margination to remove bacteria. As less deformable RBCs transverse to the side channels, it causes the margination of bacteria and leukocytes to the peripheral outlets as well, leaving the center outlet bacteria-free. Reprinted with permission from Hou et al., (2013). (F) Massively parallel arrangement of 40 straight channels utilizing inertial microfluidic for filtration of bacteria at a flow rate of 240 mL/h. ~ 80% in pathogen depletion efficiency was achieved with two cycles of processing. Reprinted with permission from Mach and di Carlo (2010).
Identifying the strains of sepsis-causing bacteria can guide therapeutic intervention. Recently, Patterson et al. reported a novel microfluidic chip-based detection and strain discrimination of microbial pathogens, particularly for Salmonella serovars derived from whole blood of septic mice (Patterson et al., 2013). Mahalanabis et al. also created a microfluidic based detection system for cell lysis and DNA extraction of gram-positive and gram-negative bacteria, known causative of sepsis that may be coupled with PCR to construct a μTAS (Mahalanabis et al., 2009). An improved version would be to create capabilities to differentiate even bacteria strains within the same gram family. STMicroelectronics and Mobidiag have unveiled a lab-on-chip system for DNA-based detection of ten sepsis-causing bacteria as well as methicillin-resistant strains of Staphylococcus (S.) aureus from positive blood culture samples. Similarly, Hou et al. have also made use of spiral inertial microfluidic technique to isolate different strains of bacteria with numbers as low as 10–100/mL (Hou et al., 2015) (Fig. 4C). This platform has also been coupled to RNA detection to profile antibiotic resistance of pathogens. Making use of mannose binding lectin (MBL) coated magnetic beads and subsequent magnetic flux concentrator, Cooper et al. were able to spread captured Candida (C.) albicans fungi (99%) thinly to optimize optical imaging (Cooper et al., 2014). Wang et al. argued that it is also important to differentiate live from dead bacteria to prevent unnecessary administration of antibiotics (Chang et al., 2014). The group thus designed a microfluidic system based on ethidium monoazide-based assay and PCR to probe for live bacteria from fluid isolated from periprosthetic joint infection.
Several groups have also capitalized on biochemical composition in septic patients for diagnostics and monitoring. Evidence shows that excessive nitric oxide production plays a key role in the cardiovascular manifestations of sever sepsis and septic shock (Boveris et al., 2002). Hunter and colleagues fabricated an amperometric nitric oxide (NO) microfluidic sensor with low background noise to monitor changes in blood NO levels, as a potential biomarker, during the onset of sepsis, and its configuration enabled rapid analysis of NO in small sample volumes (Hunter et al., 2013). Conventional systems for antibiotic susceptibility testing is often too long (16–24 h) for the timely treatment of sepsis. In their study, Choi and co-workers introduced a microfluidic based system that reduced anti-microbial susceptibility testing (AST) assay time for determining minimal inhibitory concentrations, by single bacterial time lapse imaging (Choi et al., 2012). Schotter et al. also reported the incorporation of magnetic markers with magnetoresistive sensors applied in the microfluidic device for cytokine-based diagnosis of sepsis (Van Der Wouden et al., 2011). One potential complication of this diagnostic method is that increased production of NO and cytokines could be triggered by other diseases and not necessarily a response to sepsis. Furthermore, individuals are likely to differ by their norm NO levels and the minimal inhibitory concentrations for AST. This may make this diagnostic technique become expensive due to a need to establish personalized reference/standard.
As sepsis can be associated with systemic intravascular activation of coagulation (Aird, 2005), it is useful to learn about the spatial distribution and location of tissue factor (TF) and the geometry of the vasculature that regulate coagulation. Shen et al. developed microfluidic systems with surfaces of phospholipid bilayers patterned with TF to demonstrate experimentally the threshold responses of initiation of coagulation to the size and shape of surfaces presenting TF (Shen et al., 2008). Thermal injury can trigger an inflammatory cascade that heralds shock, SIRS and even death (D'Avignon et al., 2010). To address the prognostic and diagnostic significance of chemotaxis after burn injury, microfluidic devices have been exploited to measure neutrophil directional migration speed in response to chemo-attractant gradients that can be established in response to infection (Butler et al., 2010). Using this assay, Butler et al. established a reference set of migration speed values for neutrophils from healthy subjects in comparisons with samples from burn patients. Boneschansker et al. also quantified leukocyte migration patterns (chemoattraction, repulsion, kinesis and inhibition) using their microfluidic device when different chemokines were added (Boneschansker et al., 2014). The group has extensively quantified the persistence and speed of migration with different dosage and receptor expression.
Based on the knowledge that bacteria have rigid cell wall and can withstand harsher chemical treatment as compared to blood cells, Zelenin and colleagues also introduced selective blood cell lysis, a sample preparation strategy to continuously isolate microorganisms from whole blood for downstream analysis (Zelenin Ramachandraiah et al., 2011). This method is useful as a sample preparation unit for molecular-based POC sepsis diagnostics. Li et al. were also able to enrich S. aureus by 107 fold using an agarose gel that is permeable to water but not bacteria (Li et al., 2014). The device was also clinically validated to facilitate sample preparation in a rapid and sensitive manner.
The current methods for sepsis diagnostic which is based on cellular and molecular analysis on bodily fluids are time consuming, costly and less sensitive can impede timely clinical interventions. Microfluidic tools have been developed to target various stages of sepsis to provide a more comprehensive picture of this disease. For instance, there are microfluidic tools that quickly identify infection by offering highly sensitive bacterial isolation (1 E. coli in 1 μL of undiluted blood sample) in less than an hour. There are also microfluidic platforms developed to monitor sepsis progression by monitoring biochemical changes in patients' bodies that are induced by sepsis. Furthermore, microfluidic tools providing information on antibiotic susceptibility and possibility of mixed infections have also been created to guide septic treatments. All in all, these microfluidic techniques can facilitate faster, more sensitive, cheaper way of targeting different stages of sepsis (infection, progression, mixed infection, antibiotic resistance etc.) that can guide clinical interventions. Nonetheless, a key challenge now will be to understand the limitations of these different diagnostic techniques and possibly integrate all these capabilities to create a platform that can provide bacterial/fungi strain identification and AST assay with high sensitivity of < 10 bacteria/mL and in a timely (< 30 min) manner.
3.2. Sepsis treatment
Current sepsis treatments usually involve antibiotics therapy with patients in critical conditions start antibiotic treatment immediately, even before the test results that identify the bacteria strain come back from the laboratory (Yung et al., 2009). The complexity and variability of the sepsis are more devastating in patients with antibiotic-resistant pathogens because of the lack of effective drugs (Kang et al., 2014b). Extracorporeal blood purification (i.e., hemofiltration) was suggested by Gotloib et al. in 1994 to be useful in septic lung injury resulting from the removal of inflammatory mediators (Gotloib et al., 1984). Novel theories have been proposed in order to explain the beneficial effects of high-volume hemofiltration (HV-HF) in severe sepsis or systemic inflammatory response syndrome (SIRS) therapy, although this hypothesis remains a subject of significant debate (Bouman, 2007). In general, there is no universal consensus on which components of the blood should be removed for better clinical outcome. To address the root of the problem, extracorporeal blood cleansing therapy that can rapidly remove microorganisms and endotoxins from blood has been proposed by Xia and colleagues in 2006 (Xia et al., 2006). Using a simple microfluidic system, they have shown successful removal of living E. coli bacteria bound to magnetic nanoparticles from solutions containing densities of red blood cells similar to that found in blood. The team further enhanced the throughput of their system using a multiplexed version of their earlier design with over 80% bacteria capture efficiency at a flow rate of 20 mL/h in a single pass (Yung et al., 2009). More recently, the same group came up with another design where they made use of magnetic beads coated with MBL to cleanse blood of bacteria, fungi, endotoxins without activating the complement cascade and coagulation (Fig. 4F). 90% of E. coli and S. aureus was reportedly depleted that also helped reduce immune cell response and inflammation (Kang et al., 2014b). Nonetheless, one main concern of using this technique is the lack of full understanding of nano-toxicity induced by the magnetic nanoparticles. This is because the biocompatible coatings on the nanoparticles can be degraded over time, exposing healthy cells to toxic free metal ions from the core. Furthermore, the method uses low flow rate (20 mL/h) in order to achieve high bacterial capture efficiency while the average body normally houses 5000 mL of blood. An extracorporeal blood cleansing procedure of 500 mL would require 25 h which may infeasible to implement continuously due to easy bubble formation and clogging in microfluidic channels.
Using the concept of inertial microfluidics in straight microchannels, Di Carlo and his team developed a massively parallel microfluidic device that passively separates pathogenic bacteria cells from diluted blood (1% hematocrit) with flow rate of around 240 mL/h and over 80% removal efficiency (Fig. 4D) (Mach and di Carlo, 2010). In addition, margination, the natural phenomenon where leukocytes move toward the sides of blood vessels (Goldsmith and Spain, 1984), has inspired novel method for treating sepsis. For instance, based on margination phenomenon, a high-throughput and label-free microfluidic blood filtration technique for microbial removal from whole blood was introduced by Hou et al. (2012). They have demonstrated high removal efficiencies of ~ 80% and ~ 90%, for E. coli and S. cerevisiae pathogens, respectively. More recently, the same team has reported successful removal of pathogens from a murine model of sepsis at flow rate of ~ 90–150 mL/h using a 32-channel parallelized platform, demonstrating the feasibility of their systems as a blood cleanser in clinical settings (Fig. 4E) (Hou et al., 2015). As aforementioned, a huge issue in using the microfluidic channels is the possibility of clogging especially when whole blood with natural coagulants is being filtered. In addition, while these techniques offer high throughput in a few hundred mL/h, there is still insufficient characterization on the effects of high shear forces on the integrity of RBCs and potential immune activation of leukocytes.
In summary, for septic blood cleansing using microfluidic platforms to be clinically useful, they must be able to perform blood cleansing in higher flow rates in the range of mL/min as human patients have much larger volume of blood than murine models while understanding the effects of shear forces on endothelial cells lining vessel walls, RBCs and leukocytes. It is also necessary to pursue higher bacterial isolation efficiency as there is the danger that the microfluidic platforms may propagate the spread of bacteria in the patients' bodies. In addition, these platforms generally lack comprehensive long-term characterization of immune response to potential debris from microfluidic substrates and other materials used such as magnetic beads.
4. HIV
HIV affects more than 40 million individuals worldwide, with about 85% of affected patients living in developing nations who are in need of an ASSURED diagnostic and antiretroviral therapy (ART) monitoring platform (UNAIDS, 2013). HIV infection and AIDS are broad terms that describe a range of symptoms caused by a retrovirus (Fauci, 2003). The retrovirus is primarily transmitted to the host via fluid exchanges during sexual contact or contaminated needles (horizontal epidemic), but can also spread from an infected mother to her child (vertical epidemic). Despite the lack of any obvious signs, the infected individual actually undergoes continuous loss of CD4 + T cells, eventually crippling the immune system and allowing the patient to succumb to opportunistic infections (e.g. pneumonia). This later stage is termed as AIDS (Dalgleish et al., 1984).
There is currently no established cure for HIV infection, but several anti-retrovirus drugs have shown efficacy in suppressing late-stage symptom onset (Deeks et al., 2012). These drugs are more effective when the disease is detected in the earlier stages (Kitahata et al., 2009).
HIV infection can lower the number of immune cells such as CD4 + T-lymphocytes. During ART, virus replication is suppressed, leading to gradual increase in CD4 + T-lymphocytes (Simon and Ho, 2003). Therefore, the enumeration of CD4 + T-lymphocytes and HIV viral load is useful for diagnostic and prognosis purposes. Typically, for effective clinical application, around 200 CD4 + cells/μL and 400 HIV virus/mL need to be detectable from whole blood (Lee et al., 2010).
