Abstract
We have completed the genetic characterization of all eight gene segments for four low pathogenic avian influenza (LPAI) viruses. The objective of this study was to detect the presence of novel signatures that may serve as early warning indicators of the conversion of LPAI viruses to high pathogenic avian influenza (HPAI) viruses. This study included three H5N2 and one H5N3 viruses that were isolated from live poultry imported into Singapore as part of the national avian influenza virus (AIV) surveillance program. Based on the molecular criterion of the World Organisation for Animal Health (OIE), sequence analysis with the translated amino acid (aa) sequence of the hemagglutinin (HA) gene revealed the absence of multibasic aa at the HA cleavage site, identifying all four virus isolates as LPAI. Detailed phylogenetic tree analyses using the HA and neuraminidase (NA) genes clustered these isolates in the Eurasian H5 lineage, but away from the HPAI H5 subtypes. This analysis further revealed that the internal genes clustered to different avian and swine subtypes, suggesting that the four isolates may possibly share their ancestry with these different influenza subtypes. Our results suggest that the four LPAI isolates in this study contained mainly avian signatures, and the phylogenetic tree for the internal genes further suggests the potential for reassortment with other different circulating avian subtypes. This is the first comprehensive report on the genetic characterization of LPAI H5N2/3 viruses isolated in South-East Asia.
Keywords: Low pathogenic avian influenza virus, H5N2, H5N3, Imported poultry, Singapore
1. Introduction
Avian influenza viruses (AIVs) are members of the Orthomyxoviridae family, Influenzavirus A genus. Their genomes comprise eight segments of single-stranded negative-sense RNA that give rise to 11 known virus proteins. These include three internal membrane proteins, the hemagglutinin (HA), neuraminidase (NA), and matrix-2 protein; four polymerase-associated proteins, the NP, PA, PB1, and PB2 proteins; the matrix-1 protein and two non-structural proteins, NS1 and NS2 proteins. The HA protein is initially expressed as an inactive precursor HA0, which must undergo post-translational cleavage to generate the HA1 and HA2 subunits. To date, sixteen different HA (HA1 to HA16) and nine NA (NA1 to NA9) subtypes have been identified, giving rise to an array of different viruses with different HA and NA combinations that have been isolated from a variety of avian species (Wright and Webster, 2007). New subtypes are continuously being reported, for example, the new H16N3 subtype was recently detected in black headed gulls in Sweden (Fouchier et al., 2005) and another new H5N7 subtype from Danish mallard ducks (Bragstad et al., 2005).
In the past 10 years, outbreaks of AIV infections in wild and domestic birds, as well as in other non-avian species, have been the focal point of heightened surveillance and research (see review by Capua and Alexander, 2007). In most AIV surveillance programs, particular emphasis has been placed on H5 and H7 subtypes since these subtypes have shown to be capable of undergoing mutation from low pathogenic avian influenza (LPAI) to high pathogenic avian influenza (HPAI). For example, outbreaks caused by HPAI H7 isolates were reported in Italy (Capua et al., 2003), Chile (Suarez et al., 2004), Canada (Pasick et al., 2005), British Columbia (Hirst et al., 2004) and in Netherlands (Fouchier et al., 2004). Emergence of HPAI from the LPAI H5N2 subtype in domesticated poultry was reported in North America (Bean et al., 1985, Kawaoka and Webster, 1985), and next in Mexico (Horimoto et al., 1995, Lee et al., 2005) and Europe (Donatelli et al., 2001). Although LPAI H5N2 have been reported in poultry in several Asian countries (Cheng et al., 2005, Okamatsu et al., 2007), there has been no reported cases of the emergence of HPAI H5N2 in South-East Asia.
Since 2005, the molecular criterion defining the pathogenicity of an AIV has been formally classified by the World Organisation for Animal Health (OIE) (World Organisation for Animal Health, 2008). This classification is based on the amino acid (aa) sequence of the cleavage site of the HA precursor protein, with the presence of multiple basic residues defining a particular AIV as HPAI. It is believed that LPAI H5 and H7 viruses are the ancestral viruses for HPAI isolates in poultry, but the mechanism for the emergence from LPAI to HPAI is currently unclear. It was reported that HPAI arose by the introduction of LPAI from wild birds into poultry (Garcia et al., 1997, Perdue et al., 1997). Other reports described the presence of mutations taking place gradually in LPAI which has been circulating in poultry for some time, before the virus finally emerges as HPAI (Horimoto et al., 1995, Lee et al., 2005). In the case of the LPAI strain Chicken/Pennsylvannia/1/83 isolated during the Pennsylvania outbreak in 1983, the conversion from LPAI to HPAI was attributed to the cleavability of the HA protein, which was influenced by the loss of an N-linked glycosylation site located near the vicinity of the HA cleavage site (Kawaoka et al., 1984, Webster et al., 1986, Deshpande et al., 1987). For the Chilean (Suarez et al., 2004) and Canadian H7N3 HPAI (Pasick et al., 2005), the authors hypothesized that the emergence to HPAI may be due to the recombination of the HA gene with its internal genes (Perdue et al., 1997, Alexander, 2006).
Between 1993 and 2007 four H5 influenza viruses were isolated from live broiler ducks imported into Singapore. Preliminary results from both the antigenic and partial sequence analyses of the HA and NA genes identified three of these as H5N2 subtypes, and one as the subtype H5N3. In addition, molecular analysis of the aa sequence of the HA cleavage site suggested that these were LPAI isolates, as defined by OIE criterion. In response to the recent reports that virus virulence is not solely related to the structure of the HA cleavage site (Kawaoka et al., 1984, Webster et al., 1986, Deshpande et al., 1987, Perdue et al., 1997, Alexander, 2006), we performed complete molecular characterization of all the eight genes for these four LPAI isolates. The objective was to detect the presence of known signatures that are associated with virulence and that could serve as predictors for the potential conversion of these viruses to HPAI. The sequence analyses in this study suggest that the four virus isolates contained mainly avian virulence signatures. However, the phylogenetic tree for the internal genes suggests the possibility of further reassortment with other different avian subtypes. This is the first comprehensive report on the genetic characterization of LPAI H5 subtypes isolated in South-East Asia.
