Abstract
Bovine herpesvirus-1 infected cell protein 27 (BICP27) was detected predominantly in the nucleolus. The open reading frame of BICP27 was fused with the enhanced yellow fluorescent protein (EYFP) gene to investigate its subcellular localization in live cells and BICP27 was able to direct monomeric, dimeric or trimeric EYFP exclusively to the nucleolus. By constructing a series of deletion mutants, the putative nuclear localization signal (NLS) and nucleolar localization signal (NoLS) were mapped to 81RRAR84 and 86RPRRPRRRPRRR97 respectively. Specific deletion of the putative NLS, NoLS or both abrogated nuclear localization, nucleolar localization or both respectively. Furthermore, NLS was able to direct trimeric EYFP predominantly to the nucleus but excluded from the nucleolus, whereas NoLS targeted trimeric EYFP primarily to the nucleus, and enriched in the nucleolus with faint staining in the cytoplasm. NLS + NoLS directed trimeric EYFP predominantly to the nucleolus with faint staining in the nucleus. Moreover, deletion of NLS + NoLS abolished the transactivating activity of BICP27 on gC promoter, whereas deletion of either NLS or NoLS did not. The study demonstrated that BICP27 is a nucleolar protein, adding BICP27 to the growing list of transactivators which localize to the nucleolus.
Keywords: Nuclear localization signal, Nucleolar localization signal, Bovine herpesvirus-1, BICP27, Transactivation
1. Introduction
Bovine herpesvirus type 1 (BHV-1) is a typical alphaherpesvirus that causes significant economic losses to the cattle industry (Turin et al., 1999). The viral genome is believed to encode at least 70 polypeptides, among of which 25–33 are structural components of the virion (Misra et al., 1981). It also encodes regulatory proteins and enzymes involved in DNA metabolism (Schwyzer and Ackermann, 1996). During the lytic cycle of BHV-1 infection, the viral proteins are expressed in a cascade of three temporally distinct and functionally interdependent phases termed immediate-early (IE), early and late phase (Wirth et al., 1989). BHV-1 encodes three major IE proteins (BICP0, BICP4, and BICP22). They are the structural and functional homologues to infected cell protein ICP0, ICP4, and ICP22 from herpes simplex virus-1 (HSV-1) (Fraefel et al., 1994, Schwyzer et al., 1993, Wirth et al., 1992). The infected cell protein 27 (BICP27) is the fourth regulatory protein of BHV-1, and a structural homologue of HSV-1 ICP27 (Singh et al., 1996). ICP27 is a multifunctional regulator of gene expression which assumes different roles during the course of infection (Sandri-Goldin, 2008). ICP27 and its other herpesviral homologues, such as ORF57 of herpesvirus saimiri (HVS) (Boyne et al., 2008) and UL69 of human cytomegalovirus (HCMV) (Toth and Stamminger, 2008), have been extensively studied. However, there is still limited knowledge on BICP27.
It has been reported that BICP27 is present primarily in the nuclei of infected cells and expressed with early kinetics (Singh et al., 1996), whereas its homologous ICP27 from HSV-1 belongs to the IE kinetic class. Furthermore, there is only 32% similarity between ICP27 and BICP27 in amino acid sequence. Although ICP27 and BICP27 both have a zinc-finger-like domain (CCHC), BICP27 does not have an RGG motif. The transcriptional and translational expression profiles of ICP27 and BICP27 were significantly different, suggesting that their functions may be partially different (Chalifour et al., 1996). The exact role of BICP27 in virus infection is still unclear, but it has several properties that make it of particular interest. Besides its transactivating activity on gC promoter region (Hamel and Simard, 2003), it has been demonstrated that BICP27 may be involved in 3′ processing of mRNA (Singh et al., 1996), like its HSV-1 counterpart.
The nucleolus is the center of ribosomal biogenesis, which is a highly complex process leading to the production of pre-ribosomal particles that are then released to the nucleoplasm and exported to the cytoplasm as mature ribosomal subunits (Carmo-Fonseca et al., 2000). Exogenous non-ribosomal proteins have been demonstrated to be transported to the nucleolus under certain conditions. Proteins from porcine reproductive and respiratory syndrome virus (Rowland et al., 1999), Semliki Forest virus (Favre et al., 1994), coronavirus (Hiscox et al., 2001), and Borna disease virus (Pyper et al., 1998) have been reported to localize in the nucleolus and be involved in various aspects of virus replication. A cluster of basic amino acids is responsible for targeting proteins to the nucleus or nucleolus (Kaffman and O'Shea, 1999).
In previous studies, we have characterized the subcellular localization of BHV-1 VP8 and VP22 and identified their nuclear localization and export signals (Zheng et al., 2004, Zheng et al., 2005). The intracellular distribution of BICP27 protein has not been well characterized. As an important step toward understanding the detailed functions of BICP27 in vivo is to determine its precise subcellular localization, a study has been undertaken to characterize the exact subcellular localization of BICP27. Immunofluorescence and cell fractionation methods revealed that BICP27 was located predominantly in the nucleolus with a faint staining in the nucleus in BHV-1 infected cells and transient transfected cells. By sequence analysis and constructing mutants, the putative nuclear localization signal (NLS) and nucleolar localization signal (NoLS) of BICP27 were identified and confirmed by functional analysis.
2. Materials and methods
2.1. Cells and viruses
Madin Darby bovine kidney (MDBK) cells were grown in minimal essential medium (MEM; Gibco-BRL) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL). COS-7 cells (a monkey kidney cell line) and 3T3 (a mouse embryo cell line) cells were grown in Dulbecco's modified MEM (DMEM; Gibco-BRL) supplemented with 10% FBS. A BHV-1 virulent isolated strain (wild type) obtained from Dr. Liu Zhengfei (College of Veterinary Medicine, Huazhong Agriculture University) was used for infections and viral genomic DNA purification.
2.2. Plasmid construction
All enzymes used for cloning procedures were purchased from Amersham Pharmacia Biotech (GE). The BICP27 ORF (Singh et al., 1996) and the minimal gC promoter sequence (Hamel and Simard, 2003) were amplified from BHV-1 genomic DNA by PCR using Deep Vent DNA polymerase (New England BioLabs). The primers for constructing all the recombinant plasmids are listed in Table 1 . The deletion mutants of putative NLS, NoLS or NLS + NoLS of BICP27 were generated by ligating two PCR fragments with vector pEYFP-N1 (Clontech), in which one with EcoRI site in N-terminus and a blunt end in C-terminus, and another one with BamHI site in C-terminus and a blunt end in N-terminus. The BICP27 ORF and its NLS, NoLS or both deletion mutants were amplified from the respective EYFP fusion constructs by PCR into pcDNA3.1(+) (Invitrogen) to generate respective eukaryotic expression plasmids. The gC promoter sequence was inserted into the BglII and HindIII sites of pGL3 (Promega) to generate a luciferase reporter gene plasmid pGL-gCp-Luc. Each construct was confirmed by sequencing. Enhanced cyan fluorescence protein (ECFP) and ribosomal protein L23 fusion protein expression plasmid pECFP-L23 was obtained from Dr. Johannes A. Schmid at University of Vienna, Austria. Plasmid DNA was purified by QIAGEN plasmid Mini kits (QIAGEN).