Current strategies for disease detection include measurement of viral load (O'Gorman and Gelman, 1997) and determination of CD4 + cell counts (Ho et al., 1995) from blood or oral fluids. Conventional techniques such as polymerase chain reaction (PCR) runs or western blots are rapid but are limited by the inability to detect low viral load or gradual fluctuations in CD4 + cell counts, especially in the early stages of symptomless HIV infection. For infants within 11/2 year old, rapid antibody assays are also not sufficient to validate the presence of HIV infection due to persisting maternal antibodies (Kellerman and Essajee, 2010). Shafiee et al. have comprehensively reviewed the HIV disease, its diagnostics and also management. In their review, they present the state-of-the-art HIV diagnostic technologies for CD4 + T lymphocyte count, viral load measurement, and drug resistance testing highlighting the role of microfluidics in HIV (Shafiee et al., 2015).
Currently, immune-capture of CD4 + T cells is the most prevalent method. Cheng and co-workers made use of cell affinity chromatography (CD4 antibody functionalized microchannel surfaces) under controlled shear stress to isolate CD4 + lymphocytes from 10 μL whole blood to for HIV diagnosis (Cheng et al., 2007). Differential surface CD4 expressions and sizes influence cell binding on antibody-functionalized surface, allowing lymphocytes and monocytes to be differentiated. The platform has a specificity of ~ 95% in regard to CD4 + lymphocytes (versus monocytes) capture and can perform the detection in less than 10 min. However, this platform relies heavily on the coating of antibody on the microchannel surfaces and with a lack of technology for strict quality control, reproducibility of well-coated platforms with antibodies may be compromised. Furthermore, the orientation of the CD4 + lymphocytes as they flow through the microchannels may not expose them to the coated surfaces, thus reducing diagnostic sensitivity.
Other groups further integrated immune-affinity with antibody functionalized surfaces with automated imaging to quantify cell count. Alyassin et al. developed a microfluidic platform coupled to automated optical imaging that is able to perform CD3+CD4+ lymphocytes counting 100 times faster than manual counting (Alyassin et al., 2009) (Fig. 5A). Their designed software is able to account for uneven background, overlapping cell images and detect for cells with multiple stains, offering great flexibility to HIV detection. Kim and colleagues also made use of optimal imaging with quantum dots to identify HIV (Kim et al., 2009). Dual-labeled quantum dots were able to recognize external envelope gp120 protein and high mannose glycans at 20 ng/μL, the lowest dilution limit, and with high sensitivity. Ramachandraiah and co-workers made use of centrifugal microfluidic on a functionalized CD platform for automated counting of CD4 + lymphocytes using microscope (Ramachandraiah et al., 2013). Wang et al. also developed an m-ELISA to detect CD4 + cell in unprocessed blood. Each step in ELISA was spatially confined and cellphone was used to quantify the number of CD4 + cells (Wang et al., 2014). To overcome potential loss of signal, Chin et al. capitalized on the effects of reduction of silver ions onto gold nanoparticles (Sia et al., 2004) and meandering channels for signal amplification (Chin et al., 2011) (Fig. 5B). Their fully-functional m-chip requires only 1 μL of unprocessed blood for HIV and syphilis diagnosis with near 100% sensitivity and specificity. Microfluidic platforms integrated with imaging capabilities facilitate automated diagnostic results that are not subject to users' interpretations and biases. However, to make these integrated platforms accessible to developing nations where there is much higher HIV infection rates, the imaging components must be cheap and portable like the cell-phone based system Wang et al. developed but yet with comparable spatial resolution sensitivity.
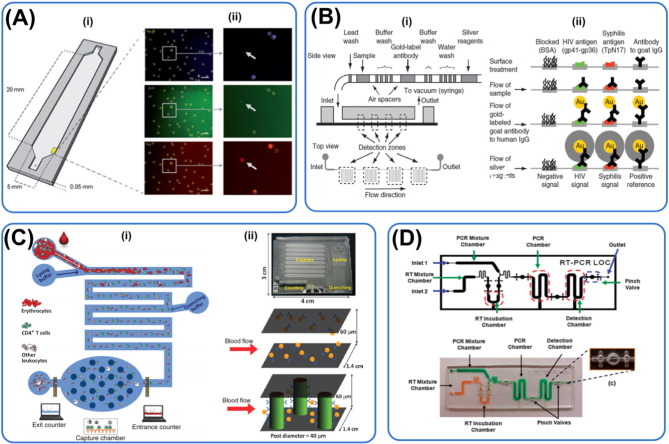
Fig. 5.
Microfluidic technologies for HIV diagnosis. Principles of diagnosis: (A & B) Immunoaffinity, (C) electrical impedance and (D) RT-PCR. (A) Captured CD3 + CD4 + lymphocytes were stained and counted automatically by the designed software. This device allows 100 × faster speed in identifying immuno-stained lymphocytes for HIV detection. Reprinted with permission from Alyassin et al. (2009). (B) Left panel shows the sequence of steps the sample undergoes as it moves through the equipment-free microfluidic device (m-chip). Right panel illustrates the various steps of the immunoassay. The reduction of silver ions on gold nanoparticle conjugated with specific antibodies for signal amplification, facilitating readout without the use of expensive optics. Reprinted with permission from Chin et al. (2011). (C) Whole blood is introduced and RBCs are lysed, leaving a supposedly pure population of white blood cells. Total number of lymphocytes is counted followed by capture of CD4 + and CD8 + lymphocytes with microposts. Differential electrical impedance signals of the cells provide information on the degree of contamination and number of target lymphocytes. Reprinted with permission from Watkins et al. (2013). (D) RT-PCR integrated into microfluidic channel. Top panel shows the schematic and the bottom panel shows the actual device. Valves are created to isolate different steps of the process such as incubation, reaction and detection. Reprinted with permission from Lee et al. (2008).
Song and co-workers presented a methodology which improved the speed of the ELISA assay by bypassing the blocking step while keeping the limit of detection of conventional ELISA (Song et al., 2012). In a recent work presented by the same group (Laksanasopin et al., 2015), a full laboratory immunoassay was run on a smartphone accessory using microfluidic technology and miniaturized hardware. The phone accessory incorporated the aforementioned ELISA assay using the power generated from the phone. To perform the test, 2 μL of diluted blood sample was added to the cassette, which was then inserted into the dongle. Absorbance readings of the level of silver precipitation were performed 15 min later and the results for all markers were displayed on the smartphone's interface. The device performed the assay with a sensitivity of 92 to 100% and a specificity of 79 to 100%.
Watkins et al. developed a biochip for CD4 + and CD8 + lymphocyte counting based on differential electrical impedance (Watkins et al., 2013) (Fig. 5C). Whole blood is introduced and RBCs are lysed, leaving a supposedly pure population of white blood cells. Target lymphocytes flowing through a microfluidic channel can generate a spike in impedance with an amplitude and width proportional to their sizes and translocation velocities as they block the flow of current. The CD4 + and CD8 + lymphocyte count closely matched results from flow cytometer over the range of 40–1000 cells/μL and the entire process can be performed in about 20 min. This technique, however, requires on chip lysis which may also lyse target cells non-specifically and generate lysis debris that causes clogging and interferes with readouts. The authors also did not demonstrate the spike in impedance caused by RBCs that escape lysing to avoid no-specific readouts.
Glynn et al. specifically isolated magnetically tagged CD4 + cells directly from patient blood (Glynn et al., 2014). The low-cost and disposable microfluidic chip was operated by dual-force CD4 + cell magnetophoresis; whereby the interplay of flow and magnetic fields governed the trajectory of target cells depending on whether the cell bound to a magnetic microbead. Instrument-free pumping was implemented by a finger-actuated elastic membrane; tagged beads were laterally deflected by a small and reusable permanent magnet. The use of nanoparticles have enhanced the sensitivity of the technique as compared to antibody functionalized microchannel surfaces as nanoparticles are smaller and are capable of binding to target lymphocytes. Although this method is made cheaper without the use of instrument for generating flow, finger-actuated pressure may not generate steady flow, making diagnostic results non-reproducible.
Nonetheless, the common limitation of these techniques is the need for blood lysis that can also lyse target lymphocytes and create cell debris that lead to microchannel clogging. Furthermore, as these microfluidic platforms tend to rely on antibodies for HIV detection, and knowing that HIV progression can influence CD4 + expressions on T-lymphocytes (Trial et al., 1995) and non-specific binding to antibody (Anderson et al., 1993), key parameters such as specificity and limit of detection can be adversely affected.
To overcome the heavy reliance on antibodies, Lee et al. thus developed a disposable polymer for reverse transcriptase (RT)-PCR and chemiluminescence assay for HIV diagnostic that is not constraints by the limitations of the antibodies used (Lee et al., 2008) (Fig. 5D). Primers targeting p24 (a major core protein) and gp120 are utilized in this biochip. This platform can perform diagnosis in less than an hour and is supposedly more accurate than the use of antibodies. However, in this device, the highly unstable RNA is currently isolated externally and may introduce inconsistencies and contamination due to technicians' skills. It is worth exploring the possibility of on-chip RNA isolation and amplification as discussed in Section 2.3 molecular analysis for malaria diagnosis.
Emerging HIV diagnostics methods such as nanoplasmonic detection (Inci et al., 2013) and nanostructured photonic crystals (Shafiee et al., 2014) may also be integrated with microfluidic for use in resource-scarce communities where diagnostics is most needed. Nonetheless the described technological advances, HIV management in least developed region remain a major challenge as expensive and bulky machines are needed for the readout.
In summary, unlike malaria and sepsis where iRBCs and bacteria can be isolated based on their physical properties respectively, HIV viruses are too small for isolation using microfluidic devices. Therefore, the current technique for microfluidic diagnostic for HIV depends highly on the use of antibodies targeting CD4 + and CD8 + lymphocytes. However, HIV stages and progression, can reduce the specificity of binding. Furthermore, when target lymphocytes flow through the microchannels, due to unsteady flow rates and differential cell sizes, they may not be oriented properly to be exposed to the antibody coatings on the microchannels, hence reducing diagnostic sensitivity. The cost for antibody coating can also be prohibitive as antibody solution is usually used in excess. This can creates an issue of irreproducible manufacturing. While DNA/RNA based technique circumvents the problem of antibodies reliance and can offer higher sensitivity and specificity, no group has yet to create a fully integrated system that is free from users' skills and biases. Nonetheless, interested groups can learn from research in the malaria field where on-chip nucleic acid isolation and amplification have been demonstrated.
5. Other infectious diseases (SARS, dengue, tuberculosis)
SARS which is caused by a new coronavirus (CoV) started to spread around the world in 2003. The outbreak of SARS was eventually controlled through management strategies such as isolation of suspects and their close contacts. Due to the absence of effective cure, SARS has claimed 744 lives in 2003 mainly in the Asian-Pacific region (Hui et al., 2004). Conventional means that include determining antibody level and isolating SARS virus have been employed for diagnosis. Nonetheless, antibody levels start increasing only about 10 days after infection while cell culture of SARS virus is tedious and time-consuming (Zhou et al., 2004).