2. Materials and methods
2.1. Viruses
The four virus isolates, named as A/duck/Singapore/F119/3/97 (isolated in 1997), A/duck/Malaysia/F59-04/98 (isolated in 1998), A/duck/Malaysia/F118/08/04 (isolated in 2004) and A/duck/Malaysia/F189/07/04 (isolated in 2004), were obtained from the Agri-Food and Veterinary Authority of Singapore (AVA). For this manuscript, the names of the isolates have been shortened and designated as F119, F59, F118 and F189, respectively. These viruses were isolated as part of the routine surveillance for AIVs in avian species imported from Malaysia, by direct inoculation of cloacal or tracheal swabs into 9–11-day special pathogen-free embryonated eggs. Viral isolates detected were sent to Weybridge, United Kingdom, for confirmation by subtyping with specific antisera to HA and NA.
2.2. RNA extraction and sequencing of the influenza virus genes
Viral RNAs (vRNAs) were extracted from infected allantoic fluid of embryonated chicken eggs, using the RNeasy mini kit (Qiagen Inc., Valencia, CA, USA). Reverse transcription (RT) of the vRNA was performed with SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen Corporation, CA, USA) according to manufacturer's instructions, using the Uni12 primer (5′ AGCRAAAGCAGG 3′) as described in Hoffmann et al. (2001). Each of the eight gene segments was PCR-amplified with 5 μl of the cDNA and 0.5 μM of gene-specific primers (Hoffmann et al., 2001) with the Expand Long Template PCR system (Roche Diagnostics, Mannheim, Germany), according to manufacturer's instructions. PCR reactions were subjected to an initial denaturation at 95 °C for 2 min and 30 cycles consisting of 94 °C for 15 s, 50 °C for 30 s, and 72 °C for 2 min. The final extension step was extended to 7 min at 72 °C. The HA and NA PCR-amplified fragments were purified using QIAquick Gel Extraction Kit (Qiagen Inc., Valencia, CA, USA), cloned into vectors pCR2.1-TOPO or pCR4-TOPO (Invitrogen Corporation, CA, USA) and sequenced with M13 reverse and forward primers. The remaining PCR-amplified PB1, PB2, PA, NP, M, and NS1 fragments were directly sequenced with gene-specific primers. The partial sequence information obtained from sequencing the 5′ and 3′ ends of these gene fragments were used to design new primers (Primer Select, DNASTAR, Lasergene Version 7) for sequencing the internal regions. All primers were synthesized by Proligo Singapore, and the details of primer sequences are shown in Table 1 . Sequencing was performed with the ABI Prism BigDye (Applied Biosystems, USA) in the presence of 1 μM of each respective sequencing primer. Each region of the PCR product and plasmids were sequenced at least twice.
Table 1.
Details of primer sequences used for PCR-amplifications and sequencing of the AIV genes.
| Primer names | Gene targets/virus subtypes | Sequence 5′ to 3′ | References |
|---|---|---|---|
| Primers for PCR amplification of the full-length gene fragments | |||
| Ba-PB2-1F | PB2/all | TATTGGTCTCAGGGAGCGAAAGCAGGTC | Hoffmann et al. (2001) |
| Ba-PB2-2341R | ATATGGTCTCGTATTAGTAGAAACAAGGTCGTTT | ||
| Bm-PB-1F | PB1/all | TATTCGTCTCAGGGAGCGAAAGCAGGCA | |
| Bm-PB1-2341R | ATATCGTCTCGTATTAGTAGAAACAAGGCATTT | ||
| Bm-PA-1F | PA/all | TATTCGTCTCAGGGAGCGAAAGCAGGTAC | |
| Bm-PA-2233R | ATATCGTCTCGTATTAGTAGAAACAAGGTACTT | ||
| Bm-HA-1F | HA/all | TATTCGTCTCAGGGAGCAAAAGCAGGGG | |
| Bm-NS-890R | ATATCGTCTCGTATTAGTAGAAACAAGGGTGTTTT | ||
| Bm-NP-1F | NP/all | TATTCGTCTCAGGGAGCAAAAGCAGGGTA | |
| Bm-NP1565R | ATATCGTCTCGTATTAGTAGAAACAAGGGTATTTTT | ||
| Ba-NA-1F | NA/N1 and N2 | TATTGGTCTCAGGGAGCAAAAGCAGGAGT | |
| Ba-NA-1413R | ATATGGTCTCGTATTAGTAGAAACAAGGAGTTTTTT | ||
| Bm-N3-1F | NA/N3 | TATTCGTCTCAGGGAGCAAAAGCAGGTGC | |
| Bm-N3-1420R | ATATCGTCTCGTATTAGTAGAAACAAGGTGCTTTTT | ||
| Bm-M-1 | M/all | TATTCGTCTCAGGGAGCGAAAGCAGGTAG | |
| Bm-M-1027R | ATACGTCTCGTATTAGTAGAAACAAGGTAGTTTTT | ||
| Bm-NS1 | NS/all | TATTCGTCTCAGGGAGCGAAAGCAGGGTG | |
| Bm-NS-890R | ATATCGTCTCGTATTAGTAGAAACAAGGGTGTTTT | ||
| Primers for sequencing | |||
| H5N2_PB2_225F | PB2/H5N2 | TCCCTGAAAGGAATGAGCAAGGC | In-house |
| H5N2-PB2-1020R | GAGCTGATTCTTAGACCCATTG | ||
| H5N2_PB2_1370F | GGGGAATTGAACCCATCGACAA | ||
| H5N2-PB2-1480R | CTCTCTCAGTACTGGAATATTCATC | ||
| H5N2-PB2-1800R | TTGGCCTCGGGCAGCCTTAG | ||
| H5N2-PB1-580F | PB1/H5N2 | GGAGAGTAAGGGACAACATGACC | |
| H5N2-PB1-1850R | GTCTACCCTGGTAATCTTCATCC | ||
| N3-PB1-740R | PB1/H5N2 and 3 | CATTCCAGGTGTTGCAATTGCCC | |
| N3-PB1-1550R | ATCCCAGATACTCCGAAGCTGGG | ||
| H5N2-PA-475F | PA/H5N2 | ACCGGAGAGGAAATGGCCACC | |
| H5N2-PA-1800R | GGACTCAGCTTCAATCATGCTCTC | ||
| N3-PA-620R | PA/H5N2 and 3 | TCTCTTCGCCTCTCTCGGACTGA | |
| N3-PA-1280R | CCGTCAATTCGCATGCCTTGTTG | ||
| HA5-896F | HA/H5 | ACTCCAATGGGGGCGATAAAC | Poddar (2002) |
| FluN2-779F | NA/N2 | GGAAATCGTTCATATTAGCCCATTG | |
| N3_internalF | NA/H5N3 | AGATATGTGTTGCTTGGTCAAGT | In-house |
| H5N2-NP-320F | NP/H5N2 | CTGGAGGTCCAATTTACCGAAGG | |
Nt position for each primer designed in-house is relative to the gene target from H5N2 subtype (A/duck/Malaysia/F118/08/04) or H5N3 subtype (A/duck/Singapore/F119/3/97), and the accession numbers are described in Section 2. F denotes primer in the forward or positive strand, and R denotes primer in the reverse or negative strand of each gene.