Table 1.
Primers for constructing recombinant plasmids and deletion mutants.
| Plasmids’ name | Forward primer | Reverse primer |
|---|---|---|
| pcDNA3.1–BICP27 | CGGAATTCATGGCGGACCCCGAGATCGC | GCTCTAGATTACGGAAGATCTCGTTGCAGTAAA |
| pBICP27–EYFP | CGGAATTCATGGCGGACCCCGAGATCGC | CGGGATCCCGGAAGATCTCGTTGCAGTAAA |
| pEYFP–BICP27 | CGGAATTCATGGCGGACCCCGAGATCGC | CGGGATCCTTACGGAAGATCTCGTTGCAGTAAA |
| pBICP27–dEYFP | CGGGATTCATGGTGAGCAAGGGCGAGGA | CGGGATCCCTTGTACAGCTCGTCCATGC |
| pBICP27–tEYFP | CGGGATTCATGGTGAGCAAGGGCGAGGA | CGGGATCCCTTGTACAGCTCGTCCATGC |
| paa1–50–EYFP | CGGAATTCATGGCGGACCCCGAGATCGC | CGGGATCCCAGCTTGCGCTTGCGAACCT |
| paa1–85–EYFP | CGGAATTCATGGCGGACCCCGAGATCGC | CGCGGATCCACGCGAGCCCGCCGCGCCG |
| paa1–91–EYFP | CGGAATTCATGGCGGACCCCGAGATCGC | CGCGGATCCCGCGGCCGCCGCGGCCGCA |
| paa1–100–EYFP | CGGAATTCATGGCGGACCCCGAGATCGC | CGCGGATCCGCGGGCTGTCTTCGGCGCG |
| paa1–214–EYFP | CGGAATTCATGGCGGACCCCGAGATCGC | CGGGATCCATGAGCGTCTCCCAGGTGAC |
| paa101–400–EYFP | CGGAATTCATGGGAGAGCAGCGGTCCCGCGG | CGACCGGTGGATCCCGGAAG |
| paa92–400–EYFP | CGGAATTCATGAGGCGCCCGCGCCGAAGACA | CGACCGGTGGATCCCGGAAG |
| paa86–400–EYFP | CGGAATTCATGCGGCCGCGGCGGCCGCGCAG | CGACCGGTGGATCCCGGAAG |
| paa81–400–EYFP | CGGAATTCATGCGGCGGGCTCGCGTGCGGCC | CGACCGGTGGATCCCGGAAG |
| NLS-del | CGCCGGAGGTGCTTCGGCGG | CGGCCGCGGCGGCCGCGCAG |
| NoLS-del | CACGCGAGCCCGCCGCGCCG | CAGCCCGCCGGAGAGCAGCG |
| NLS + NoLS-del | CGCCGGAGGTGCTTCGGCGG | CAGCCCGCCGGAGAGCAGCG |
| ptEYFP | CGGAATTCATGGTGAGCAAGGGCGAGGA | GAAGATCTTTGTACAGCTCGTCCATGC |
| paa81–97–tEYFP | AATTCATGCGGCGGGCTCGCGTGCGGCCGCGGCGGCCGCGCAGGCGCCCGCGCCGAAGG | GATCCCTTCGGCGCGGGCGCCTGCGCGGCCGCCGCGGCCGCACGCGAGCCCGCCGCATG |
| paa86–97–tEYFP | AATTCATGCGGCCGCGGCGGCCGCGCAGGCGCCCGCGCCGAAGACAG | GATCCTGTCTTCGGCGCGGGCGCCTGCGCGGCCGCCGCGGCCGCATG |
| paa81–84–tEYFP | AATTCATGCGGCGGGCTCGCG | GATCCGCGAGCCCGCCGCATG |
| pGL-gCp-Luc | TTAGATCTTGAACGGCTGCCGCCGAC | AAAAGCTTGGCGGACGCGCGGCGAGG |
2.3. Transfection and luciferase assays
COS-7, 3T3 or MDBK cells were plated onto six-well plates at a density of 2.5 × 105 cells per well. Cells were grown overnight to 60–80% confluency before transfection. COS-7 or 3T3 cells were tranfected with 1.0 μg plasmid DNA using Lipofectamine 2000 plus reagent according to the instruction of the manufacturer (Invitrogen). Monolayer of MDBK cells was tranfected with 2.0 μg plasmid DNA using SuperFect transfection reagent (QIAGEN) according to the instruction of the manufacturer.
For luciferase assay, COS-7 cells were transfected with 2 μg of pGL-gCp-Luc and 0.7 μg of eukaryotic expression plasmids encoding BICP27 and its deletion mutants. Forty-eight hours after transfection, cells were harvested and the expression of Luc was determined using a luciferase assay system (Promega) according to the manufacturer's instructions with a Luminat LB 9507 (Berthold, Wildbad, Germany). Relative fold induction is calculated as light units of the test sample divided by the pGL-3 alone transfected cells.
2.4. Western blot analysis
Protein samples were separated by sodium docecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membrane (Bio-Rad). The membranes were blocked with 3% skim milk in phosphate-buffered saline (PBS) with 0.05% Tween 20 (PBST) overnight at 4 °C, washed with PBST once, incubated with antipeptide serum 50 (Singh et al., 1996) directed against the amino terminus of BICP27 or antibodies against EYFP (Clontech) at room temperature for 2 h, then washed with PBST three times and incubated with secondary antibody conjugated with alkaline phosphatase (Kirkegaard & Perry Laboratories). Reactive bands were revealed with nitro blue tetrazolium bromochlorindodyl phosphate tablets (Sigma–Aldrich). Images were scanned and processed using Adobe Photoshop.
2.5. Immunofluorescence microscopy
Cells were fixed in 4% paraformaldehyde for 20 min, washed three times with PBS and permeabilized with 0.5% Triton X-100 for 10 min. The cells were rinsed with PBS and then blocked with PBS containing 10% FBS for 20 min at room temperature. Subsequently, BICP27 specific antibody diluted in PBS containing 10% FBS was added to the cells, which were again incubated for 20 min at room temperature. Finally, FITC-conjugated goat anti-rabbit IgG (Zymed Laboratories) in PBS containing 10% FBS was added, followed by 20 min incubation at room temperature. After each incubation step, cells were washed extensively with PBS. The cells were mounted in Vectashield (Vector Laboratories). Fixed or live cells were analyzed using fluorescence microscopy. All the photomicrographs were taken under a magnification of 400×. Each photomicrograph represents a vast majority of the cells with similar subcellular localization. Images were processed using Adobe Photoshop.