Zhou et al. created a diagnostic platform for SARS-CoV via multiplex real time (RT) PCR and capillary electrophoresis integrated on a microfluidic chip. The system was able to provide positive results from 17 (out of 18) of the clinically diagnosed SARS patients while conventional RT-PCR with agarose gel electrophoresis gave only 12 positive results (Zhou et al., 2004). Huh et al. also developed a microfluidic platform that integrated magnetic force-activated micromixer system and functionalized surfaces for cell lysis, purification of intracellular proteins and SARS-CoV detection (Choi et al., 2002). Similarly, Ramalingan and colleagues made use of isothermal helicase-dependent amplification method to detect for SARS virus DNA on an integrated microfluidic PCR chip (Ramalingam et al., 2009). Similar to HIV, as SARS is caused by a virus, there is limited techniques for diagnosis as HIV viruses are difficult to isolate. Currently, there are only a few microfluidic-based diagnostic platforms for SARS detection. The platforms that make use of antibody (against SARS-CoV) functionalized surfaces face the problems of non-specific binding due to disease progression and reproducible manufacturing of functionalized surfaces. The antibodies can also be expensive and be used in excess in order to fully functionalize the microchannel surfaces. This can increase the cost of the microfluidic device and makes it less accessible to patients in the developing nations. On the other hand, nucleic acid-based method offers higher sensitivity and specificity as compared to antibody method. However, this latter technique requires a longer diagnostic time for nucleic acid isolation and amplification. There must also be precise temperature control for efficient amplification to occur which may add manufacturing complexity to the diagnostic chip. Respiratory infectious diseases represent the one of the largest cause of deaths and DALYs lost (Yager et al., 2006). Therefore, techniques in areas of immune-affinity, optics are encouraged to be applied to this area to boost disease diagnosis.
Dengue virus is propagated by the mosquito vector and the majority of its infections is concentrated in Latin America and Asia (Rigauperez et al., 1998). There are four serotypes of dengue virus that can produce mild dengue fever to severe dengue shock syndrome in humans (Gubler, 2002). Due to the small sizes of the dengue viruses, the main diagnostic methods are still nucleic acid or protein/serological based. The clinical tests for dengue such as hemagglutination inhibition and neutralization can be non-specific and prohibitively expensive. Teles argue that there is thus a role that microfabricated devices can play to facilitate dengue diagnosis (Teles, 2011). Readers can refer to more extensive review by Darwish et al. on conventional means of dengue diagnostic (Darwish et al., 2015).
Su and colleagues coated the two surfaces of an immunochip with glycoprotein-E and non-structural protein 1 (NS1) and analyzed signals this piezotransducer-like arrangement to diagnose for dengue (Su et al., 2003). Kumbhat et al. utilized surface plasmon resonance for diagnose as deposition of dengue virus increased the resonance angle (Kumbhat et al., 2010). Zaytseva and colleagues made use of fluorescent liposomes conjugated with reporter probes that binds to dengue virus RNA for diagnosis (Zaytseva et al., 2005). Weng et al. report the development of a microfluidic device functionalized for antibodies specific to dengue virus and equipped with a suction unit to create negative pressure for the transport and mixing of fluids (Weng et al., 2011). The entire duration needed for diagnosis can be reduced from 4 h to ~ 30 min with their device. Similarly, Gunda and colleagues made use of dengue virus NS1 bound to micropillars to improve the surface area for binding and subsequently, enhance detection sensitivity (Gunda et al., 2013). Aeinehvand and team incorporated a microballoon mixer (that expands and contracts) with centrifugal microfluidic platform to improve the sensitivity of ELISA detection of dengue virus due to improved mixing (Aeinehvand et al., 2015). Recently, Zhang and colleagues developed a multi-stack paper immunoassay that removes proteinaceous substance in the saliva before detection of target antigen with adsorbed antibody (Zhang et al., 2015). This device obviates the steps of centrifugation and boost detection of dengue-specific immunoglobins (IgG) in the serum that can be used to differentiate primary from secondary infection.
In addition to diagnosis, there have also been efforts to improve our understanding of the biology of the dengue virus. For instance, Huang et al. also made use of microfluidic device coupled with optics to measure the effects of carrageenans to inhibit the life cycle of the dengue viruses. The group measured the cellular oxygen consumption rates (OCRs) that can reflect cellular metabolic rates and found that treatment with carrageenans maintained the OCRs of cells indicating high cell viability (Huang et al., 2014). Their technology is also applicable for drug screening/toxicity for a range of infectious diseases.
Microfluidic devices for dengue diagnosis have demonstrated higher sensitivity with shorter processing time as compared to conventional methods which can be useful for timely clinical interventions. Other than the aforementioned problems with the antibody functionalized surface method for diagnostic, one key limitation in all these studies is that the microfluidic platforms cannot differentiate different types and stages of dengue infection. Dengue can be caused be 4 different dengue viruses. In addition, severity of disease is greater with co-infection and re-infection with another virus. Hence, the challenge now is to create microfluidic device useful for clinical dengue diagnosis by offering results that can differentiate virus strains and disease stages.
WHO estimated that tuberculosis claims about 1.3 million deaths annually (WHO, 2013). Although this is a largely treatable diseases, the large number of undiagnosed cases (global detection rate ~ 63%) prevent timely therapeutic interventions (McNerney and Daley, 2011). Readers are encouraged to refer to review by Dheda et al. for a more comprehensive discussion on available diagnostic tools such as liquid culture, smear microscopy and the Xpert MTB/RIF assay for tuberculosis (Dheda et al., 2013). Conventional diagnostic means for tuberculosis face the same limitations as other infectious diseases diagnosis such as poor specificity, high costs and large volume of sample needed (Mani et al., 2014). To this note, microfabricated devices may potentially fill some of the gaps in diagnostic efforts for tuberculosis. For instance, Wojcik and co-workers designed a colorimetric DNA test based on conjugation between gold nanoparticles covalently bound to the gene fragment of the Mycobacterium (M.) tuberculosis (Bernacka-Wojcik et al., 2013). This test requires only 90 ng of target DNA and 30 min for completion. Multiplexed kits using multiple antigens specific for M. tuberculosis have also been developed (Dheda et al., 2013). Liong et al. made use of magnetic barcoding technique to detect DNA of M. tuberculosis in 2.5 h (Liong et al., 2013). The team describes a partially integrated microfluidic platform where the DNA of M. tuberculosis is first extracted externally and then captured onto polymeric beads with complimentary capture DNA. The beads are then coupled to probe-DNA bound magnetic nanoparticles which render the MNP-loaded beads paramagnetic. As the paramagnetic MNP-loaded beads can produce local magnetic fields, beads loaded with different quantity of M. tuberculosis DNA produce different decay rate when subject to nuclear magnetic resonance readings, allowing diagnosis to be performed. A portable microfluidic device capable of performing on-chip loop-mediated isothermal nucleic acid amplification (LAMP) with manual inspection has also been designed by Fang and colleagues for tuberculosis detection in 1 h (Fang et al., 2012). Recently, a label-free assay for detection of DNA of M. tuberculosis and related drug resistance has been designed (Domínguez et al., 2015). This platform makes use of the quantification of hydration induced stress on micro-cantilever functionalized oligonucleotide probes before and after hybridization with specific nucleic acid targets. The total analysis time is 1.5 h and a sensitivity of 2 pg/mL was reported. However, in these described methods, the platforms are not fully integrated and require off-chip processing steps such as nucleic acid extraction which may affect the reproducibility of the diagnostic results due to skills of the technicians/users.
Wang and co-workers thus developed a fully integrated (individual chambers for cell lysis, DNA isolation, amplification etc.) microfluidic cartridge to interrogate single-base variations in codons for multi-drug resistant form of M. tuberculosis (Wang et al., 2012a). Micropillars are also used to enhance the density of DNA adsorption to enhance the colorimetric readout. This platform can be fully automated to prevent users' biases, mass-produced as it is made of thermoplastic which ensures production quality and low cost and disposable useful for one-time use and potential infections with contaminated samples.
As tuberculosis-causing bacteria can be isolated from pleural fluids, microfluidic devices making use of inertial focusing, DEP and magnetic beads (as described in malaria and sepsis diagnosis) can be used. Technologies with their strengths and weaknesses described in the previous sections can be used as a reference for creating microfluidic platforms for tuberculosis diagnosis. Although there are currently not many microfluidic devices created for tuberculosis diagnostic, there are other point-of-care assays that may be potentially integrated with microfluidics such as those making use of mass/piezoelectric (Ren et al., 2008), optical (Buijtels et al., 2008), enzymatic (Xie et al., 2012) and acoustic techniques (He et al., 2003). Nanotechnologies are also emerging as highly useful tools to enhance the sensitivity of signals for diagnostic results (Vandan et al., 2009). However, one important note is that as tuberculosis has largely been eliminated from developed nations, the devices should be mainly targeted at developing nations and should have low cost and high portability (Fig. 6 ) (Table 1 ).
Fig. 6.
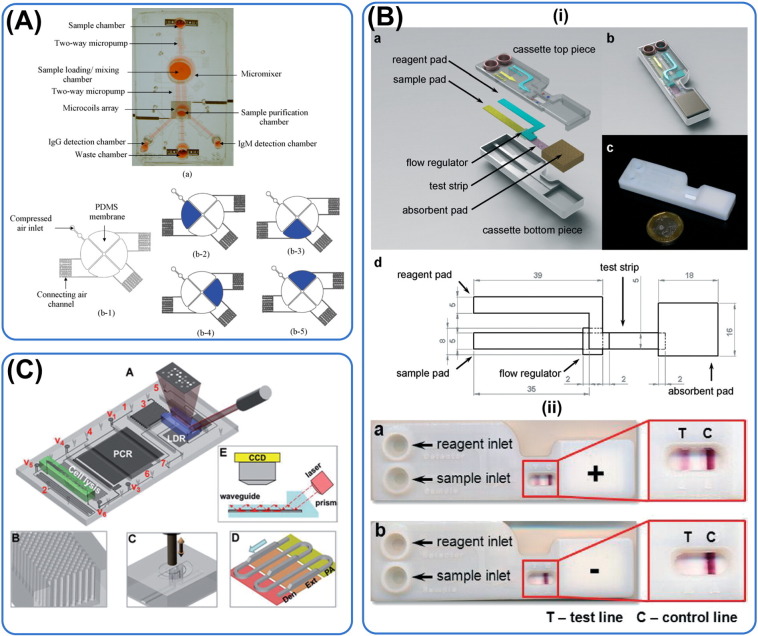
Microfluidic platforms for the diagnostic of dengue and tuberculosis. (A) Top: dengue virus-bound magnetic beads in the sample loading/mixing chamber are used to identify dengue patients' samples containing IgG and IgM. Magnetic coils are then turned on to collect the IgG/M-bound magnetic beads followed by purification and subsequent fluorescent readouts at the detection chambers loaded with antibodies. Bottom: design of micro-mixer for efficient mixing of magnetic beads and biological samples. Reprinted with permission from Lee et al. (2009). (B) Design of a stacking lateral flow paper microfluidic assay for dengue diagnostic. Saliva from patients are filtered through a paper layer made of fiber glass to remove proteinaceous substances. A detection sensitivity of 20 ng/mL of α-fetoprotein in the saliva serum is achieved. A control line is also present in the device as a positive control. Reprinted with permission from (Zhang et al., 2015). (C) A fully integrated thermoplastic microfluidic device for detection of DNA of M. tuberculosis and drug resistance with fluidic path controlled by electrically actuated solenoid. Steps such as cell lysis, DNA isolation and PCR can be performed fully on the microfluidic chip. Micro-pillars are also employed to enhance the density of adsorbed DNA for colorimetric detection. Reprinted with permission from Wang et al. (2012a).
Table 1.