2.3. Sequence and phylogenetic analyses
Sequence information was compiled using Seqman (DNASTAR, Lasergene Version 7) and the sequences were deposited into GenBank. The accession numbers are as follows: DQ122147, DQ104701–DQ104703, DQ092869–DQ092870, EU249545–EU249550, and EU271936–EU271953. Nucleotide and translated amino acid (aa) sequences for each gene segment were compared between the four virus isolates and other published sequences from GenBank (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) with Megalign (DNASTAR, Lasergene Version 7) using the Clustal W algorithm. Percent (%) sequence homology was calculated for each of the full-length gene. Phylogenetic tree was constructed using the neighbour-joining method at the nt level, with bootstrap analysis performed on 1000 replicates. Phylogenetic trees were viewed with TreeExplorer (v2.12, http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html).
2.4. 3D modelling
Alignment of the aa sequences was performed using MUSCLE (Edgar, 2004). Three-dimensional (3D) model of the F118-HA protein was generated by Modeller (Sali and Blundell, 1993), using the coordinates from the crystal structures acquired from the H3N2 isolate A/Aichi/2/1968 [Protein Data Bank (PDB) entry 1HA0] and H5N3 isolate A/Duck/Singapore/3/97 (PDB entry 1JSM) as the templates. The sequence identity between the HA1 subunit of F118 and A/Aichi/2/1968, and that between F118 and A/Duck/Singapore/3/97 are 44% and 94%, respectively. The models are visualized using PyMOL (http://www.pymol.or).
3. Results
3.1. Surveillance on AIV in Singapore
The Agri-Food and Veterinary Authority (AVA) conducts a routine national surveillance program for the detection of AIVs in live poultry imported into Singapore. Although between 1993 and 2007 a total of 50 LPAI viruses of different subtypes were isolated in Singapore, no HPAI viruses have been isolated (Lin, 2007; unpublished results). These LPAI viruses were isolated from a variety of free-living and captive birds, and domestic poultry, confirming the findings from previous reports which described the presence of AIV infections in a wide range of avian hosts (Hindshaw et al., 1981, Alexander, 2000, Alexander, 2001, Capua and Alexander, 2007).
In this study we have examined four of these isolates in more detail, which included three H5N2 isolates (F118, F119 and F189) and one H5N3 isolate (F59). In the case of F118 and F189 isolates, these AIVs were isolated during the heightened surveillance for the AIV subtype H5N1 in year 2004. Both isolates tested positive for preliminary H5 antigen testing, using the Directigen™ Flu A test kit (Diagnostics Systems, Becton, Dickinson and Company, USA). PCR testings and DNA sequencing on partial HA and NA genes (Yuen et al., 1998, Hoffmann et al., 2001, Spackman et al., 2002) identified both virus isolates to be of the same subtype, H5N2 (results not shown). These results were verified using antigenic neutralization assays from Weybridge, United Kingdom. We next proceeded on to perform complete sequence analysis for both the structural and internal genes for the two isolates. In order to track the genetic changes over time, we also sequenced the complete genomes of two other LPAIVs, F119 (isolated in 1997) and F59 (isolated in 1998), isolated from poultry imported from the same geographical region, in this case Malaysia.
3.2. Sequence and phylogenetic analyses of the HA genes
The classification of an AIV as being either LPAI or HPAI is defined by the sequence of the HA cleavage site (World Organisation for Animal Health, 2008). Therefore, the full-length HA genes for each of the four viral isolates were analysed, and compared with each other and that of other published AIV sequences. We observed that both the F118 and F189 showed a high degree of sequence identity (at both the nt and aa levels) with an Italian H5N3 strain (Table 2 ). Both F119 and F59 were more closely related to a Japanese subtype H5N3 (at the nt level), and to the Netherland subtype H5N2 (at the aa level) (Table 2). The structure of the HA cleavage site in virulent H5N2 subtypes contains approximately four basic amino acid residues, for example the RRRK and RKRK motifs represented by strains A/chicken/Italy/312/1997 and A/chicken/Queretaro/14588-19/1995. The structure of the HA cleavage site in all four isolates was RETR (Fig. 1 ), which is a motif observed in several LPAI isolates, which was consistent with all four viruses being LPAI H5 subtypes.
Table 2.