2.6. Subcellular fractionation
Nuclear and cytoplasmic fractions were isolated as described previously (Sandri-Goldin, 1998, Sandri-Goldin and Mendoza, 1992). Briefly, cells were scraped into ice-cold PBS, centrifuged at 3000 × g, and resuspended in a lysis buffer consisting of 10 nm Tris (pH 7.4), 3 mM CaCl2, 2 mM MgCl2, and 0.5% NP-40. The cells were lysed by five strokes of a Dounce tissue homogenizer (Bellco Glass). The nuclei were pelleted by centrifugation at 2000 × g at 4 °C. The supernatant was collected as the cytoplasmic fraction. For further subcellular fraction (Siomi et al., 1988), the nuclear pellet mentioned above was suspended in 0.34 M sucrose containing 0.05 mM MgCl2 and 0.5 mM PMSF, and then sonically disrupted until >99% of nuclei were broken and nucleoli were released as monitor by azure C staining technique. The sonicate was layered over two volumes of 0.88 M sucrose containing 0.05 mM MgCl2 and 0.5 mM PMSF and centrifuged at 2000 × g for 20 min. The supernatant was used as the nucleoplasmic fraction, and the pellet was used as the nucleolar fraction. To make sure that the subcellular fractions were separated properly, subcellular lysates were verified by the antibodies against the corresponding fractions. These antibodies (Abcam) include anti-nucleolin against nucleolin for the nucleoli, anti-calreticulin against ER for the cytoplasm and lamin A antibody against lamin for the nucleoplasm.
3. Results
3.1. Localization of BICP27 in MDBK cells infected with BHV-1 and in MDBK, COS-7 and 3T3 cells transfected with pcDNA3.1-BICP27
To determine the exact subcellular localization during infection, the localization of BICP27 in MDBK cells infected with BHV-1 was investigated. The ECFP-tagged ribosomal protein L23 (ECFP-L23) encoded by plasmid pECFP-L23, was used as a nucleolar marker. The ECFP-L23 has been demonstrated to localize to the nucleoli upon overexpression (Birbach et al., 2004, Gleizes et al., 2001). Immediately after transfection with pECFP-L23, MDBK cells were infected with BHV-1 at a multiplicity of infection (MOI) of 0.1. The cells were fixed 16 h post-infection and analyzed by an immunofluorescence assay using antipeptide serum 50 (Singh et al., 1996) against BICP27. BICP27 staining exhibited predominant nucleolus localization with a faint staining in the nucleus (Fig. 1A). Based on their size and numbers, these subnuclear structures appeared to be nucleoli (Fig. 1A). To confirm this assumption we cotransfected virus infected cells with the pECFP-L23. Fluorescence microscopy showed clear co-localization of BICP27 and ECFP-L23 (Fig. 1A). This also demonstrated that the dense, dark-staining, irregular shaped subcellular organelles in the nucleus are nucleoli (Fig. 1A, transmitted and ECFP-L23). Thus phase-contrast images were taken to indicate the nucleolus in the following experiments. To further confirm this observation, subcellular fractions were subjected to Western blotting analysis. Fig. 1B showed that BICP27 was enriched in the nucleolus-containing fraction. No fluorescence was observed when preimmune serum was used (Fig. 1C).
Fig. 1.
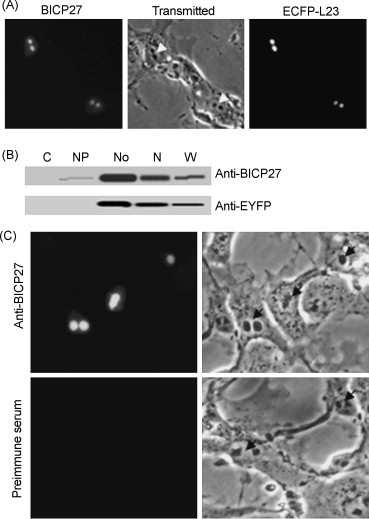
Subcellular localization of BICP27 in BHV-1 infected MDBK cells and transiently transfected cells. (A) MDBK cells were infected with BHV-1 at MOI of 0.1 immediately after transfection with pECFP-L23. 16 h after infection immunofluorescence staining of BICP27 was performed using antipeptide serum 50. Immunofluorescence photomicrograph of BICP27, the corresponding phase-contrast image and ECFP-L23 location photomicrographs are shown. Arrows indicated the nucleoli. (B) Subcellular localization of BICP27 and ECFP-L23. Twelve micrograms of protein was applied to lanes 1–5. The antibodies for BICP27 and ECFP were indicated on the right margin. Lane C, cytoplasmic fraction; lane NP, nucleoplasmic fraction; lane No, nucleolar fraction; lane N, nuclear fraction; lane W, whole cell extract. (C) Immunofluorescence analysis of COS-7 cells expressing BICP27. Cells transfected with pcDNA3.1-BICP27 were fixed 24 h post-transfection and immunofluorescent assay was carried out to detect the expression and subcellular localization of BICP27. Immunofluorescence photomicrograph (left) and the corresponding phase-contrast photomicrograph (right) are shown. Arrows indicated the nucleoli. Each image is representative of the vast majority of the cells observed.
To further investigate the subcellular localization of BICP27 in the absence of other viral proteins, the ORF of BICP27 was cloned into the eukaryotic expression vector pcDNA3.1(+) to generate pcDNA3.1–BICP27. To confirm the expression of BICP27 in transfected cells, proteins synthesized in these cells were examined by Western blot. The BICP27 expressed in MDBK, COS-7 or 3T3 cells migrated on SDS-PAGE at the same rate as BICP27 synthesized in BHV-1-infected cells (data not shown). In all three cell lines tested, BICP27 was found to localize predominantly to the nucleoli with a faint staining in the nucleus, indicating that there is a conserved mechanism for nucleolar localization in all three lines of cells. The immunofluorescent micrograph of transfected COS-7 cells was shown in Fig. 1C as a representative. As BICP27 showed similar distribution in all cell types tested, the following transfection experiments were performed in COS-7 cell, which has the highest transfection efficiency among the three cell lines. No fluorescence was observed in mock transfected cells or when preimmune serum was used (data not shown). To address whether the nucleolar accumulation of the BICP27 was an artifact of overexpression, a time course of expression showed that BICP27 accumulated in the nucleolus as early as 3 h after transfection when expression levels were low (data not shown).
3.2. Localization of BICP27–EYFP fusion protein in transfected cells
It is well known that some fixation protocols may alter the localization of proteins, resulting in misleading conclusions in the analysis of the intracellular distribution of a specific protein. To avoid this problem, the fluorescence microscopy technique was applied using live cells expressing EYFP. To investigate whether BICP27 plays a role in transporting a heterogenous protein to the nucleolus, ORF of BICP27 was inserted into pEYFP-N1 to generate pBICP27–EYFP. BICP27 fusions with EYFP dimer (dEYFP) or trimer (tEYFP) were also constructed as shown in Fig. 2A. Western blotting analysis showed that BICP27–EYFP, BICP27–dEYFP and BICP27–tEYFP were expressed at the expected molecular sizes 78, 106 and 134 kDa respectively using antipeptide serum 50 against BICP27 (Singh et al., 1996) (Fig. 2B) or polyclonal antibodies against EYFP (data not shown), indicating that they were fusions of BICP27 (50 kDa) and different copies of EYFP (28 kDa). The subcellular localization of BICP27–EYFP fusion protein in live cells was visualized by fluorescence microscopy. Low level expression of BICP27–EYFP was observed in the nucleolus right after removing the transfection mixtures (about 3 h after transfection), and the localization of BICP27–EYFP does not change after 48 h transfection (data not shown). This also confirmed that the nucleolar localization was not an artifact of high expression levels.