Examples of microfluidic devices for diagnosing infectious diseases and their characteristics.
| Infectious diseases | Principle of diagnosis | Advantages | Disadvantages | LOD | Sample | Reproducibility | Level of training for users | Processing time | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Malaria | Deformability: isolate less deformable iRBCs |
|
|
– | Diluted and whole blood |
|
Low to moderate: External pump, microscope | 5 μL/min | Hou et al. (2010) |
| Malaria | Inertial focusing: remove leukocytes |
|
|
2–10 parasites/μL | Lysed 0.25 × blood |
|
Low to moderate: External pump, centrifuge, microscope | 400 μL/min | Warkiani et al. (2015) |
| Malaria | DEP | • Optimization by tuning frequencies • Can isolate ring stage iRBCs |
• Joule heating • Variation in cell sizes • Heat-related cell death • Incompatible with areas with no access to stable electricity |
– | Diluted blood | • PDMS are deformable at high flow rate • Bubble generation • Channel clogging |
Moderate: External pump, function generator, microscope | Up to 1500 μL/min | Gascoyne et al. (2004) |
| Malaria | PCR |
|
|
500 parasite/μL | Diluted blood |
|
High: Pipetting, PCR machine | Close to an hour (lysis, mixing, reaction) | Marshall et al. (2011) |
| Malaria | Droplet microfluidic + imaging |
|
|
< 1 parasite/μL | Diluted blood |
|
Moderate to hard: External pump, droplet formation, fluorescence microscope | 2.5 h from sample preparation to readout | Juul et al. (2012) |
| Malaria | Magnetic |
|
|
– | 90% diluted blood |
|
Moderate: External pump, magnets | 1.6 μL/min | Nam et al. (2013) |
| Sepsis | Droplet microfluidic + particle counter |
|
|
1 E. coli bacterium/mL | Diluted blood |
|
Moderate: External pump, droplet formation, fluorescence microscope | 1.5–4 h | Kang et al. (2014a) |
| Sepsis | Magnetic |
|
|
1 C. albicians fungus/mL | Diluted blood |
|
Moderate: External pump, magnets, fluorescence microscope | < 3 h | Cooper et al. (2014) |
| HIV | Immunoassay |
|
|
– | Diluted blood |
|
Moderate to hard: Techniques for ELISA | 15 min | Chin et al. (2011) |
| HIV | Immunoassay + electrical impedance |
|
|
– | Diluted blood |
|
Moderate: External pump, sensors | < 20 min | Watkins et al. (2013) |
| SARS | RT-PCR |
|
|
– | Autopized lung tissue |
|
High: Pipetting, complex setup for readout | < 1 h | Zhou et al. (2004) |
| Dengue | Immunoassay |
|
|
20 ng/mL | Saliva |
|
Easy to moderate: need to train to remove bubbles in sample | 20 min | Zhang et al. (2015) |
| Tuberculosis | DNA primer |
|
|
90 ng DNA | Synthetic DNA |
|
Easy: need training for using LED and external pumps | 30 min | Bernacka-Wojcik et al. (2013) |
6. Commercialization
Commercial developments play a key role in making POC technologies accessible to resource-scarce communities. Table 2 shows the list of some microfluidic devices for diagnosis of various infectious diseases. As it can be seen from Table 2, most of the commercial technologies are either antibody or DNA primer although other means of diagnostic are available. These platforms which are based on these analytes are more adaptable to diagnose a wider range of diseases. For instance, the platform developed by Micronics were used for malaria initially but has expanded also to sepsis.
Table 2.
List of microfluidic-based diagnostic companies.
| Infectious diseases | Technology | Technical features | Name of device | Company | Website |
|---|---|---|---|---|---|
| Infectious diseases | Primer or antibody | Integrated platform, small sample volume, 1 h test time | PanNAT® | Micronics | micronics.net |
| Infectious diseases | Antibody | Disposable, compact, small sample volume, 10 min test time | N.A. | OPKO diagnostics | opko.com |
| Infectious diseases | Antibody | Piezofilm technology, 10 min test time, | N.A. | Vivacta (part of Novartis) | novartis.com |
| Infectious diseases | Antibody | Self-contained waste reservoir, small sample volume, multiplexed detection | Asklepios | Genefluidics | genefluididcs.com |
| Infectious diseases | Primer | Lab in a tube platform, 30 min test time | Liat™ | IQumm (part of Roche) | roche.com |
| HIV/AIDS | Antibody | Differentiate between HIV 1 and 2, 100% sensitivity, 99.75% specificity, 15 min test time | Alere Determine™ | Alere | alere.com |
| HIV/AIDS | Primer or antibody | Electrowetting, digital microfluidics, | N.A. | Advanced Liquid Logic (part of Illumina) | liquid-logic.com |
| Sepsis, HIV/AIDS | Antibody | Disposable, integrated platform with positive and negative controls, | Spinit® CRP/BC | Biosurfit | biosurfit.com |
| HIV/AIDS | Electrical impedance | Count cells by analyzing intracellular content electrically, | CD4 system | Daktari Diagnostics | daktaridx.com |
| HIV/AIDS | Primer | CD4 + cells monitoring, compact, quantum dot detection | N.A. | LabNow | labnow.com |
| HIV/AIDS, sepsis | Antibody | CD4 + counting, CD64 sepsis marker monitoring, small sample volume | Accellix | LeukoDx | leukodx.com |
| Tuberculosis | Antibody | Low cost, low power fluorescence, | LightDeck® | Mbio Diagnostics | mbiodx.com |
| Tuberculosis | Primer | Can detect drug resistance, compact time, 3 h detection time, detect multiple gene targets, disposable | VereMTB | Veredus Laboratories | veredus.com |
| Tuberculosis | Primer | Detect for DNA of Mycobacterium tuberculosis and its drug resistance, 91% sensitivity, 94% specificity, | GeneXpert MTB/RIF | Cepheid | cepheid.com |
| Sepsis | Primer | Integrated PCR platform, detect multiple bacteria strains | Jaguar™ | HandyLab (part of BD) | bd.com |
Information was obtained from the website of the various companies. There may be further developments since the time of sourcing these information and readers are encouraged to refer to the original websites of the companies to learn about the various technologies.
Search function can be used on the company's website to look for the respective products.
There are now greater number of publications to guide innovators to commercialize their microfluidic technologies (Chin et al., 2012, Su et al., 2015). However, some of the challenges deserve to be highlighted for innovators and we will briefly mention two of them. (1) Financing. Diagnostics targeted for resource-scarce communities have to be priced low to be competitive and accessible. This creates pressure for profit generation and may not appeal to private venture capitalists. Therefore, it is more likely that companies that can serve both developed and developing nations flourish in the market. Governments and philanthropic foundations such as Bill and Melinda Gates have helped commercialize POC diagnostics for resource-scarce communities and we expect them to continue playing significant role in financing. (2) Integration. As mentioned earlier in Section 2.3, the various technologies such as for fluid delivery, mixing and on-chip heating and cooling for microfluidic PCR has to be combined to create a functional product. Another example is the acquisition of Advanced Liquid Logic which specializes in electrowetting and droplet microfluidic by Illumina for its molecular diagnostics business arm. There is a need for collaboration amongst different teams or even companies to capitalize on one another's expertise.
7. Conclusions and future outlook
Conventional diagnostic tools for infectious diseases are limited by their long processing time, high costs, non-user-friendliness, technical complexity, poor sensitivity and specificity. Microfluidic devices have demonstrated that even with small sample volume, they are able to offer high accuracy in detection, and at a cost compatible with the average income in resource-scarce communities. Many innovative ideas have been generated for malaria diagnosis such as use of dielectrophoretic properties and lower deformability of iRBCs. Droplet microfluidics has also been integrated with software to analyze for bacteria contamination and sepsis. Other than diagnostic tools, therapeutic equipments such as blood cleansing devices have also been created from advances in microfabrication technologies.
Nonetheless, it should be stressed that diseases such as SARS and dengue which are concentrated in developing nations are still very much neglected by the scientific communities in developed nations. Governments, non-governmental organizations and philanthropic foundations play important roles to finance projects in these neglected infectious diseases for improvements to be made. Additionally, companies commercializing POC devices also face challenges in raising funds from private sectors due to their target markets in resource-scarce settings.
We foresee that mobile technology will play a key role together with microfluidic diagnostic devices for remote health monitoring, especially in resource-scarce regions where healthcare is poorly accessible. With that in mind, electrochemical, electrical impedance and colorimetric assays will be very much useful as means for readout using cell phones. We believe continued efforts by scientists and biomedical industry in developing microfluidic technology can benefit the millions of people who are disproportionately affected by infectious diseases.
References
- Aceti A., Bonincontro A., Cametti C., Celestino D., Leri O. Electrical conductivity of human erythrocytes infected with Plasmodium falciparum and its modification following quinine therapy. Trans. R. Soc. Trop. Med. Hyg. 1990;84(5):671–672. doi: 10.1016/0035-9203(90)90140-a. ([Internet]. Jan [cited 2015 Mar 1] Available from: http://www.ncbi.nlm.nih.gov/pubmed/2278066) [DOI] [PubMed] [Google Scholar]
- Adamo A., Sharei A., Adamo L., Lee B., Mao S., Jensen K.F. Microfluidics-based assessment of cell deformability. Anal. Chem. 2012;84:6438–6443. doi: 10.1021/ac300264v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeinehvand M.M., Ibrahim F., Harun S.W., Djordjevic I., Hosseini S., Rothan H.A. Biosensing enhancement of dengue virus using microballoon mixers on centrifugal microfluidic platforms. Biosens. Bioelectron. 2015;67:424–430. doi: 10.1016/j.bios.2014.08.076. ([Internet]. May 15 [cited 2015 May 6] Available from: http://www.sciencedirect.com/science/article/pii/S0956566314006721) [DOI] [PubMed] [Google Scholar]
- Aird W.C. Sepsis and coagulation. Crit. Care Clin. 2005:417–431. doi: 10.1016/j.ccc.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Alizadehrad D., Imai Y., Nakaaki K., Ishikawa T., Yamaguchi T. Quantification of red blood cell deformation at high-hematocrit blood flow in microvessels. J. Biomech. 2012;45:2684–2689. doi: 10.1016/j.jbiomech.2012.08.026. [DOI] [PubMed] [Google Scholar]
- Alyassin M.A., Moon S., Keles H.O., Manzur F., Lin R.L., Hæggstrom E. Rapid automated cell quantification on HIV microfluidic devices. Lab Chip. 2009;9:3364–3369. doi: 10.1039/b911882a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S., Shugars D.C., Swanstrom R., Garcia J.V. Nef from primary isolates of human immunodeficiency virus type 1 suppresses surface CD4 expression in human and mouse T cells. J. Virol. 1993;67:4923–4931. doi: 10.1128/jvi.67.8.4923-4931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K., Tilley L., Vennerstrom J.L., Roberts D., Rogerson S., Ginsburg H. Oxidative stress in malaria parasite-infected erythrocytes: host–parasite interactions. Int. J. Parasitol. 2004:163–189. doi: 10.1016/j.ijpara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Beebe D.J., Mensing G.A., Walker G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- Bell D., Wongsrichanalai C., Barnwell J.W. Ensuring quality and access for malaria diagnosis: how can it be achieved? Nat. Rev. Microbiol. 2006;4:S7–20. doi: 10.1038/nrmicro1474. [DOI] [PubMed] [Google Scholar]