Nucleotide and translated amino acid comparisons with the most similar isolats from Genbank.
| Avian genes | Duck/Malaysia/F118/08/04 H5N2, Duck/Malaysia/F189/08/04 H5N2 |
Duck/Malaysia/F59-04/98 H5N2 |
Duck/Singapore/F119/3/97 H5N3 |
|||
|---|---|---|---|---|---|---|
| Isolate | Max identity | Isolate | Max identity | Isolate | Max identity | |
| PB2 | Mallard/Gurjev/244/1982 H14 (Duck/Hokkaido/Vac-3/2007 H5N1) | 93% (98%) | Chicken/Iran/11T/1999 H9N2 (Duck/HK/147/1977 H9N6) | 95% (98%) | Turkey/Italy/4169/1999 H7N1 (Duck/Nanchang/1904/1992 H7N1) | 96% (99%) |
| PB1 | Duck/Nanchang/4-165/2000 H4N6 (Environment/Delaware/232/2005 H11N8) | 95% (98%) | Duck/Nanchang/1941/1993 H4N4 (Mallard/Postdam/178-4/83 H2N2) | 96% (98%) | Duck/Nanchang/1941/1993 H4N4 (Pintail/Ohio/73/1989 H6N2) | 96% (98%) |
| PA | Swan/Hokkaido/51/1996 H5N3 (Turkey/Italy/4169/1999 H7N1) | 96% (99%) | Wild duck/Shantou/19988/2000 H4N9 (Mallard/Netherlands/12/2000 H7N3) | 96% (99%) | Wild duck/Shantou/19988/2000 H4N9 (Duck/Denmark/65047/04 H5N2) | 97% (99%) |
| HA | Mallard/Italy/208/2000 H5N3 (Mallard/Italy/208/2000 H5N3) | 94% (96%) | Swan/Hokkaido/51/1996 H5N3 (Mallard/Netherlands/3/99 H5N2) | 96% (97%) | Swan/Hokkaido/51/96 H5N3 (Mallard/Netherlands/3/99 H5N2) | 98% (98%) |
| NP | Mallard/Xuyi/8/2004 H11N (Duck/Nanchang/1904/1992 H7N1) | 96% (99%) | Poultry/Italy/373/1997 H5N2, Poultry/Italy/330/1997 H5N2, Chicken/Italy/312/1997 H5N2 (Duck/Hong Kong/3461/99 H6N1) | 95% (97%) | Duck/Singapore/645/97 H5N3 (Duck/England/1/1956 H11N6) | 99% (99%) |
| NA | Duck/Mongolia/54/2001 H5N2 (Duck/Eastern China/848/2003 H3N2) | 97% (97%) | Duck/Nanchang/1749/1992 H11N2 (Duck/Denmark/65047/04 H5N2) | 94% (96%) | Mallard/Italy/208/2000 H5N3 (Mallard/Italy/208/00 H5N3) | 95% (96%) |
| M | Duck/Jiang Xi/6146/2003 H5N3 (M1: Chicken/Korea/S6/2003 H3N2) (M2: A/swan/Guangxi/307/2004 H5N1) | 97% (100%) (100%) | Duck/Hokkaido/95/2001 H2N2 (M1: Duck/Germany/113/95 H9N2) (M2: Chicken/Singapore/98 H5N2) | 97% (100%) (100%) | Duck/Hokkaido/120/2001 H6N2 (M1: Duck/Germany/113/95 H9N2) (M2: Chicken/Singapore/1997 H5N3) | 98% (100%) (100%) |
| NS | Duck/Taiwan/WB459/2004 H6N5 (NS1: Duck/NY/14933/95 H6N8) (NS2: duck/Hong Kong/278/1978 H2N9) | 96% (96%) (96%) | Duck/Hong Kong/610/79 H9N2 (NS1: Duck/NY/14933/95 H6N8) (NS2: Chicken/Korea/LPM88/2006 H3N2) | 96% (98%) (100%) | Duck/Hong Kong/610/79 H9N2 (NS1: Duck/NY/14933/95 H6N8) (NS2: Herring gull/DE/677/1988 H2N8) | 97% (98%) (100%) |
The percent maximum identities at the aa level are indicated within brackets.
Fig. 1.
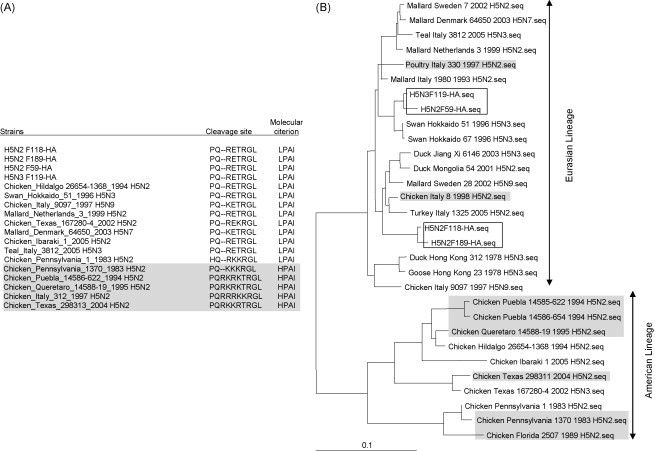
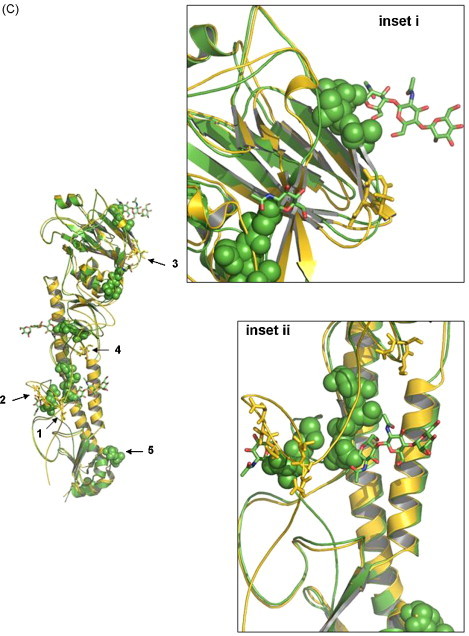
(A) Comparison of the HA cleavage site of various low and high pathogenic H5 influenza A viruses. The four low pathogenic AIV isolates that we have sequenced in this study are highlighted in bold and various HPAI H5N2 are highlighted in italics. (B) Phylogenetic trees of the full-length HA genes based on nt sequence alignment. The four LPAI H5 isolates F118, F189, F59 and F119 are boxed, while high pathogenic influenza isolates are highlighted in grey. (C) Overall structure of F118 monomer. The 3D models of F118 (H5) and 1HA0 (H3) were superimposed. Gold, F118; Green, 1HA0. The potential glycosylation sites postulated for F118 are shown in sticks and highlighted for regions 1–5, representing amino acids located at position 26 (NNST), 39 (NVT), 181 (NNT), 302 (NSS), and 496 (NGT) on the F118 HA model. Region 5 is overlapping with, and covered by aa residues from 1HA0. Amino acid located at 555 is not shown as the region has not been crystallized for the template we chose for the modelling. Regions 1–4 are found in the HA1, and region 5, in the HA2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Phylogenetic analysis of the full-length HA genes from the new LPAI were performed to examine the evolutionary relationship with several H5 AIV isolates. As shown in Fig. 1B, the phylogenetic tree of the HA gene is distinctly split into Eurasian and American lineages, and was consistent with previous reports (Lee et al., 2004a, Lee et al., 2005, Okamatsu et al., 2007, Pillai et al., 2008). All the new isolates characterized in this study were grouped under the Eurasian lineage, which comprises both LPAI and HPAI H5N2 isolates from Europe and other Asian countries, and other H5 subtypes (e.g. H5N9 and H5N7). The American lineage on the other hand, comprises the contemporary LPAI/HPAI H5N2 and H5N3 subtypes from countries in America. The F119 and F59 clustered with the Swan/Hokkaido H5N3 subtypes and other H5 subtypes from Europe, whereas the isolates F189 and F118 grouped by themselves. Our findings suggest that there is an active exchange of avian genes among the different geographical areas of the Eurasian region occurs, which is consistent with recent observations (Duan et al., 2007).