Fig. 2.
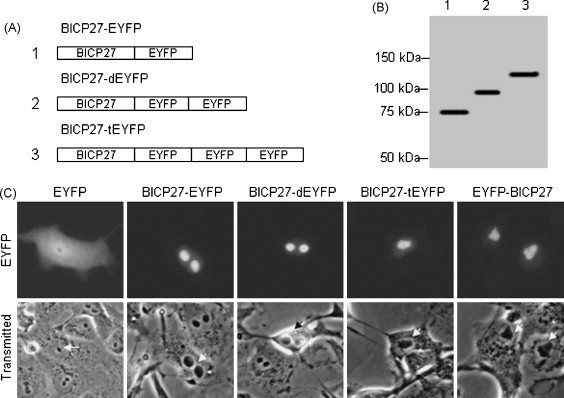
Localization of BICP27 and EYFP fusion protein in transfected cells. (A) Schematic diagram of the BICP27 fusion with EYFP monomer, dimer and trimer in its C-terminus; (B) Western blotting analysis of expression of fusion proteins using antipeptide serum 50 (Singh et al., 1996). The lane number corresponds to the constructs in Fig. 2A. (C) Fluorescence microscopy analysis of the COS-7 cells expressing BICP27–EYFP, BICP27–dEYFP, BICP27–tEYFP and EYFP–BICP27 in comparison with phase-contrast photomicrographs of the same cells. Arrows indicate the nucleoli. Each image is representative of the vast majority of the cells observed.
Fig. 2C showed micrographs of COS-7 cells 24 h after transfection with plasmids encoding BICP27 and EYFP fusion proteins. BICP27 fused with one, two or three copies of EYFP showed similar patterns of localization. The fluorescence was predominantly restricted to the nucleolus by BICP27 (Fig. 2C). On the contrary, the fluorescence was evenly distributed throughout the cytoplasm and the nucleoplasm but not the nucleolar structures in cells transfected with plasmids encoding EYFP monomer (pEYFP-N1) or dimer (pdEYFP-N1) (Fig. 2C). In cells transfected with plasmid ptEYFP-N1 encoding EYFP trimer, the fluorescence was localized exclusively to the cytoplasm (Fig. 5C, tEYFP), due to the molecular size of EYFP trimer (84 kDa) which prevents passive trafficking between the cytoplasm and the nucleus (Gorlich and Mattaj, 1996). Western blotting analysis of subcellular fractions of BICP27–EYFP showed similar results to that of BICP27 in BHV-1 infected cells (data not shown). No fluorescence was observed in non-transfected cells (data not shown). In order to investigate whether the orientation of BICP27 affects its localization in cells, DNA construct was also made to express BICP27 fused to the C-terminus of EYFP (EYFP–BICP27) (Fig. 2C). The expression of EYFP–BICP27 was confirmed by Western blotting (data not shown). The fluorescence microscopy demonstrated identical subcellular distribution patterns of BICP27–EYFP and EYFP–BICP27, thus the following experiments were performed only for EYFP fusion to C-terminus of BICP27. The subcellular distribution of BICP27–EYFP was also probed with a BICP27 antibody. The result of indirect immunofluorescence was consistent with that of EYFP autofluorescence, indicating that BICP27–EYFP remained intact and localized to the nucleolus. Taken together, these results indicated that BICP27 was responsible for driving EYFP into the nucleolus.
Fig. 5.
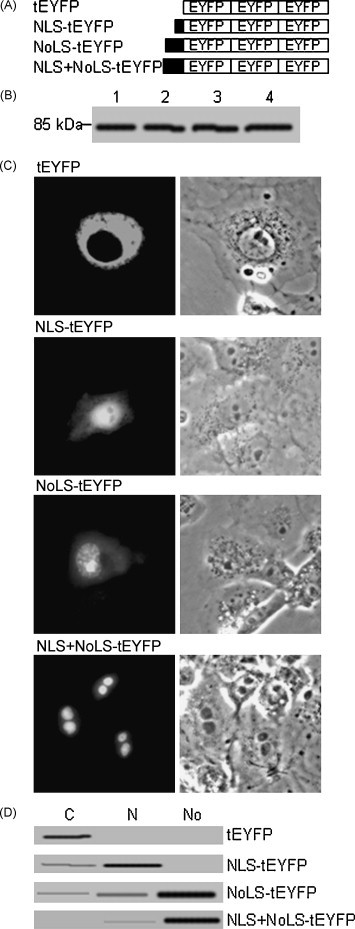
81RRAR84 and 86RPRRPRRRPRRR97 are functional NLS and NoLS respectively. (A) Schematic diagram of the putative NLS and NoLS fusion with trimeric EYFP (tEYFP). (B) Western blotting analysis of the different deletion mutants of arginine-rich domain using EYFP polyclonal antibodies. (C) Subcellular localization of the putative NLS, NoLS, or NLS + NoLS fusion with tEYFP. COS-7 cells were transfected with plasmids for indicated EYFP fusion proteins and representative EYFP fluorescence images of the vast majority living cells expressing indicated fusion protein were shown. (D) Western blotting analysis of the subcellular localization of NLS–tEYFP, NoLS–tEYFP and NLS + NoLS–tEYFP using EYFP polyclonal antibodies.
3.3. Mapping the nuclear and nucleolar localization signal in the arginine-rich region of BICP27
From the above results we hypothesize that there is a nucleolar localization or targeting signal in BICP27 which can guide exogenous protein to the nucleolus. Since BICP27 fusions with EYFP monomer, dimer or trimer showed similar nucleolus localization patterns, the following experiments were performed with monomer EYFP fusion. To investigate the amino acid sequence of BICP27 that is responsible for nucleolar localization, plasmids were constructed to encoding BICP27 deletion mutants in frame with EYFP (Fig. 3A). Subcellular localization of a series of deletion mutants encompassing amino acids 1–50, 1–85, 1–91, 1–100, 1–214, 101–400, 92–400, 86–400 and 81–400 fused with EYFP was investigated. The expressions of all these mutants were confirmed by Western blot analysis using anti-EYFP antibody, which detected the proteins of the expected sizes (Fig. 3B). COS-7 cells were transfected with these constructs and the subcellular localization of each mutant was viewed by fluorescence microscopy at 24 h post-transfection. aa1–50–EYFP showed similar distribution patterns as EYFP, with fluorescence in both the nucleus and cytoplasm, but not in the nucleolus (Fig. 3C). aa1–85–EYFP and aa1–91–EYFP were demonstrated to be located in the nucleus, but completely excluded from the nucleolus (Fig. 3C). The cellular distribution patterns of aa1–100–EYFP and aa1–214–EYFP were similar to wild type BICP27–EYFP fusion protein, with the fluorescence enriched to the nucleoli. aa101–400–EYFP was distributed exclusively in the cytoplasm (Fig. 3C). aa92–400–EYFP was found predominantly in the cytoplasm with faint staining in the nucleoli (Fig. 3C). aa86–400–EYFP localized predominantly to the nucleolus with cytoplasmic staining and background staining in the nucleus (Fig. 3C). However, aa81–400–EYFP was localized predominantly in the nucleolus with background staining in the nucleus. The pattern was identical to that of wild type BICP27 and BICP27–EYFP (Fig. 3C, comparing with Fig. 1, Fig. 2 panel EYFP). To confirm this observation, subcellular fractions were subjected to Western blot analysis as shown in Fig. 3D. Taken together, these results indicated that the putative nuclear localization signal (NLS) and nuleolar localization signal (NoLS) of BICP27 were located in its N-terminus arginine-rich domain from amino acids 81 to 85 and 86 to 97 respectively.