- Bernacka-Wojcik I., Lopes P., Catarina Vaz A., Veigas B., Jerzy Wojcik P., Simoes P. Bio-microfluidic platform for gold nanoprobe based DNA detection-application to Mycobacterium tuberculosis. Biosens. Bioelectron. 2013;48:87–93. doi: 10.1016/j.bios.2013.03.079. [DOI] [PubMed] [Google Scholar]
- Bhagat A.A.S., Bow H., Hou H.W., Tan S.J., Han J., Lim C.T. Microfluidics for cell separation. Med. Biol. Eng. Comput. 2010:999–1014. doi: 10.1007/s11517-010-0611-4. [DOI] [PubMed] [Google Scholar]
- Bhakdi S.C., Ottinger A., Somsri S., Sratongno P., Pannadaporn P., Chimma P. Optimized high gradient magnetic separation for isolation of Plasmodium-infected red blood cells. Malar. J. 2010;9:38. doi: 10.1186/1475-2875-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisceglia E., Cubizolles M., Trainito C.I., Berthier J., Pudda C., Français O. A generic and label free method based on dielectrophoresis for the continuous separation of microorganism from whole blood samples. Sensors Actuators B Chem. 2015;212:335–343. ([Internet]. Feb [cited 2015 Feb 16] Available from: http://www.sciencedirect.com/science/article/pii/S0925400515001860) [Google Scholar]
- Boneschansker L., Yan J., Wong E., Briscoe D.M., Irimia D. Microfluidic platform for the quantitative analysis of leukocyte migration signatures. Nat. Commun. 2014;5:4787. doi: 10.1038/ncomms5787. ([Internet]. Jan [cited 2015 Feb 18] Available from. http://www.ncbi.nlm.nih.gov/pubmed/25183261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman S.C.C. High volume hemofiltration as adjunctive therapy for sepsis and systemic inflammatory response syndrome. background, definition and a descriptive analysis of animal and human studies [Internet] Adv. Sepsis. 2007 ([cited 2015 May 10]. Available from: http://www.advancesinsepsis.com/pdfs/4339.pdf) [Google Scholar]
- Boveris A., Alvarez S., Navarro A. The role of mitochondrial nitric oxide synthase in inflammation and septic shock. Free Radic. Biol. Med. 2002:1186–1193. doi: 10.1016/s0891-5849(02)01009-2. [DOI] [PubMed] [Google Scholar]
- Bow H., Pivkin I.V., Diez-Silva M., Goldfless S.J., Dao M., Niles J.C. A microfabricated deformability-based flow cytometer with application to malaria. Lab Chip. 2011;11:1065–1073. doi: 10.1039/c0lc00472c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer D.N., Maamari R.N., Switz N.A., Lam W.A., Fletcher D.A. Mobile phone based clinical microscopy for global health applications. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijtels P.C.A.M., Willemse-Erix H.F.M., Petit P.L.C., Endtz H.P., Puppels G.J., Verbrugh H.A. Rapid identification of mycobacteria by Raman spectroscopy. J. Clin. Microbiol. 2008;46(3):961–965. doi: 10.1128/JCM.01763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler K.L., Ambravaneswaran V., Agrawal N., Bilodeau M., Toner M., Tompkins R.G. Burn injury reduces neutrophil directional migration speed in microfluidic devices. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Gonzalez A., Spearman P.W., Stoll B.J. Neonatal infectious diseases. Evaluation of neonatal sepsis. Pediatr. Clin. N. Am. 2013:367–389. doi: 10.1016/j.pcl.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.-H., Wang C.-H., Yang S.-Y., Lin Y.-C., Wu J.-J., Lee M.S. Rapid isolation and diagnosis of live bacteria from human joint fluids by using an integrated microfluidic system. Lab Chip. 2014;14(17):3376–3384. doi: 10.1039/c4lc00471j. ([Internet] Royal Society of Chemistry Sep 7 [cited 2015 Mar 1] Available from: http://pubs.rsc.org/en/Content/ArticleHTML/2014/LC/C4LC00471J) [DOI] [PubMed] [Google Scholar]
- Cheng X., Irimia D., Dixon M., Sekine K., Demirci U., Zamir L. A microfluidic device for practical label-free CD4(+) T cell counting of HIV-infected subjects. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.D., Laksanasopin T., Cheung Y.K., Steinmiller D., Linder V., Parsa H. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- Chin C.D., Linder V., Sia S.K. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip. 2012:2118. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- Chiodini P.L., Bowers K., Jorgensen P., Barnwell J.W., Grady K.K., Luchavez J. The heat stability of Plasmodium lactate dehydrogenase-based and histidine-rich protein 2-based malaria rapid diagnostic tests. Trans. R. Soc. Trop. Med. Hyg. 2007;101:331–337. doi: 10.1016/j.trstmh.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Choi J.-W., Oh K.W., Thomas J.H., Heineman W.R., Halsall H.B., Nevin J.H. An integrated microfluidic biochemical detection system for protein analysis with magnetic bead-based sampling capabilities. Lab Chip. 2002;2:27–30. doi: 10.1039/b107540n. [DOI] [PubMed] [Google Scholar]
- Choi J., Jung Y.-G., Kim J., Kim S., Jung U., H Na. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip. 2012 doi: 10.1039/c2lc41055a. [DOI] [PubMed] [Google Scholar]
- Cojoc D., Finaurini S., Livshits P., Gur E., Shapira A., Mico V. Toward fast malaria detection by secondary speckle sensing microscopy. Biomed. Opt. Express. 2012:991. doi: 10.1364/BOE.3.000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R.M., Leslie D.C., Domansky K., Jain A., Yung C., Cho M. A microdevice for rapid optical detection of magnetically captured rare blood pathogens. Lab Chip. 2014;14:182–188. doi: 10.1039/c3lc50935d. ([Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/24169822) [DOI] [PubMed] [Google Scholar]
- Cranston H.A., Boylan C.W., Carroll G.L., Sutera S.P., Williamson J.R., Gluzman I.Y. Plasmodium falciparum maturation abolishes physiologic red cell deformability. Science. 1984;223:400–403. doi: 10.1126/science.6362007. [DOI] [PubMed] [Google Scholar]
- D'Avignon L.C., Hogan B.K., Murray C.K., Loo F.L., Hospenthal D.R., Cancio L.C. Contribution of bacterial and viral infections to attributable mortality in patients with severe burns: an autopsy series. Burns. 2010:773–779. doi: 10.1016/j.burns.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Dalgleish A.G., Beverley P.C., Clapham P.R., Crawford D.H., Greaves M.F., Weiss R.A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312(5996):763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Damhorst G.L., Murtagh M., Rodriguez W.R., Bashir R. Microfluidics and nanotechnology for detection of global infectious diseases. Proc. IEEE. 2015;103(2):150–160. ([Internet]. Feb [cited 2015 May 6] Available from: http://ieeexplore.ieee.org/lpdocs/epic03/wrapper.htm?arnumber=7067024) [Google Scholar]
- Darwish N.T., Alias Y.B., Khor S.M. An introduction to dengue-disease diagnostics. TrAC Trends Anal. Chem. 2015;67:45–55. ([Internet]. Apr [cited 2015 Mar 11]. Available from: http://www.sciencedirect.com/science/article/pii/S0165993615000333) [Google Scholar]
- Deeks S.G., Autran B., Berkhout B., Benkirane M., Cairns S., Chomont N. Towards an HIV cure: a global scientific strategy. Nat. Rev. Immunol. 2012;12(8):607–614. doi: 10.1038/nri3262. ([Internet] Available from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K., Ruhwald M., Theron G., Peter J., Yam W.C. Point-of-care diagnosis of tuberculosis: past, present and future. Respirology. 2013;18(2):217–232. doi: 10.1111/resp.12022. ([Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/23190246) [DOI] [PubMed] [Google Scholar]
- Di Carlo D., Jeong K.-H., Lee L.P. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip. 2003;3(4):287–291. doi: 10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- Domínguez C.M., Kosaka P.M., Sotillo A., Mingorance J., Tamayo J., Calleja M. Label-free DNA-based detection of Mycobacterium tuberculosis and rifampicin resistance through hydration induced stress in microcantilevers. Anal. Chem. 2015;87(3):1494–1498. doi: 10.1021/ac504523f. ([Internet]. American Chemical Society; Feb 3 [cited 2016 Jan 2] Available from:) [DOI] [PubMed] [Google Scholar]
- Dondorp A.M., Angus B.J., Chotivanich K., Silamut K., Ruangveerayuth R., Hardeman M.R. Red blood cell deformability as a predictor of anemia in severe falciparum malaria. Am.J.Trop. Med. Hyg. 1999;60:733–737. doi: 10.4269/ajtmh.1999.60.733. [DOI] [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361(5):455–467. doi: 10.1056/NEJMoa0808859. ([Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3495232&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du E., Dao M., Suresh S. Quantitative biomechanics of healthy and diseased human red blood cells using dielectrophoresis in a microfluidic system. Extreme Mech. Lett. 2014 doi: 10.1016/j.eml.2014.11.006. ([Internet]. Dec [cited 2015 Mar 1]; Available from: http://www.sciencedirect.com/science/article/pii/S2352431614000091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmond K., Zaidi A. New approaches to preventing, diagnosing, and treating neonatal sepsis. PLoS Med. 2010;7(3):1–8. doi: 10.1371/journal.pmed.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman L.K., Kain K.C. Molecular diagnostic and surveillance tools for global malaria control. Travel Med. Infect. dis. 2008;6:82–99. doi: 10.1016/j.tmaid.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Fang X., Chen H., Xu L., Jiang X., Wu W., Kong J. A portable and integrated nucleic acid amplification microfluidic chip for identifying bacteria. Lab Chip. 2012;12(8):1495. doi: 10.1039/c2lc40055c. [DOI] [PubMed] [Google Scholar]
- Fauci A.S. HIV and AIDS: 20 years of science. Nat. Med. 2003;9(7):839–843. doi: 10.1038/nm0703-839. [DOI] [PubMed] [Google Scholar]
- Furdui V.I., Harrison D.J. Immunomagnetic T cell capture from blood for PCR analysis using microfluidic systems. Lab Chip. 2004;4:614–618. doi: 10.1039/b409366f. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P., Mahidol C., Ruchirawat M., Satayavivad J., Watcharasit P., Becker F.F. Microsample preparation by dielectrophoresis: isolation of malaria. Lab Chip. 2002;2:70–75. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P., Satayavivad J., Ruchirawat M. Microfluidic approaches to malaria detection. Acta Trop. 2004;89:357–369. doi: 10.1016/j.actatropica.2003.11.009. ([Internet] Available from: http://www.sciencedirect.com/science/article/pii/S0001706X03003164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geislinger T.M., Chan S., Moll K., Wixforth A., Wahlgren M., Franke T. Label-free microfluidic enrichment of ring-stage Plasmodium falciparum-infected red blood cells using non-inertial hydrodynamic lift. Malar. J. 2014;13(1):375. doi: 10.1186/1475-2875-13-375. ([Internet] Jan [cited 2015 Feb 6]. Available from: http://www.malariajournal.com/content/13/1/375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genton B., D'Acremont V., Rare L., Baea K., Reeder J.C., Alpers M.P. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med. 2008;5(6):e127. doi: 10.1371/journal.pmed.0050127. ([Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2429951&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebreyesus T.A., Witten K.H., Getachew A., Yohannes A.M., Tesfay W., Minass M. The community-based malaria control programme in Tigray, northern Ethiopia. A review of programme set-up, activities, outcomes and impact. Parassitologia. 2000:255–290. [PubMed] [Google Scholar]