3.3. Potential glycosylation sites for HA
The glycosylation pattern on influenza viral HA protein has been shown to have three biological functions (reviewed by Reading et al., 2007), which include the maintenance and stability of the protein structure for virus assembly (Wilson et al., 1981), recognition of antigens (Wiley et al., 1981) and to protect the antigenic epitopes on the globular head against proteolytic degradation (Nakamura and Compans, 1978). The potential glycosylation sites in the HA proteins of the newly isolated AIVs were identified at six location, namely N26, N39, N181, N302, N496 and N555. We noted that these six sites were different from the contemporary Mexican (Garcia et al., 1997) and American H5N2 viruses (Lee et al., 2004a), and that the potential glycosylation site at N26 in the new AIVs was in close proximity to the HA cleavage site. In the H5N2 strain Chicken/Pennsylvannia/1/83, isolated in the Pennsylvania outbreak in 1983, virulence was contributed by the presence of site-specific glycosylation near the HA cleavage site, with additional influence from its internal genes (Kawaoka et al., 1984, Webster et al., 1986, Deshpande et al., 1987). Molecular modelling was next employed to investigate the possibility of steric hindrance offered by the potential glycosylation at N26 for the new AIVs. Structural models of the HA protein were generated using the coordinates from the H3N2 isolate A/Aichi/2/1968 and the H5N3 isolate A/Duck/Singapore/3/97 as templates (Fig. 1C). The H3N2 isolate A/Aichi/2/1968 was the only high resolution HA protein structure with an intact cleavage site, that is, at the precursor HA0 (Chen et al., 1998), while the H5N3 isolate A/Duck/Singapore/3/97 was the first available crystal structure for the H5 subtype (Ha et al., 2001). Due to the similarity of the HA protein model for all four AIVs examined, only the structural model for the F118-HA protein was shown (Fig. 1C). We observed that the potential glycosylation sites at N26 was the nearest thus far to the HA cleavage loop (Fig. 1C (inset ii)). This analysis suggests that position N26 is too remote to offer any steric influence for proteolytic processing of the HA cleavage site. Many HPAI isolates possess an additional glycosylation site on the globular head of the HA (Bender et al., 1999). For example, the presence of glycosylation at N158 for the chicken isolate of H5N1 decreased the binding affinity of the virus to soluble sialyglycoproteins in in vitro (Matrosovich et al., 1999). The potential glycosylation at position N181 of the HA protein model for the new AIVs (indicated by the sphere) is within the proximity of the receptor binding region (Fig. 1C (inset i)). This suggests that the glycosylation may possibly assert some influence on the virus binding. However, this possibility needs to be confirmed experimentally as both the Italian LPAI and HPAI H5N2s contained a potential glycosylation site at N165, which is located on the HA globular heads (Donatelli et al., 2001).
3.4. Sequence and phylogenetic analyses of the NA2 and NA3 genes
The full-length NA gene sequences from the new AIVs were next analysed, and a close examination of the aa sequence revealed that all four isolates did not contain the stalk deletion that was reported to be associated with some of the H5N2 HPAI American isolates in chickens (data not shown). In virulent H5N1 isolates, this feature was reported to be associated with virus adaptation to the chicken host (Matrosovich et al., 1999, Li et al., 2004). The absence of the stalk region is consistent with the isolation of the new H5 viruses from ducks. The phylogenetic analyses further showed that the H5N2 isolates F118 and F189 clustered with an assortment of N2 subtypes isolated from ducks, subtypes H6N2 and H3N2 from Eastern China and subtype H5N2 from Mongolia (Fig. 2A). The F59 isolated about more than 8 years ago, surprisingly clustered with a chicken and swine H9N2 from Korea. The F119 clustered with an assortment of N3 subtypes such as the H9N3 from Ireland, H2N3 from Potsdam, and H7N3 from Pakistan; all isolated in the pre-2000 years (1980s to the 1998s) (Fig. 2B). The results from this analysis confirmed that the LPAIs studied were of the subtype H5N2 (F118, F189 and F119) and H5N3, respectively (F59).
Fig. 2.