Fig. 3.
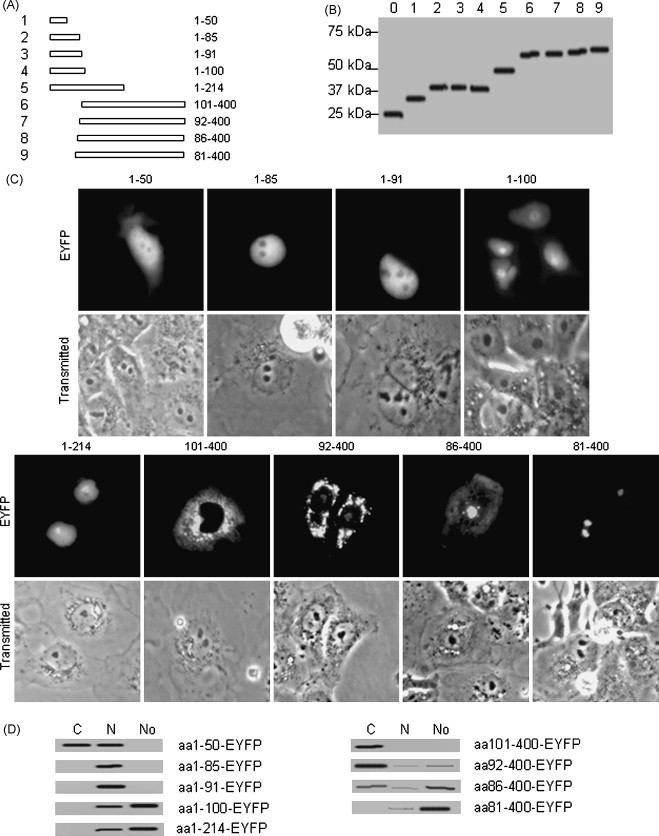
The nucleolar localization signal resides in the arginine-rich region of BICP27. (A) Schematic diagram of BICP27 mutants fused with EYFP. (B) Western blotting analysis of the expression of the truncation mutants using the anti-EYFP antibody. The lane number corresponds to the constructs in Fig. 3A. Lane 0 indicates EYFP alone. (C) Subcellular localization of BICP27 mutants fused with EYFP. Representative fluorescence images of the vast majority of living cells for indicated EYFP fusion proteins and EYFP fluorescence was analyzed in living cells 24 h after transfection. Each image is representative of the vast majority of the cells observed. (D) Western blotting analysis of the subcellular localization of different BICP27 mutants using EYFP polyclonal antibodies.
3.4. Identification of functional NLS and NoLS in BICP27
The function of a putative NLS or NoLS can be tested either by fusion with a cytoplasmic reporter protein or by mutation. If the sequence is an NLS or NoLS, it will drive the cytoplasmic reporter protein into the nucleus or nucleolus respectively. In the latter case, a protein containing a defective NLS or NoLS will fail to enter the nucleus or nucleolus respectively. To identify the functional NLS and/or NoLS in BICP27, the specific deletion of NLS, NoLS or NLS/NoLS from BICP27 was made as shown in Fig. 4A. The expression of BICP27–EYFP-NLS-del, BICP27–EYFP-NoLS-del and BICP27–EYFP-NLS/NoLS-del was confirmed by Western blotting (Fig. 4B). The NLS, NoLS, NLS/NoLS deletions significantly attenuated protein accumulation in the nucleus, the nucleolus or both respectively. The subcellular localization pattern of BICP27–EYFP-NLS-del was almost identical to that of aa86–400–EYFP (Fig. 4C, comparing with Fig. 3B, panel 86–400). The subcellular localization pattern of BICP27–EYFP-NoLS-del was identical to that of aa1–85–EYFP and aa1–91–EYFP (Fig. 4C, comparing with Fig. 3B panels 1–85 and 1–91) and the subcellular localization pattern of BICP27–EYFP-NLS/NoLS-del was identical to that of aa101–400–EYFP (Fig. 4C, comparing to Fig. 3B panel 101–400). To confirm this observation, subcellular fractions were subjected to Western blot analysis as shown in Fig. 4D. Furthermore, mutation of arginine to alanine in the NLS, NoLS, or NLS/NoLS resulted in identical subcellular localization pattern to the respective deletion construct (data not shown). Therefore aa81–84 (RRAR) and aa86–97 (RPRRPRRRPRRR) were identified as the functional NLS and NoLS respectively in the context of wild type BICP27.
Fig. 4.
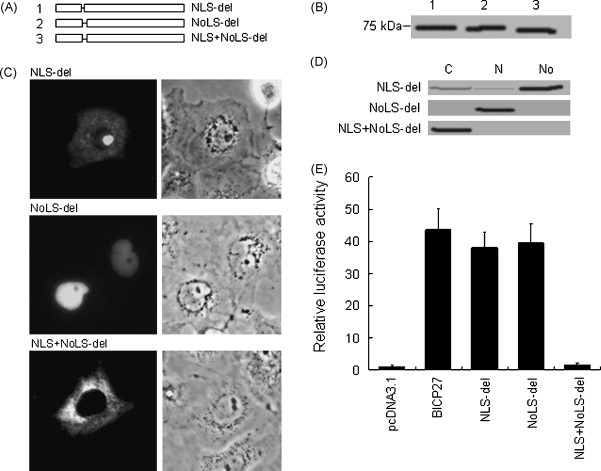
Mutation of arginine-rich amino acids residues abrogates the nuclear or nucleolar localization of BICP27. (A) Schematic diagram of deletion of arginine-rich domain in BICP27. (B) Western blotting analysis of the different deletion mutants of arginine-rich domain using EYFP polyclonal antibodies. The lane number corresponds to the constructs in Fig. 4A. (C) Subcellular localization of BICP27 arginine-rich domain deletion mutants fused with EYFP. Representative fluorescence images of the vast majority living cells expressing indicated EYFP fusion proteins and EYFP fluorescence was analyzed in living cells 24 h after transfection. (D) Western blotting analysis of the subcellular localization of different BICP27 deletions using EYFP polyclonal antibodies. (E) Transactivation analysis of gC promoter by BICP27 and its deletion mutants. Cells were harvested 48 h post-transfection and assayed for luciferase activity. Relative fold induction is calculated as light units of the test sample divided by the pGL-3 transfected cells. Standard deviations from the mean of three independent experiments are indicated.