- Glynn M.T., Kinahan D.J., Ducrée J. Rapid, low-cost and instrument-free CD4 + cell counting for HIV diagnostics in resource-poor settings. Lab Chip. 2014;14(15):2844–2851. doi: 10.1039/c4lc00264d. ([Internet]. Royal Society of Chemistry; Aug 7 [cited 2015 Mar 1]. Available from: http://pubs.rsc.org/en/Content/ArticleHTML/2014/LC/C4LC00264D) [DOI] [PubMed] [Google Scholar]
- Golden J.P., Kim J.S., Erickson J.S., Hilliard L.R., Howell P.B., Anderson G.P. Multi-wavelength microflow cytometer using groove-generated sheath flow. Lab Chip. 2009;9:1942–1950. doi: 10.1039/b822442k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith H.L., Spain S. Margination of leukocytes in blood flow through small tubes. Microvasc. Res. 1984;27:204–222. doi: 10.1016/0026-2862(84)90054-2. [DOI] [PubMed] [Google Scholar]
- Gossett D.R., Tse H.T.K., Lee S.A., Ying Y., Lindgren A.G., Yang O.O. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl. Acad. Sci. 2012:7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotloib L., Barzilay E., Shustak A., Lev A. Sequential hemofiltration in nonoliguric high capillary permeability pulmonary edema of severe sepsis: preliminary report. Crit. Care Med. 1984;12(11):997–1000. doi: 10.1097/00003246-198411000-00018. [DOI] [PubMed] [Google Scholar]
- Gubler D.J. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10(2):100–103. doi: 10.1016/s0966-842x(01)02288-0. ([Internet] Feb [cited 2015 Mar 7]. Available from: http://www.sciencedirect.com/science/article/pii/S0966842X01022880) [DOI] [PubMed] [Google Scholar]
- Gunda N.S.K., Singh M., Purwar Y., Shah S.L., Kaur K., Mitra S.K. Micro-spot with integrated pillars (MSIP) for detection of dengue virus NS1. Biomed. Microdevices. 2013;15(6):959–971. doi: 10.1007/s10544-013-9787-3. [DOI] [PubMed] [Google Scholar]
- Guo Q., Reiling S.J., Rohrbach P., Ma H. Microfluidic biomechanical assay for red blood cells parasitized by Plasmodium falciparum. Lab Chip. 2012:1143. doi: 10.1039/c2lc20857a. [DOI] [PubMed] [Google Scholar]
- Handayani S., Chiu D.T., Tjitra E., Kuo J.S., Lampah D., Kenangalem E. High deformability of Plasmodium vivax-infected red blood cells under microfluidic conditions. J Infect Dis. 2009;199(3):445–450. doi: 10.1086/596048. ([Internet] Feb 1 [cited 2015 Mar 1]. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4337984&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänscheid T. Diagnosis of malaria: a review of alternatives to conventional microscopy. Clin. Lab. Haematol. 1999;21(January):235–245. doi: 10.1046/j.1365-2257.1999.00220.x. [DOI] [PubMed] [Google Scholar]
- Hänscheid T., Grobusch M.P. How useful is PCR in the diagnosis of malaria? Trends Parasitol. 2002:395–398. doi: 10.1016/s1471-4922(02)02348-6. [DOI] [PubMed] [Google Scholar]
- He F., Zhao J., Zhang L., Su X. A rapid method for determining Mycobacterium tuberculosis based on a bulk acoustic wave impedance biosensor. Talanta. 2003;59(5):935–941. doi: 10.1016/S0039-9140(02)00643-4. [DOI] [PubMed] [Google Scholar]
- Ho D.D., Neumann A.U., Perelson A.S., Chen W., Leonard J.M., Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Hopkins H., González I.J., Polley S.D., Angutoko P., Ategeka J., Asiimwe C. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J. Infect. dis. 2013;208(4):645–652. doi: 10.1093/infdis/jit184. ([Internet] Aug 15 [cited 2015 Feb 24]. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3719898&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horning M.P., Delahunt C.B., Singh S.R., Garing S.H., Nichols K.P. A paper microfluidic cartridge for automated staining of malaria parasites with an optically transparent microscopy window. Lab Chip. 2014;14:2040–2046. doi: 10.1039/c4lc00293h. ([Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/24781199) [DOI] [PubMed] [Google Scholar]
- Hosseini S.M., Feng J.J. How malaria parasites reduce the deformability of infected red blood cells. Biophys. J. 2012;103:1–10. doi: 10.1016/j.bpj.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss R.S., Karl I.E. The pathophysiology and treatment of sepsis. N. Engl. J. Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- Hou H.W., Bhagat A.A.S., Chong A.G.L., Mao P., Tan K.S.W., Han J. Deformability based cell margination—a simple microfluidic design for malaria-infected erythrocyte separation. Lab Chip. 2010;10:2605–2613. doi: 10.1039/c003873c. [DOI] [PubMed] [Google Scholar]
- Hou H.W., Bhattacharyya R.P., Hung D.T., Han J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip. 2015;15(10):2297–2307. doi: 10.1039/c5lc00311c. ([Internet]. Royal Society of Chemistry; Apr 17 [cited 2015 Apr 19]. Available from: http://pubs.rsc.org/en/Content/ArticleHTML/2015/LC/C5LC00311C) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H.W., Vera M.P., Levy B.D., Baron R.M., Han J. MicroTAS Conf. 2013 Freiburg, Ger. 2013. A novel microfluidic “cell-based” blood dialysis platform for murine model of sepsis [Internet] ([cited 2015 May 10]. Available from: http://www.rsc.org/images/loc/2013/PDFs/Papers/617_0927.pdf) [Google Scholar]
- Huang S., Undisz A., Diez-Silva M., Bow H., Dao M., Han J. Dynamic deformability of Plasmodium falciparum-infected erythrocytes exposed to artesunate in vitro. Integr. Biol. (Camb) 2013;5:414–422. doi: 10.1039/c2ib20161e. ([Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23254624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.-H., Lin Y.-S., Wu C.-W., Wu C.-J. Assessment of the inhibition of Dengue virus infection by carrageenan via real-time monitoring of cellular oxygen consumption rates within a microfluidic device. Biomicrofluidics. 2014;8(2):024110. doi: 10.1063/1.4870772. ([Internet]. Available from: http://scitation.aip.org/content/aip/journal/bmf/8/2/10.1063/1.4870772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S.C., Chan M.C.H., Wu A.K., Ng P.C. Severe acute respiratory syndrome (SARS): epidemiology and clinical features. Postgrad. Med. J. 2004;80(945):373–381. doi: 10.1136/pgmj.2004.020263. ([Internet] Jul [cited 2015 Mar 1] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1743054&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt H.C., Wilkinson J.S. Optofluidic integration for microanalysis. Microfluid. Nanofluid. 2008:53–79. doi: 10.1007/s10404-007-0223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter R.A., Privett B.J., Henley W.H., Breed E.R., Liang Z., Mittal R. Microfluidic amperometric sensor for analysis of nitric oxide in whole blood. Anal. Chem. 2013;85:6066–6072. doi: 10.1021/ac400932s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inci F., Tokel O., Wang S., Gurkan U.A., Tasoglu S., Kuritzkes D.R. Nanoplasmonic quantitative detection of intact viruses from unprocessed whole blood. ACS Nano. 2013;7(6):4733–4745. doi: 10.1021/nn3036232. ([Internet]. American Chemical Society; Jun 25 [cited 2015 Mar 1] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J., Hira P.R., Sher A., Al-Enezi A.A. Diagnosis of imported malaria by Plasmodium lactate dehydrogenase (pLDH) and histidine-rich protein 2 (PfHRP-2)-based immunocapture assays. Am J Trop Med Hyg. 2001;64(1–2):20–23. doi: 10.4269/ajtmh.2001.64.20. ([Internet]. Jan [cited 2015 Mar 1]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11425156) [DOI] [PubMed] [Google Scholar]
- Ji Q.Q., Du G.S., Van Uden M.J., Fang Q., Den Toonder J.M.J. Microfluidic cytometer based on dual photodiode detection for cell size and deformability analysis. Talanta. 2013;111:178–182. doi: 10.1016/j.talanta.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Jung W., Han J., Choi J.-W., Ahn C.H. Point-of-care testing (POCT) diagnostic systems using microfluidic lab-on-a-chip technologies. Microelectron. Eng. 2015;132:46–57. ([Internet] Jan [cited 2015 Jan 4]. Available from: http://www.sciencedirect.com/science/article/pii/S0167931714004316) [Google Scholar]
- Juul S., Nielsen C.J.F., Labouriau R., Roy A., Tesauro C., Jensen P.W. Droplet microfluidics platform for highly sensitive and quantitative detection of malaria-causing plasmodium parasites based on enzyme activity measurement. ACS Nano. 2012;6:10676–10683. doi: 10.1021/nn3038594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.-K., Ali M.M., Zhang K., Huang S.S., Peterson E., Digman M.A. Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nat. Commun. 2014;5:5427. doi: 10.1038/ncomms6427. ([Internet]. Nature Publishing Group; Jan 13 [cited 2014 Nov 15] Available from: http://www.nature.com/ncomms/2014/141113/ncomms6427/full/ncomms6427.html) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.H., Super M., Yung C.W., Cooper R.M., Domansky K., Graveline A.R. An extracorporeal blood-cleansing device for sepsis therapy. Nat. Med. 2014;20(10):1211–1216. doi: 10.1038/nm.3640. ([Internet]. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; Sep 14 [cited 2014 Sep 15] Available from:) [DOI] [PubMed] [Google Scholar]
- Kellerman S., Essajee S. HIV testing for children in resource-limited settings: what are we waiting for? PLoS Med. 2010;7(7):e1000285. doi: 10.1371/journal.pmed.1000285. ([Internet] Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Hee Jang S., Jia G., Zoval J.V., Da Silva N.A., Madou M.J. Cell lysis on a microfluidic CD (compact disc) Lab Chip. 2004;4(5):516–522. doi: 10.1039/b401106f. [DOI] [PubMed] [Google Scholar]
- Kim Y.G., Moon S., Kuritzkes D.R., Demirci U. Quantum dot-based HIV capture and imaging in a microfluidic channel. Biosens. Bioelectron. 2009;25:253–258. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.B., Koo K., Bae H., Dokmeci M.R., Hamilton G.A., Bahinski A. A mini-microscope for in situ monitoring of cells. Lab Chip. 2012:3976. doi: 10.1039/c2lc40345e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahata M.M., Gange S.J., Abraham A.G., Merriman B., Saag M.S., Justice A.C. Effect of early versus deferred antiretroviral therapy for HIV on survival. N. Engl. J. Med. 2009;360(18):1815–1826. doi: 10.1056/NEJMoa0807252. ([Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2854555&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Roberts D., Wood K.E., Light B., Parrillo J.E., Sharma S. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006;34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- Kumbhat S., Sharma K., Gehlot R., Solanki A., Joshi V. Surface plasmon resonance based immunosensor for serological diagnosis of dengue virus infection. J. Pharm. Biomed. Anal. 2010;52(2):255–259. doi: 10.1016/j.jpba.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Laksanasopin T., Guo T.W., Nayak S., Sridhara A.A., Xie S., Olowookere O.O. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 2015;7(273) doi: 10.1126/scitranslmed.aaa0056. (273re1-273re1 [Internet] Feb 4 [cited 2015 Feb 4]. Available from: http://stm.sciencemag.org/content/7/273/273re1.short) [DOI] [PubMed] [Google Scholar]
- Lee S.W., Tai Y.C. Micro cell lysis device. Sensors Actuators A Phys. 1999;73(1–2):74–79. [Google Scholar]
- Lee W.G., Bang H., Yun H., Lee J., Park J., Kim J.K. On-chip erythrocyte deformability test under optical pressure. Lab Chip. 2007;7:516–519. doi: 10.1039/b614912j. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Kim S.-W., Kang J.Y., Ahn C.H. A polymer lab-on-a-chip for reverse transcription (RT)-PCR based point-of-care clinical diagnostics. Lab Chip. 2008;8(12):2121–2127. doi: 10.1039/b811131f. ([Internet] Dec [cited 2015 Mar 1]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19023475) [DOI] [PubMed] [Google Scholar]