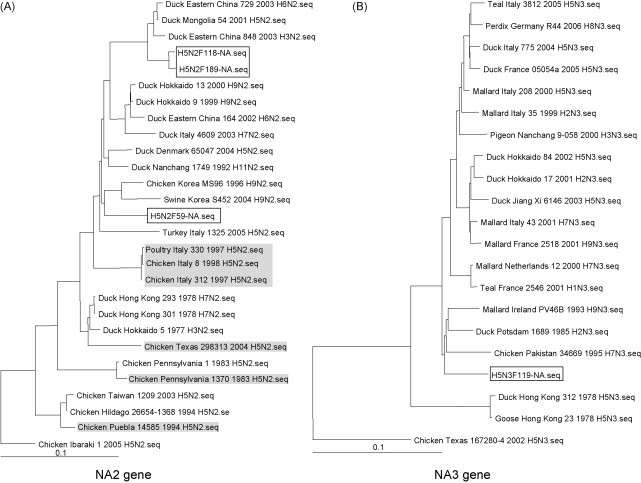
(A) Comparison of the neuraminidase of various N2 subtypes. A typical 20 aa deletion is observed in the stalk region for chicken isolates, although the A/chicken/Texas/298313/04 (H5N2) isolate has a distinct 13 aa deletion slightly upstream from the rest. (B) Phylogenetic trees of the full-length NA genes based on nt sequence alignment. The four LPAI H5 isolates F118, F189, F59 and F119 are boxed while high pathogenic influenza isolates are highlighted in grey.
3.5. Sequence and phylogenetic analyses of the internal genes
It was reported that the proteins encoded by the internal genes can also play a significant role in AIV pathogenicity. For example, the increased virulence in the H7N3 HPAI outbreak in 2004 in Canada was attributed to the insertion of seven amino acid sequence from the M gene to the HA cleavage site (Hirst et al., 2004). It is therefore important to sequence the internal genes of AIV isolates to identify mutations or recombinations that may have implications for virus transmission and pathogenicity. The full-length sequences of the six segments that give rise to the internal virus proteins were compared with various AIV subtypes from different geographical origins. The sequence analyses showed that the sequence of these segments were closely related to the corresponding segments from a variety of AIV subtypes isolated from different avian hosts, including mallards and ducks (Table 2). The PB2 gene showed the lowest nt sequence identity (93%) whereas the M gene had the highest nt sequence identity (97–98%). The aa sequence identity was 100% for the M1 and M2, and NS2 proteins with assorted Genbank isolates, suggesting that the two genes faced the least evolutionary constraints. The five aa deletion at positions 80–84 of the NS1 protein reported in several HPAI H5N1 isolates since 2001 (Viseshakul et al., 2004), was not observed in any of the NS1 genes for F118, F189, F59 and F 119, nor any of the contemporary H5N2/3 subtypes (results not shown).
To examine the evolutionary relationship with existing AIVs we performed phylogenetic analysis for each of the genes that encode the internal proteins with that in selected LPAI and HPAI AIVs (Fig. 3 ). Most of the genes in the four AIVs in this study did not cluster with any of the Eurasian H5N2 or H5N3 viruses. For PB2, PB1, PA and NS1 genes, the AIVs clustered into two groups, the F118 and F189 in one group, and the F59 and F119 in a second group (Fig. 3A and B). The PB2 genes for the F118 and F189 clustered with one HPAI H5N2 and one LPAI H5N2 from Mexico (isolated before the use of H5N2 vaccine). This PB2 gene cluster contained European H7 subtypes, including one H9N2 from Germany, and one HPAI H5N1 from China. Interestingly, the PB1 gene clustered with more HPAI H5N1, which was mainly avian in origin although there was at least one swine H5N1 from China (Fig. 3B). The tree for the PA gene showed that the two AIVs again fall into two sub-groups, similar to that observed for the PB2 and PB1 genes, but in this case they clustered away from all the H5N2 and H5N3, including the Mexican H5N2s (Fig. 3C). The PA genes for the new AIVs clustered with Chinese H5N1 isolates, including those of swine and avian origin, and other subtypes. The NP gene for F59 clustered closely with both swine and chicken H9N2 isolated from China and a chicken H5N1 from China (Fig. 3D), although, the NP genes from F118, F189, and F119 were closely clustered (Fig. 3D). The Italian HPAI H5N2 viruses were also found in this cluster. The M and NS1 genes for H5N2 F118 and H5N2 F189 clustered with an assortment of subtypes from Eurasia isolated after 2000 (Fig. 3E and F). Our phylogenetic findings suggest that the internal genes for the AIVs isolated in South-East Asia in this study share a common ancestry with avian viruses of different subtypes and from different geographical locations. The data further confirmed what was reported previously (Duan et al., 2007), that there is a dynamic and continuous reassortment of different gene pools within the Eurasia region.
Fig. 3.

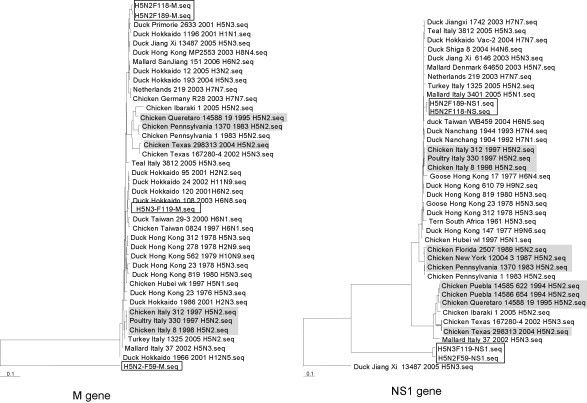
Phylogenetic trees of full-length genes for (A) PB2, (B) PB1, (C) PA, (D) NP, (E) M, and (F) (NS), based on their nt sequence alignment. The four LPAI H5 isolates F118, F189, F59 and F119 are boxed while high pathogenic influenza isolates are highlighted in grey.