3.5. Regulation of gC promoter by BICP27 and its NLS, NoLS or both deletion mutants
To address whether NLS, NoLS or both is critical for the transactivation activity of BICP27 on gC gene promoter, a PCR-based approach was used to isolate the BHV-1 gC promoter (Hamel and Simard, 2003). The promoter sequence was cloned into the pGL3 vector to generate pGL-gCp-Luc. The NLS, NoLS or both deletion mutants were subcloned to generate respective eukaryotic expression plasmids. Then COS-7 cells were cotransfected with reporter plasmid pGL-gCp-Luc and effector plasmid expressing BICP27 and its deletion mutants. Cells were harvested 48 h post-transfection and the relative luciferase activity was determined. As shown in Fig. 4E, either NLS or NoLS deletion did not abrogate the transactivation activity of BICP27. However, the NLS and NoLS double deletion mutant lost the transactivating function.
3.6. Verification of the function of the NLS and NoLS in BICP27
To validate the function of the identified NLS and NoLS, DNA fragments corresponding to the amino acid sequences 81RRAR84, 86RPRRPRRRPRRR97 or 81RRARVRPRRPRRRPRRR97 were synthesized, containing the putative NLS, NoLS or NLS + NoLS. The fragments were inserted to the 5′ terminus of tEYFP gene to encode NLS–tEYFP, NoLS–tEYFP or NLS/NoLS–tEYFP fusion proteins (Fig. 5A). The expressions of NLS–tEYFP, NoLS–tEYFP or NLS + NoLS–tEYFP were corroborated by Western blotting (Fig. 5B). In COS-7 cells transfected with ptEYFP-N1, tEYFP was localized exclusively to the cytoplasm (Fig. 5C). When NLS was fused to tEYFP, it directed tEYFP predominantly to the nucleus but not the nucleolus. When NoLS was fused to tEYFP, it targeted tEYFP primarily to the nucleus and accumulated in the nucleolus with faint fluorescence in the cytoplasm. When both NLS and NoLS were fused to tEYFP, they targeted the tEYFP predominantly to the nucleolus with faint staining in the nucleus and this localization pattern was identical to wild type BICP27 and BICP27–EYFP (Fig. 5C, comparing with Fig. 1, Fig. 2 panel EYFP). To confirm this observation, subcellular fractions were subjected to Western blotting analysis as shown in Fig. 5D. To test whether the orientation of NLS or NoLS would affect the protein localization, NLS, NoLS or NLS + NoLS were also placed at the C-terminus of tEYFP, similar observations were obtained (data not shown), indicating that the orientation of the NLS, NoLS or NLS + NoLS in the sequence did not affect its function. Taken together, a functional NLS and a functional NoLS in BICP27 were identified as amino acid residues 81RRAR84 and 86RPRRPRRRPRRR97 respectively.
In summary, we mapped the functional NLS and NoLS of BICP27 to its N-terminal basic residues between amino acids 81–84 and 86–97 respectively. This makes BICP27 the first bovine herpesvirus-1 protein to be identified in the nucleoli.
4. Discussion
BICP27 is an early protein of BHV-1 which may stimulate mRNA 3′ processing (Singh et al., 1996). In this study we examined the subcellular localization of BICP27 and found that BICP27 protein localized predominantly to the nucleolus with faint staining in the nucleus. Chemical fixation may lead to the artifact of the protein localization, so the EYFP was introduced in this study. EYFP was used as a marker to visualize the subcellular localization of BICP27 in live cells and to identify the NLS and NoLS. BICP27–EYFP fusion protein with a molecular mass of 78 kDa was not expected to passively diffuse into the nucleoplasm without an NLS/NoLS. A previous study has shown that BICP27 targeted mainly to the nucleus (Singh et al., 1996), though the protein was obviously enriched in the nucleolus too. This is different from the results in this research (Fig. 1A). This might be due to the different fixation reagent. The former applied permeable reagent methanol, whereas the non-permeable reagent paraformaldehyde was used in this study.
Amino acid sequence analysis demonstrated that two clusters of arginine-rich sequence in N-terminus of BICP27 represented the potential NLS/NoLS. The arginine-rich region between amino acids 86 and 97 resembled functional NoLS described in several other viral proteins (Kubota et al., 1996, Liu et al., 1997, Mears et al., 1995, Siomi et al., 1990, Siomi et al., 1988) (Table 2 ). Similar sequences were also detected in cellular proteins (Henderson et al., 1995, Stegh et al., 1998). Although BICP27 and ICP27 are homologous proteins, the amino acid sequence for their NoLS is quite different, in which ICP27's NoLS has RGG motif, which is an RNA binding motif, whereas BICP27's NoLS only contains arginine residues.
Table 2.
Alignment of NoLS sequencesa of identified nucleolar proteins and BICP27.
| Protein | NoLS | Reference |
|---|---|---|
| HTLV-1 Rex | MPKTRRRPRRSQRKRPPTP | Siomi et al. (1988) |
| HIV-1 Tat | GRKKRRQRRRP | Siomi et al. (1990) |
| HIV-1 Rev | RQARRNRRRRWRERQR | Kubota et al. (1996) |
| MDV MEQ | RRRKRNRDAARRRRKQ | Liu et al. (1997) |
| PTHrPb | GKKKGKPGKRREQEKKKRRT | Henderson et al. (1995) |
| HSV-1 ICP27 | RGGRRGRRRGRGRGG | Mears et al. (1995) |
| BICP27 | RPRRPRRRPRRR |
The Arg and Lys residues are indicated by boldface type.
PTHrP, parathyroid hormone-related peptide.
The potential NLS and NoLS domains in BICP27 were further investigated and confirmed by deletion–mutation analysis. Deletion of the NLS-containing region (NLS-del) resulted in the presence of BICP27 predominantly in the nucleolus with only faint staining in the nucleus, whereas deletion of the NoLS-containing region (NoLS-del) resulted in the presence of BICP27 in the nucleus, but excluded from the nucleolus. This suggested that NoLS does not display nuclear localization function in the context of wild type of BICP27. However, similar to previous reports, the NoLS motif is arginine-rich (Table 2). This suggested that NoLS plays nuclear localization function due to its arginine-rich residues only when there is no functional NLS present in the protein and translocates some of BICP27-NLS-del through NPC to the nucleus and targets to the nucleolus (Fig. 4 panel NLS-del and Fig. 5 panel NoLS–tEYFP). This also suggested that NLS and NoLS function differently when expressed in the full-length protein. Furthermore, NoLS–tEYFP has stronger nuclear localization than BICP27–EYFP-NLS-del, this might be due to a potential NES in the C-terminus of BICP27. Taken together, we presented compelling evidence that 81RRAR84 and 86RPRRPRRRPRRR97 represent the functional NLS and NoLS of BICP27 respectively.