- Lee Y.-F., Lien K.-Y., Lei H.-Y., Lee G.-B. An integrated microfluidic system for rapid diagnosis of dengue virus infection. Biosens. Bioelectron. 2009;25:745–752. doi: 10.1016/j.bios.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.G., Kim Y.-G., Chung B.G., Demirci U., Khademhosseini A. Nano/microfluidics for diagnosis of infectious diseases in developing countries. Adv. Drug Deliv. Rev. 2010;62:449–457. doi: 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.A., Leitao R., Zheng G., Yang S., Rodriguez A., Yang C. Color capable sub-pixel resolving optofluidic microscope and its application to blood cell imaging for malaria diagnosis. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A., Mackenzie I. Sepsis: definition, epidemiology, and diagnosis. BMJ. 2007;335:879–883. doi: 10.1136/bmj.39346.495880.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M.M., Fink M.P., Marshall J.C., Abraham E., Angus D., Cook D. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit. Care Med. 2003:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- Li Y., Yan X., Feng X., Wang J., Du W., Wang Y. Agarose-based microfluidic device for point-of-care concentration and detection of pathogen. Anal. Chem. 2014;86(21):10653–10659. doi: 10.1021/ac5026623. ([Internet]. American Chemical Society; Nov 4 [cited 2015 Mar 1]. Available from:) [DOI] [PubMed] [Google Scholar]
- Lim D.V., Simpson J.M., Kearns E.A., Kramer M.F. Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin. Microbiol. Rev. 2005:583–607. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liong M., Hoang A.N., Chung J., Gural N., Ford C.B., Min C. Magnetic barcode assay for genetic detection of pathogens. Nat. Commun. 2013;4:1752. doi: 10.1038/ncomms2745. ([Internet]. Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabey D., Peeling R.W., Ustianowski A., Perkins M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- Mach A.J., di Carlo D. Continuous scalable blood filtration device using inertial microfluidics. Biotechnol. Bioeng. 2010;107(2):302–311. doi: 10.1002/bit.22833. [DOI] [PubMed] [Google Scholar]
- Mahalanabis M., Al-Muayad H., Kulinski M.D., Altman D., Klapperich C.M. Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip. 2009;9:2811–2817. doi: 10.1039/b905065p. [DOI] [PubMed] [Google Scholar]
- Maier A.G., Cooke B.M., Cowman A.F., Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat. Rev. Microbiol. 2009;7:341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- Makler M.T., Palmer C.J., Ager A.L. A review of practical techniques for the diagnosis of malaria. Ann. Trop. Med. Parasitol. 1998;92:419–433. doi: 10.1080/00034989859401. [DOI] [PubMed] [Google Scholar]
- Malleret B., Claser C., Ong A.S.M., Suwanarusk R., Sriprawat K., Howland S.W. A rapid and robust tri-color flow cytometry assay for monitoring malaria parasite development. Sci. Rep. 2011 doi: 10.1038/srep00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani V., Wang S., Inci F., De Libero G., Singhal A., Demirci U. Emerging technologies for monitoring drug-resistant tuberculosis at the point-of-care. Adv. Drug Deliv. Rev. 2014;78:1–50. doi: 10.1016/j.addr.2014.05.015. ([Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24882226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L.A., Han C.M., Santiago J.G. Extraction of DNA from malaria-infected erythrocytes using isotachophoresis. Anal. Chem. 2011;83:9715–9718. doi: 10.1021/ac202567j. [DOI] [PubMed] [Google Scholar]
- Martinez A.W., Phillips S.T., Whitesides G.M., Carrilho E. Diagnostics for the developing world: microfluidic paper-based analytical devices. Anal. Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- McNerney R., Daley P. Towards a point-of-care test for active tuberculosis: obstacles and opportunities. Nat. Rev. Microbiol. 2011;9(3):204–213. doi: 10.1038/nrmicro2521. [DOI] [PubMed] [Google Scholar]
- McWhirter J.L., Noguchi H., Gompper G. Deformation and clustering of red blood cells in microcapillary flows. Soft Matter. 2011:10967. [Google Scholar]
- Mills C.D., Burgess D.C.H., Taylor H.J., Kain K.C. Evaluation of a rapid and inexpensive dipstick assay for the diagnosis of Plasmodium falciparum malaria. Bull. World Health Organ. 1999;77:553–559. [PMC free article] [PubMed] [Google Scholar]
- Mohan K., Dubey M.L., Ganguly N.K., Mahajan R.C. Plasmodium falciparum: role of activated blood monocytes in erythrocyte membrane damage and red cell loss during malaria. Exp. Parasitol. 1995;80:54–63. doi: 10.1006/expr.1995.1007. [DOI] [PubMed] [Google Scholar]
- Murray C.K., Gasser R.A., Magill A.J., Miller R.S. Update on rapid diagnostic testing for malaria. Clin. Microbiol. Rev. 2008:97–110. doi: 10.1128/CMR.00035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers F.B., Lee L.P. Innovations in optical microfluidic technologies for point-of-care diagnostics. Lab Chip. 2008;8:2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- Nam J., Huang H., Lim H., Lim C., Shin S. Magnetic separation of malaria-infected red blood cells in various developmental stages. Anal. Chem. 2013;85:7316–7323. doi: 10.1021/ac4012057. [DOI] [PubMed] [Google Scholar]
- Naumann K.M., Jones G.L., Saul A., Smith R. A Plasmodium falciparum exo-antigen alters erythrocyte membrane deformability. FEBS Lett. 1991;292:95–97. doi: 10.1016/0014-5793(91)80842-q. [DOI] [PubMed] [Google Scholar]
- O'Gorman M.R., Gelman R. Inter- and intrainstitutional evaluation of automated volumetric capillary cytometry for the quantitation of CD4- and CD8-positive T lymphocytes in the peripheral blood of persons infected with human immunodeficiency virus. Site Investigators and the NIAI. Clin. Diagn. Lab. Immunol. 1997;4(2):173–179. doi: 10.1128/cdli.4.2.173-179.1997. ([Internet]. Mar [cited 2015 Oct 28]. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=170497&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan A., Demirci U. Ultra wide-field lens-free monitoring of cells on-chip. Lab Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- Park S., Zhang Y., Lin S., Wang T.H., Yang S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011:830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson A.S., Heithoff D.M., Ferguson B.S., Tom Soh H., Mahan M.J., Plaxco K.W. Microfluidic chip-based detection and intraspecies strain discrimination of Salmonella serovars derived from whole blood of septic mice. Appl. Environ. Microbiol. 2013;79:2302–2311. doi: 10.1128/AEM.03882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F., Roath S., Melville D., Warhurst D.C., Osisanya J.O. Separation of malaria-infected erythrocytes from whole blood: use of a selective high-gradient magnetic separation technique. Lancet. 1981;2:70–71. doi: 10.1016/s0140-6736(81)90414-1. [DOI] [PubMed] [Google Scholar]
- Peng W.K., Kong T.F., Ng C.S., Chen L., Huang Y., Bhagat A.A.S. Micromagnetic resonance relaxometry for rapid label-free malaria diagnosis. Nat. Med. 2014;20(9):1069–1073. doi: 10.1038/nm.3622. ([Internet]. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; Sep [cited 2015 Mar 1]. Available from:) [DOI] [PubMed] [Google Scholar]
- Pethig R. Review article—dielectrophoresis: status of the theory, technology, and applications. Biomicrofluidics. 2010;4(2):1–35. doi: 10.1063/1.3456626. ([Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2917862&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethig R., Markx G.H. Applications of dielectrophoresis in biotechnology. Trends Biotechnol. 1997;15(10):426–432. doi: 10.1016/S0167-7799(97)01096-2. [DOI] [PubMed] [Google Scholar]
- Psaltis D., Quake S.R., Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature. 2006;442:381–386. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- Quinn D.J., Pivkin I., Wong S.Y., Chiam K.-H., Dao M., Karniadakis G.E. Combined simulation and experimental study of large deformation of red blood cells in microfluidic systems. Ann. Biomed. Eng. 2011;39:1041–1050. doi: 10.1007/s10439-010-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandraiah H., Amasia M., Cole J., Sheard P., Pickhaver S., Walker C. Lab-on-DVD: standard DVD drives as a novel laser scanning microscope for image based point of care diagnostics. Lab Chip. 2013;13:1578–1585. doi: 10.1039/c3lc41360h. ([Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/23440071) [DOI] [PubMed] [Google Scholar]
- Ramalingam N., San T.C., Kai T.J., Mak M.Y.M., Gong H.Q. Microfluidic devices harboring unsealed reactors for real-time isothermal helicase-dependent amplification. Microfluid. Nanofluid. 2009;7:325–336. doi: 10.1007/s10404-008-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo M., Shapiro H.M., Amaral T., Melo-Cristino J., Hänscheid T. Haemozoin detection in infected erythrocytes for Plasmodium falciparum malaria diagnosis—prospects and limitations. Acta Trop. 2012;123:58–61. doi: 10.1016/j.actatropica.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Ren J., He F., Yi S., Cui X. A new MSPQC for rapid growth and detection of Mycobacterium tuberculosis. Biosens. Bioelectron. 2008;24(3):403–409. doi: 10.1016/j.bios.2008.04.018. ([Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/18539020) [DOI] [PubMed] [Google Scholar]
- Reyes D.R., Iossifidis D., Auroux P.-A., Manz A. Micro total analysis systems. 1. Introduction, theory, and technology. Anal. Chem. 2002;74(12):2623–2636. doi: 10.1021/ac0202435. [DOI] [PubMed] [Google Scholar]
- Riddle M.S., Jackson J.L., Sanders J.W., Blazes D.L. Exchange transfusion as an adjunct therapy in severe Plasmodium falciparum malaria: a meta-analysis. Clin. Infect. Dis. 2002;34:1192–1198. doi: 10.1086/339810. [DOI] [PubMed] [Google Scholar]
- Rigauperez J., Clark G., Gubler D., Reiter P., Sanders E., Vancevorndam A. Dengue and dengue haemorrhagic fever. Lancet. 1998;352(9132):971–977. doi: 10.1016/s0140-6736(97)12483-7. ([Internet] Available from: http://linkinghub.elsevier.com/retrieve/pii/S0140673697124837) [DOI] [PubMed] [Google Scholar]
- Ritzi-Lehnert M. Development of chip-compatible sample preparation for diagnosis of infectious diseases. Expert. Rev. Mol. Diagn. 2012:189–206. doi: 10.1586/erm.11.98. [DOI] [PubMed] [Google Scholar]
- Rosenbluth M.J., Lam W.A., Fletcher D.A. Analyzing cell mechanics in hematologic diseases with microfluidic biophysical flow cytometry. Lab Chip. 2008;8:1062–1070. doi: 10.1039/b802931h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio J.M., Benito A., Berzosa P.J., Roche J., Puente S., Subirats M. Usefulness of seminested multiplex PCR in surveillance of imported malaria in Spain. J. Clin. Microbiol. 1999;37:3260–3264. doi: 10.1128/jcm.37.10.3260-3264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O., Bercovici M. Microfluidic assay for continuous bacteria detection using antimicrobial peptides and isotachophoresis. Anal. Chem. 2014;86(20):10106–10113. doi: 10.1021/ac5017776. ([Internet]. American Chemical Society; Oct 21 [cited 2015 Mar 1]. Available from:) [DOI] [PubMed] [Google Scholar]