3.6. Analysis of virus signatures
Past pandemic viruses of 1957 (H2N2) and 1968 (H3N2) were believed to have arisen by reassorted viruses of human and avian origin (Scholtissek et al., 1978, Kawaoka et al., 1989). In the case of the 1918 pandemic virus, sequence and phylogenetic analyses suggested that the virus adapted entirely from an ancestral avian virus (Taubenberger et al., 2005). These evidences suggest that tracking new isolates for specific signatures at the genetic level may be a way to predict circulating viruses that have pandemic potential. In this study, the predicted aa sequences of all the virus proteins were analysed for known viral signature sequences that have been described previously (Okazaki et al., 1989, Gorman et al., 1990, Shaw et al., 2002, Taubenberger et al., 2005, Chen et al., 2006). As shown in Table 3 , the presence of several avian signature sequences was detected in all the AIV isolates. However, one novel mutation (N/S/T)375D was found in the PB1 proteins of F118 and F189, while another novel mutation L107Q was present in the NS2 protein of the F189 isolate. In the case of F59 and F119 isolates, one novel mutation V100I was observed in their PA protein, with an additional mutation R31G in the NP gene for F59. These novel mutations have not been previously associated with either human-, swine- or equine-hosts. We next examined the isolates for the presence of human-associated signature. Although these were absent in the F118 and F189 isolates, we observed two human-associated mutations, T121A for the M1 gene and L55F for the M2 gene, in F59 and F119. The F119 isolate has two additional human-associated mutations, A674T in PB2 and S20N in the M2 gene. A survey of the published literature did not yield any information on the significance of these four human-associated signatures. Although not associated with any known virulence, the M gene was examined for the presence of amantadine resistance. The anti-viral drug amantadine inhibits influenza A virus replication by blocking the ion channel activity of the M2 protein (Pinto et al., 1992). Our analysis revealed that the M2 protein sequence in F118, F189, F59 and F119-M2 proteins did not contain mutations that are associated with drug resistance (Hay et al., 1985, Sugrue and Hay, 1991) (Table 3).
Table 3.
Host-associated aa signatures.
| Protein | Position | Avian | Human | F118 | F189 | F59 | F119 | References |
|---|---|---|---|---|---|---|---|---|
| PB2 | 44 | A | S | A | A | A | A | Shaw et al. (2002), Taubenberger et al. (2005) and Chen et al. (2006) |
| 81 | T | M | T | T | T | T | ||
| 199 | A | S | A | A | A | A | ||
| 271 | T | A | T | T | T | T | ||
| 475 | L | M | L | L | L | L | ||
| 567 | D | N | D | D | D | D | ||
| 588 | A | I | A | A | A | A | ||
| 613 | V | T | V | V | V | V | ||
| 627 | E | K | E | E | E | E | ||
| 661 | A | T | A | A | A | A | ||
| 674 | A | T | A | A | T | A | ||
| 702 | K | R | K | K | K | K | ||
| PB1 | 327 | R | K | R | R | R | Taubenberger et al. (2005) and Chen et al. (2006) | |
| 336 | V | I | V | V | V | V | ||
| 375 | N/S/T | S | N | N | ||||
| PA | 28 | P | L | P | P | P | P | Okazaki et al. (1989), Shaw et al. (2002), Taubenberger et al. (2005) and Chen et al. (2006) |
| 55 | D | N | D | D | D | D | ||
| 57 | R | Q | R | R | R | R | ||
| 65 | S | L | S | S | S | S | ||
| 100 | V | A | V | V | ||||
| 225 | S | C | S | S | S | S | ||
| 241 | C | Y | C | C | C | C | ||
| 268 | L | I | L | L | L | L | ||
| 312 | K | R | K | K | K | K | ||
| 356 | K | R | K | K | K | K | ||
| 382 | E | D | E | E | E | E | ||
| 400 | Q/T/S | L | S | S | S | S | ||
| 404 | A | S | A | A | A | A | ||
| 409 | S | N | S | S | S | S | ||
| 552 | T | S | T | T | T | T | ||
| HA | 237 | N | R | N | N | N | N | Chen et al. (2006) |
| 389 | D | N | D | D | D | D | ||
| NP | 16 | G | D | G | G | G | G | Gorman et al. (1990), Shaw et al. (2002) and Chen et al. (2006). |
| 31 | R | K | R | R | R | |||
| 33 | V | I | V | V | V | V | ||
| 61 | I | L | I | I | I | I | ||
| 100 | R | V | R | R | R | R | ||
| 109 | I | V | I | I | I | I | ||
| 127 | E | D | E | E | E | E | ||
| 136 | L | M | L | L | L | L | ||
| 214 | R | K | R | R | R | R | ||
| 283 | L | P | L | L | L | L | ||
| 293 | R | K | R | R | R | R | ||
| 305 | R | K | R | R | R | R | ||
| 313 | F | Y | F | F | F | F | ||
| 357 | Q | K | Q | Q | Q | Q | ||
| 372 | E | D | E | E | E | E | ||
| 375 | D | G/E | D | D | D | D | ||
| 422 | R | K | R | R | R | R | ||
| 442 | T | A | T | T | T | T | ||
| 455 | D | E | D | D | D | D | ||
| M1 | 115 | V | I | V | V | V | V | Shaw et al. (2002) and Chen et al. (2006) |
| 121 | T | A | T | T | A | A | ||
| 137 | T | A | T | T | T | T | ||
| M2 | 11 | T | I | T | T | Shaw et al. (2002) and Chen et al. (2006) | ||
| 16 | E | G | E | E | E | E | ||
| 20 | S | N | S | S | N | S | ||
| 55 | L | F | L | L | F | F | ||
| 57 | Y | H | Y | Y | Y | |||
| 78 | Q | K | Q | Q | ||||
| 86 | V | A | V | V | V | V | ||
| NS1 | 227 | E | R | E | E | E | E | Chen et al. (2006) |
| NS2 | 70 | S | G | S | S | S | S | Chen et al. (2006) |
| 107 | L | F | L | L | L | |||
Residues that are novel are bold and highlighted in grey; residues that are associated with human host are boxed.
4. Discussion
During the epidemic with the severe acute respiratory syndrome (SARS), Singapore was able to rally her resources rapidly and contain the disease within 3 months (Tan et al., 2005). The lessons from the SARS crisis clearly highlighted the potential for human carriers to bring new and emerging pathogens into Singapore. Singapore is a busy global trading city and has not been spared from the presence of infectious diseases in the past. For example, the 1918 influenza pandemic resulted in an estimation of 3500 deaths (Lee et al., 2007). Currently the mortality rate caused by the seasonal influenza infection has been estimated to be at 588 per year (Chow et al., 2006). The introduction of new human influenza virus subtypes into Singapore will result in a large number of people being at risk from the infection. An active surveillance is urgently required to build up the influenza sequence database in the healthcare sector. This should help to mitigate the risk of a potential pandemic by active monitoring of genetic changes in influenza strains circulating in the human population.