As demonstrated in previous study, BICP27 has transactivation activity on gC promoter (Hamel and Simard, 2003). Our results confirmed this activity and extended the study. BICP27 with either NLS or NoLS deletion alone did not abrogate the nuclear localization and affect its transactivation activity, whereas BICP27 with double deletion of NLS and NoLS targeted to the cytoplasm and lost the transactivation activity. Accordingly, the nuclear localization is very critical for BICP27 to perform its regulationary function.
The results shown in this study indicated that the identified NLS or NoLS can direct trimeric EYFP into the nucleus or nucleolus but not as efficiently as full-length BICP27. This might be explained by which, in the context of the full-length BICP27, the NLS may adopt a conformation recognized by the nuclear or nucleolar translocation machinery. In the context of wt BICP27, the NLS or NoLS maybe located on the outer surface of BICP27. However, in the NLS–tEYFP or NoLS–tEYFP fusion protein, the basic cluster of NLS or NoLS may not be accessible or in the same conformation as they are in the wt BICP27 or BICP27 fused to EYFP, thereby resulting in less efficient targeting.
Recently, growing attention has been drawn to the interrelationship between nuclear structure and nuclear functions. Many viral and cellular proteins are expressed not only in the nucleoplasm but also in specific subnuclear compartments. Localization to the nucleolus has been proved to be part of the strategy for virus to control both host cell and virus subgenomic RNA translation. The coronavirus N protein which is located in the nucleolus disrupted host cell division (Hiscox et al., 2001), and the HIV regulatory proteins Tat and Rev made use of nucleolus for trafficking of HIV-1 RNA (Michienzi et al., 2000, Siomi et al., 1990). A number of transcription factors have also been found to be expressed in the nucleolus. Among these transcription factors, the tumor suppressor TBP (Jones et al., 1992) and Rb (Cavanaugh et al., 1995), in addition to their roles in RNA polymerase II transcription, also regulate rRNA transcription. BICP27 may be involved in 3′ processing of mRNA (Singh et al., 1996), and it might act as a transcription factor, thus its nucleolar localization is expected.
Viral proteins may serve pleiotropic roles in regulating viral and host functions. For Rev and Rex, the nucleolar localization is required for their functions in posttranscriptional regulation of viral mRNA (Dundr et al., 1995). Us11, an RNA binding protein of HSV-1, is involved in regulating mRNA accumulation in the cytoplasm and can functionally substitute for Rev and Rex (Diaz et al., 1996). The nucleolar localization of BICP27 may suggest a new function for this protein.
In this study, we clearly demonstrated that the arginine-rich domain in N-terminus was responsible for the translocation. Based on the analysis of BICP27 deletion mutants and EYFP fusion proteins, the arginine-rich domain is a necessary signal for nucleolar localization. The identified domain has a 12-basic-residue cluster which is longer than most NLSs but similar in length to the NoLS of viral proteins such as Rex, Tat, Rev and a number of other proteins (Table 2). The alignment of identified NoLS reveals a hallmark of a long stretch of basic residues. It has been shown that Rev (Fankhauser et al., 1991), Rex (Adachi et al., 1993) and Tat (Marasco et al., 1994) interact with B23, a nucleolar shuttling protein. The interaction domain for B23 was mapped to its highly acidic domain (Adachi et al., 1993, Szebeni et al., 1995), whereas for Rev (Szekely et al., 1995) and Rex (Adachi et al., 1993) the interaction domains were mapped to their respective NoLSs. By analogy to Rev, Rex, Us11 and ICP27, BICP27 may be involved in the regulation of viral RNA processing or transport. To clearly define the role of BICP27's nucleolar localization in viral replication, it would be helpful if mutants that dissociate transcriptional function from nucleolar localization could be developed.
Taken together, this study suggested that BICP27's function as viral or host gene regulator may not be limited to its transcriptional potential. Its association with the nucleolus may provide new leads to uncovering other novel activities.
Acknowledgements
This study was supported by grants from the Start-up Fund of the Hundred Talents Program of the Chinese Academy of Science (20071010-141); the National Natural Science Foundation of China (30870120); the Open Research Fund Program of the State Key Laboratory of Virology of China (2007003 and 2009007). We thank Dr. Martin Schwyzer for the generous gift of the antibody against BICP27 and preimmune serum, Dr. Johannes A. Schmid for the generous gift plasmid pECFP-L23 and Dr. Christopher H Mody for critical review of the manuscript.
References
- Adachi Y., Copeland T.D., Hatanaka M., Oroszlan S. Nucleolar targeting signal of Rex protein of human T-cell leukemia virus type I specifically binds to nucleolar shuttle protein B-23. J. Biol. Chem. 1993;268(19):13930–13934. [PubMed] [Google Scholar]
- Birbach A., Bailey S.T., Ghosh S., Schmid J.A. Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NF-kappaB inducing kinase NIK. J. Cell Sci. 2004;117(Pt 16):3615–3624. doi: 10.1242/jcs.01224. [DOI] [PubMed] [Google Scholar]
- Boyne J.R., Colgan K.J., Whitehouse A. Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front. Biosci. 2008;13:2928–2938. doi: 10.2741/2898. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Mendes-Soares L., Campos I. To be or not to be in the nucleolus. Nat. Cell Biol. 2000;2(6):E107–112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- Cavanaugh A.H., Hempel W.M., Taylor L.J., Rogalsky V., Todorov G., Rothblum L.I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374(6518):177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- Chalifour A., Basso J., Gagnon N., Trudel M., Simard C. Transcriptional and translational expression kinetics of the early gene encoding the BICP27 protein of bovine herpesvirus type 1. Virology. 1996;224(1):326–329. doi: 10.1006/viro.1996.0536. [DOI] [PubMed] [Google Scholar]
- Diaz J.J., Dodon M.D., Schaerer-Uthurralt N., Simonin D., Kindbeiter K., Gazzolo L., Madjar J.J. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature. 1996;379(6562):273–277. doi: 10.1038/379273a0. [DOI] [PubMed] [Google Scholar]
- Dundr M., Leno G.H., Hammarskjold M.L., Rekosh D., Helga-Maria C., Olson M.O. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J. Cell Sci. 1995;108(Pt 8):2811–2823. doi: 10.1242/jcs.108.8.2811. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Izaurralde E., Adachi Y., Wingfield P., Laemmli U.K. Specific complex of human immunodeficiency virus type 1 rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol. Cell. Biol. 1991;11(5):2567–2575. doi: 10.1128/mcb.11.5.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre D., Studer E., Michel M.R. Two nucleolar targeting signals present in the N-terminal part of Semliki Forest virus capsid protein. Arch. Virol. 1994;137(1-2):149–155. doi: 10.1007/BF01311181. [DOI] [PubMed] [Google Scholar]