- Sethu P., Anahtar M., Moldawer L.L., Tompkins R.G., Toner M. Continuous flow microfluidic device for rapid erythrocyte lysis. Anal. Chem. 2004;76(21):6247–6253. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- Shafiee H., Lidstone E.A., Jahangir M., Inci F., Hanhauser E., Henrich T.J. Nanostructured optical photonic crystal biosensor for HIV viral load measurement. Sci. Rep. 2014;4:4116. doi: 10.1038/srep04116. ([Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3937800&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiee H., Wang S., Inci F., Toy M., Henrich T.J., Kuritzkes D.R. Emerging technologies for point-of-care management of HIV infection. Annu. Rev. Med. 2015;66:387–405. doi: 10.1146/annurev-med-092112-143017. ([Internet]. Annual Reviews; Jan 14 [cited 2015 Feb 13]. Available from: http://www.annualreviews.org/doi/abs/10.1146/annurev-med-092112-143017) [DOI] [PubMed] [Google Scholar]
- Shelby J.P., White J., Ganesan K., Rathod P.K., Chiu D.T. A microfluidic model for single-cell capillary obstruction by Plasmodium falciparum-infected erythrocytes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14618–14622. doi: 10.1073/pnas.2433968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Kastrup C.J., Ismagilov R.F. Using microfluidics to understand the effect of spatial distribution of tissue factor on blood coagulation. Thromb. Res. 2008 doi: 10.1016/S0049-3848(08)70015-X. [DOI] [PubMed] [Google Scholar]
- Shen F., Davydova E.K., Du W., Kreutz J.E., Piepenburg O., Ismagilov R.F. Digital isothermal quantification of nucleic acids via simultaneous chemical initiation of recombinase polymerase amplification reactions on SlipChip. Anal. Chem. 2011;83(9):3533–3540. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.K., Linder V., Parviz B.A., Siegel A., Whitesides G.M. An integrated approach to a portable and low-cost immunoassay for resource-poor settings. Angew. Chem. Int. Ed. 2004;43:498–502. doi: 10.1002/anie.200353016. [DOI] [PubMed] [Google Scholar]
- Simon V., Ho D.D. HIV-1 dynamics in vivo: implications for therapy. Nat. Rev. Microbiol. 2003;1(3):181–190. doi: 10.1038/nrmicro772. ([Internet]. Nature Publishing Group; Dec [cited 2015 Jan 25]. Available from:) [DOI] [PubMed] [Google Scholar]
- Song L., Zhang Y., Wang W., Ma L., Liu Y., Hao Y. Microfluidic assay without blocking for rapid HIV screening and confirmation. Biomed. Microdevices. 2012;14(4):631–640. doi: 10.1007/s10544-012-9644-9. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Moody A.H., Chiodini P.L. Comparison of blood-film microscopy, the OptiMAL dipstick, Rhodamine-123 fluorescence staining and PCR, for monitoring antimalarial treatment. Ann. Trop. Med. Parasitol. 2000;94:227–232. doi: 10.1080/00034980050006393. [DOI] [PubMed] [Google Scholar]
- Su C.C., Wu T.Z., Chen L.K., Yang H.H., Tai D.F. Development of immunochips for the detection of dengue viral antigens. Anal. Chim. Acta. 2003;479(2):117–123. [Google Scholar]
- Su W., Gao X., Jiang L., Qin J. Microfluidic platform towards point-of-care diagnostics in infectious diseases. J. Chromatogr. A. 2015;1377C:13–26. doi: 10.1016/j.chroma.2014.12.041. ([Internet] Jan 16 [cited 2015 Jan 15]. Available from: http://www.sciencedirect.com/science/article/pii/S0021967314019475) [DOI] [PubMed] [Google Scholar]
- Teles F.S.R.R. Biosensors and rapid diagnostic tests on the frontier between analytical and clinical chemistry for biomolecular diagnosis of dengue disease: a review. Anal. Chim. Acta. 2011:28–42. doi: 10.1016/j.aca.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R., Warner J.B., Nakano M., Libby B., Medkova M., David P.H. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat. Biotechnol. 2009;27(11):1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjitra E., Anstey N.M., Sugiarto P., Warikar N., Kenangalem E., Karyana M. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med. 2008;5(6):e128. doi: 10.1371/journal.pmed.0050128. ([Internet] Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18563962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaiuolo G., Barra M., Preziosi V., Cassinese A., Rotoli B., Guido S. Microfluidics analysis of red blood cell membrane viscoelasticity. Lab Chip. 2011;11:449–454. doi: 10.1039/c0lc00348d. [DOI] [PubMed] [Google Scholar]
- Trial J., Birdsall H.H., Hallum J.A., Crane M.L., Rodriguez-Barradas M.C., de Jong A.L. Phenotypic and functional changes in peripheral blood monocytes during progression of human immunodeficiency virus infection. Effects of soluble immune complexes, cytokines, subcellular particulates from apoptotic cells, and HIV-1-encoded proteins on mono. J. Clin. Invest. 1995;95(4):1690–1701. doi: 10.1172/JCI117845. ([Internet] Apr [cited 2015 Mar 1]. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=295681&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS . UNAIDS; 2013. Global Report: UNAIDS Report on the Global AIDS Epidemic 2013 [Internet] (Available from: www.unaids.org/…/unaids/…/2013/gr2013/UNAIDS_Global_Report_2013) [Google Scholar]
- Van Der Wouden E.J., Super M., Ingber D.E., Van Den Berg A. Detection of pathogens with impedance analysis in a lab on a chip. Procedia Eng. 2011:1465–1468. [Google Scholar]
- Vandan N., Ali M., Prasad R., Kuroiwa C. Assessment of doctors' knowledge regarding tuberculosis management in Lucknow, India: a public-private sector comparison. Public Health. 2009;123(7):484–489. doi: 10.1016/j.puhe.2009.05.004. ([Internet] Available from:) [DOI] [PubMed] [Google Scholar]
- Wang J.-H., Chien L.-J., Hsieh T.-M., Luo C.-H., Chou W.-P., Chen P.-H. A miniaturized quantitative polymerase chain reaction system for DNA amplification and detection. Sensors Actuators B Chem. 2009:329–337. [Google Scholar]
- Wang H., Chen H.-W., Hupert M.L., Chen P.-C., Datta P., Pittman T.L. Fully integrated thermoplastic genosensor for the highly sensitive detection and identification of multi-drug-resistant tuberculosis. Angew. Chem. Int. Ed. Engl. 2012:1–6. doi: 10.1002/anie.201200732. ([Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/22431490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Inci F., Chaunzwa T.L., Ramanujam A., Vasudevan A., Subramanian S. Portable microfluidic chip for detection of Escherichia coli in produce and blood. Int. J. Nanomedicine. 2012;7:2591–2600. doi: 10.2147/IJN.S29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tasoglu S., Chen P.Z., Chen M., Akbas R., Wach S. Micro-a-fluidics ELISA for rapid CD4 cell count at the point-of-care. Sci. Rep. 2014;4:3796. doi: 10.1038/srep03796. ([Internet] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3898414&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkiani M.E., Tay A.K.P., Khoo B.L., Xiaofeng X., Han J., Lim C.T. Malaria detection using inertial microfluidics. Lab Chip. 2015;15(4):1101–1109. doi: 10.1039/c4lc01058b. http://pubs.rsc.org/en/content/articlehtml/2015/lc/c4lc01058b ([Internet] The Royal Society of Chemistry; Feb 21 [cited 2015 Jul 8]. Available from: [DOI] [PubMed] [Google Scholar]
- Watkins N.N., Hassan U., Damhorst G., Ni H., Vaid A., Rodriguez W. Microfluidic CD4 + and CD8 + T lymphocyte counters for point-of-care HIV diagnostics using whole blood. Sci. Transl. Med. 2013;5(214):214ra170. doi: 10.1126/scitranslmed.3006870. ([Internet] Dec 4 [cited 2015 Mar 1]. Available from: http://stm.sciencemag.org/content/5/214/214ra170.abstract) [DOI] [PubMed] [Google Scholar]
- Wei Hou H., Gan H.Y., Bhagat A.A.S., Li L.D., Lim C.T., Han J. A microfluidics approach towards high-throughput pathogen removal from blood using margination. Biomicrofluidics. 2012;6 doi: 10.1063/1.4710992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng C.H., Huang T. Bin, Huang C.C., Yeh C.S., Lei H.Y., Lee G. Bin. A suction-type microfluidic immunosensing chip for rapid detection of the dengue virus. Biomed. Microdevices. 2011;13(3):585–595. doi: 10.1007/s10544-011-9529-3. [DOI] [PubMed] [Google Scholar]
- Whitesides G.M. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- WHO . World Heal. Organ.; 2013. Global Tuberculosis Report 2013 [Internet] (Available from: http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf) [Google Scholar]
- Wongsrichanalai C., Barcus M.J., Muth S., Sutamihardja A., Wernsdorfer W.H. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT) Am.J.Trop. Med. Hyg. 2007;77:119–127. [PubMed] [Google Scholar]
- World Health Organization World Malaria Report 2013 [Internet] Nature. 2013 (Available from: www.who.int) [Google Scholar]
- Xia N., Hunt T.P., Mayers B.T., Alsberg E., Whitesides G.M., Westervelt R.M. Combined microfluidic–micromagnetic separation of living cells in continuous flow. Biomed. Microdevices. 2006;8(4):299–308. doi: 10.1007/s10544-006-0033-0. [DOI] [PubMed] [Google Scholar]
- Xie H., Mire J., Kong Y., Chang M., Hassounah H.A., Thornton C.N. Rapid point-of-care detection of the tuberculosis pathogen using a BlaC-specific fluorogenic probe. Nat. Chem. 2012;4(10):802–809. doi: 10.1038/nchem.1435. ([Internet]. Nature Publishing Group; Oct [cited 2015 Dec 1]. Available from:) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager P., Edwards T., Fu E., Helton K., Nelson K., Tam M.R. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- Yang L., Li Y. Simultaneous detection of Escherichia coli O157:H7 and Salmonella Typhimurium using quantum dots as fluorescence labels. Analyst. 2006;131:394–401. doi: 10.1039/b510888h. [DOI] [PubMed] [Google Scholar]
- Yin X., Thomas T., Zhang J. Multiple red blood cell flows through microvascular bifurcations: cell free layer, cell trajectory, and hematocrit separation. Microvasc. Res. 2013;89:47–56. doi: 10.1016/j.mvr.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Yung C.W., Fiering J., Mueller A.J., Ingber D.E. Micromagnetic-microfluidic blood cleansing device. Lab Chip. 2009;9:1171–1177. doi: 10.1039/b816986a. [DOI] [PubMed] [Google Scholar]
- Zaytseva N.V., Montagna R.A., Baeumner A.J. Microfluidic biosensor for the serotype-specific detection of dengue virus RNA. Anal. Chem. 2005;77(23):7520–7527. doi: 10.1021/ac0509206. [DOI] [PubMed] [Google Scholar]
- Zelenin Ramachandraiah H., Hansson J., Ardabili S., Brismar H., Russom A.S. Bacteria Isolation from Whole Blood for Sepsis Diagnostics. μTAS. 2011;2011:3. doi: 10.1007/s10529-014-1734-8. [DOI] [PubMed] [Google Scholar]
- Zhang C., Xu J., Ma W., Zheng W. PCR microfluidic devices for DNA amplification. Biotechnol. Adv. 2006:243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Bai J.H., Ying J.Y. A stacking flow immunoassay for the detection of dengue-specific immunoglobulins in salivary fluid [Internet] Lab Chip. 2015 doi: 10.1039/c4lc01127a. ([cited 2015 May 10]. Available from: http://pubs.rsc.org/en/content/articlepdf/2015/lc/c4lc01127a) [DOI] [PubMed] [Google Scholar]
- Zheng Y., Nguyen J., Wang C., Sun Y. Electrical measurement of red blood cell deformability on a microfluidic device. Lab Chip. 2013;13:3275–3283. doi: 10.1039/c3lc50427a. ([Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmed/23798004) [DOI] [PubMed] [Google Scholar]
- Zhou X., Liu D., Zhong R., Dai Z., Wu D., Wang H. Determination of SARS-coronavirus by a microfluidic chip system. Electrophoresis. 2004;25:3032–3039. doi: 10.1002/elps.200305966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman P.A., Thomson J.M., Fujioka H., Collins W.E., Zborowski M. Diagnosis of malaria by magnetic deposition microscopy. Am.J.Trop. Med. Hyg. 2006;74:568–572. [PMC free article] [PubMed] [Google Scholar]