- Fraefel C., Zeng J., Choffat Y., Engels M., Schwyzer M., Ackermann M. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J. Virol. 1994;68(5):3154–3162. doi: 10.1128/jvi.68.5.3154-3162.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes P.E., Noaillac-Depeyre J., Leger-Silvestre I., Teulieres F., Dauxois J.Y., Pommet D., Azum-Gelade M.C., Gas N. Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J. Cell Biol. 2001;155(6):923–936. doi: 10.1083/jcb.200108142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Mattaj I.W. Nucleocytoplasmic transport. Science. 1996;271(5255):1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Hamel F., Simard C. Mapping of the bovine herpesvirus 1 glycoprotein C promoter region and its specific transactivation by the viral BICP27 gene product. Arch. Virol. 2003;148(1):137–152. doi: 10.1007/s00705-002-0898-z. [DOI] [PubMed] [Google Scholar]
- Henderson J.E., Amizuka N., Warshawsky H., Biasotto D., Lanske B.M., Goltzman D., Karaplis A.C. Nucleolar localization of parathyroid hormone-related peptide enhances survival of chondrocytes under conditions that promote apoptotic cell death. Mol. Cell. Biol. 1995;15(8):4064–4075. doi: 10.1128/mcb.15.8.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75(1):506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D., Lee L., Liu J.L., Kung H.J., Tillotson J.K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc. Natl. Acad. Sci. U.S.A. 1992;89(9):4042–4046. doi: 10.1073/pnas.89.9.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., O'Shea E.K. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- Kubota S., Duan L., Furuta R.A., Hatanaka M., Pomerantz R.J. Nuclear preservation and cytoplasmic degradation of human immunodeficiency virus type 1 Rev protein. J. Virol. 1996;70(2):1282–1287. doi: 10.1128/jvi.70.2.1282-1287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.L., Lee L.F., Ye Y., Qian Z., Kung H.J. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J. Virol. 1997;71(4):3188–3196. doi: 10.1128/jvi.71.4.3188-3196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco W.A., Szilvay A.M., Kalland K.H., Helland D.G., Reyes H.M., Walter R.J. Spatial association of HIV-1 tat protein and the nucleolar transport protein B23 in stably transfected Jurkat T-cells. Arch. Virol. 1994;139(1–2):133–154. doi: 10.1007/BF01309460. [DOI] [PubMed] [Google Scholar]
- Mears W.E., Lam V., Rice S.A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 1995;69(2):935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A., Cagnon L., Bahner I., Rossi J.J. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. U.S.A. 2000;97(16):8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V., Blumenthal R.M., Babiuk L.A. Proteins specified by bovine herpesvirus 1 (infectious bovine rhinotracheitis virus) J. Virol. 1981;40(2):367–378. doi: 10.1128/jvi.40.2.367-378.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyper J.M., Clements J.E., Zink M.C. The nucleolus is the site of Borna disease virus RNA transcription and replication. J. Virol. 1998;72(9):7697–7702. doi: 10.1128/jvi.72.9.7697-7702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R.R., Kervin R., Kuckleburg C., Sperlich A., Benfield D.A. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64(1):1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Sandri-Goldin R.M. Interactions between a herpes simplex virus regulatory protein and cellular mRNA processing pathways. Methods. 1998;16(1):95–104. doi: 10.1006/meth.1998.0647. [DOI] [PubMed] [Google Scholar]
- Sandri-Goldin R.M. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 2008;13:5241–5256. doi: 10.2741/3078. [DOI] [PubMed] [Google Scholar]
- Sandri-Goldin R.M., Mendoza G.E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6(5):848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- Schwyzer M., Ackermann M. Molecular virology of ruminant herpesviruses. Vet. Microbiol. 1996;53(1–2):17–29. doi: 10.1016/s0378-1135(96)01231-x. [DOI] [PubMed] [Google Scholar]
- Schwyzer M., Vlcek C., Menekse O., Fraefel C., Paces V. Promoter, spliced leader, and coding sequence for BICP4, the largest of the immediate-early proteins of bovine herpesvirus 1. Virology. 1993;197(1):349–357. doi: 10.1006/viro.1993.1596. [DOI] [PubMed] [Google Scholar]
- Singh M., Fraefel C., Bello L.J., Lawrence W.C., Schwyzer M. Identification and characterization of BICP27, an early protein of bovine herpesvirus 1 which may stimulate mRNA 3′ processing. J. Gen. Virol. 1996;77(Pt 4):615–625. doi: 10.1099/0022-1317-77-4-615. [DOI] [PubMed] [Google Scholar]
- Siomi H., Shida H., Maki M., Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J. Virol. 1990;64(4):1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Shida H., Nam S.H., Nosaka T., Maki M., Hatanaka M. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell. 1988;55(2):197–209. doi: 10.1016/0092-8674(88)90043-8. [DOI] [PubMed] [Google Scholar]
- Stegh A.H., Schickling O., Ehret A., Scaffidi C., Peterhansel C., Hofmann T.G., Grummt I., Krammer P.H., Peter M.E. DEDD, a novel death effector domain-containing protein, targeted to the nucleolus. EMBO J. 1998;17(20):5974–5986. doi: 10.1093/emboj/17.20.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni A., Herrera J.E., Olson M.O. Interaction of nucleolar protein B23 with peptides related to nuclear localization signals. Biochemistry. 1995;34(25):8037–8042. doi: 10.1021/bi00025a009. [DOI] [PubMed] [Google Scholar]
- Szekely L., Jiang W.Q., Pokrovskaja K., Wiman K.G., Klein G., Ringertz N. Reversible nucleolar translocation of Epstein-Barr virus-encoded EBNA-5 and hsp70 proteins after exposure to heat shock or cell density congestion. J. Gen. Virol. 1995;76(Pt 10):2423–2432. doi: 10.1099/0022-1317-76-10-2423. [DOI] [PubMed] [Google Scholar]
- Toth Z., Stamminger T. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 2008;13:2939–2949. doi: 10.2741/2899. [DOI] [PubMed] [Google Scholar]
- Turin L., Russo S., Poli G. BHV-1: new molecular approaches to control a common and widespread infection. Mol. Med. 1999;5(5):261–284. [PMC free article] [PubMed] [Google Scholar]
- Wirth U.V., Fraefel C., Vogt B., Vlcek C., Paces V., Schwyzer M. Immediate-early RNA 2.9 and early RNA 2. 6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 1992;66(5):2763–2772. doi: 10.1128/jvi.66.5.2763-2772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth U.V., Gunkel K., Engels M., Schwyzer M. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J. Virol. 1989;63(11):4882–4889. doi: 10.1128/jvi.63.11.4882-4889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Brownlie R., Babiuk L.A., van Drunen Littel-van den Hurk S. Characterization of nuclear localization and export signals of the major tegument protein VP8 of bovine herpesvirus-1. Virology. 2004;324(2):327–339. doi: 10.1016/j.virol.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Zheng C., Brownlie R., Babiuk L.A., van Drunen Littel-van den Hurk S. Characterization of the nuclear localization and nuclear export signals of bovine herpesvirus 1 VP22. J. Virol. 2005;79(18):11864–11872. doi: 10.1128/JVI.79.18.11864-11872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