The importation of food products is another potential route through which foreign pathogens can enter Singapore. Singapore imports much of the food products from neighbouring countries, including live animals for slaughter. The introduction of foreign zoonotic pathogens with a disease potential would potentially have serious consequences in a small densely populated country like Singapore. Therefore a routine surveillance program on different forms of imported food products has been implemented by the Agri-Food and Veterinary Authority of Singapore (http://www.ava.gov.sg/Legislation/ListOfLegislation/). In the year 2004, two AIVs of the subtype H5N2 were isolated from live poultry imported from Malaysia and molecular typing confirmed that the newly isolated AIVs (F118 and F189) were LPAI. The cleavage site in the HA0 protein contained the aa sequence observed in other LPAI H5N2 viruses. As the acquisition of virulence, inter-species transmission and pathogenicity is a polygenic process, we therefore examined the full genome sequence of the new AIVs. The whole genomes from two other AIVs, F59 and F119, isolated from the same geographical area were also investigated together. At the genetic level, the sequence analyses and phylogenetic trees showed that for the internal genes, the four AIVs clustered with an assortment of avian subtypes, away from the contemporary H5N2 and H5N3 viruses, but sharing an avian ancestry (Fig. 3). Vaccination has been used in the control of AIV in several countries, and the appearance of several H5N2 sublineages has been observed in countries where vaccination with the H5N2 strain is widely practiced (Lee et al., 2004b, Duan et al., 2007). This is in contrast to the successful vaccination strategy practiced in Italy, where a bivalent H5/H7 with different NA protein was implemented as a prophylactic vaccine (Capua and Marangon, 2007). Currently vaccination has not been practiced in Malaysian poultry farms (Lim, 2008; unpublished observations), suggesting that the four AIVs could have arisen from spontaneous reassortments with different circulating avian or swine virus subtypes. Further genetic analysis with available influenza sequence data acquired from the surveillance of different hosts from the same geographical area may thus provide answers to the exact ancestral origin of these and other circulating AIVs. Another interesting finding from the phylogenetic trees indicates that the new AIVs tend to cluster with an assortment of AIV subtypes from China. Wild birds have been identified as a reservoir to harbour both LPAI and HPAI of H5N2 viruses, suggesting a possible role for migratory birds in the dissemination of viruses from North-East to South-East Asia, although this possibility requires further examination. For example, a recent evidence for HPAI H5N2 infection in healthy wild waterfowl, suggesting for the first time that the LPAI H5N2 maybe converted to HPAI and subsequently maintained in healthy feral avian species (Gaidet et al., 2008).
The NS1 protein has been associated with virus virulence (Noah and Krug, 2005), and the NS1 proteins for the four AIVs were examined. The NS1 protein has multiple functions, and one of the functions is to act as an antagonist for the host interferon defence system in various ways (see review by Hale et al., 2008). A recent large-scale sequence analysis for the AIV-NS1 identified a PDZ domain ligand (PL) at the C-terminus of the avian NS1 protein (Obenauer et al., 2006). PDZ domains have been identified to play important roles in influencing various cellular signalling pathways (Sheng and Sala, 2001). Human viruses with avian PL motifs have been found to be highly pathogenic, while viruses with human PL motifs caused low pathogenic infections in humans (Obenauer et al., 2006), suggesting that these PL motifs are associated with virus virulence. As expected, the F118, F189, F59 and F119 NS1 proteins contained a PL sequence motif of ESEV between aa residues at positions 227–230, which is characteristic of avian viruses. At this point, it is unlikely that these AIVs will recombine with a human virus as the current sequence analysis has suggested the presence of avian signatures only. Another association with virulence in the NS1 protein is the presence of glutamic acid at position 92, and this is observed in most HPAI H5N1. A detailed study by Seo and colleagues using site-directed mutagenesis and reverse genetics demonstrated that the presence of E92 increased the resistance of AIV to the anti-viral effects of interferon (Seo et al., 2004). For example, during the HPAI H5N1 replication in human host, the AIV is believed to induce cytokine imbalance hence increasing the severity of the infection (Cheung et al., 2002). However, such an aa association with LPAI has not been made and in the current AIVs studied aspartate was present.
Another internal protein that has been associated with virulence and host tropism is the PB2 protein (Noah and Krug, 2005, Gabriel et al., 2008, Tarendeau et al., 2008), a subunit of the viral RNA polymerase. Reports have described that the substitution from E627 to K627 has been associated with increased viral replication in mammalian species (Subbarao et al., 1993). K627 is present in most HPAI H5N1, and Shinya and colleagues were able to use reverse genetics to demonstrate the increased viral replication in murine model (Shinya et al., 2004). K627 is also observed to be present in the fatal human isolate H7N7 2003 (Fouchier et al., 2004). However, K627 is not observed in any of the non-H5N1 isolates, nor the local isolates described in this study. We observed one human signature, A674T, in the F119-PB2. So far, there are no published reports associating this substitution with specific biological activities (Shaw et al., 2002, Taubenberger et al., 2005, Chen et al., 2006).
5. Conclusions
In this study, we have developed molecular strategies to rapidly characterise the HA cleavage site sequence, as well as all genes encoding the structural and internal proteins of avian influenza A virus. Our results specifically identified the three AIVs, F189, F118, and F59, as LPAI subtype H5N2, and F119 as LPAI subtype H5N3. Past reports indicate that it may be difficult to predict when a LPAI may convert to a HPAI, thus far all four AIVs showed no unique signatures that may suggest a possible mutation to HPAI. The sequence and phylogenetic data collected in this study will be useful in the epidemiological studies of the dynamics and evolution of circulating avian influenza viruses, in particular in this region of South-East Asia. A continuous surveillance with the full genome characterization of newly circulating AIVs should be strongly encouraged, especially when current evidence suggests that in addition to the structure of the HA cleavage site, genetic changes in virus genes encoding other virus proteins may play a role in AIV virulence in humans.
Acknowledgement
This project was funded by the Defence and Technology Agency of Singapore.
References
- Alexander D.J. A review of avian influenza in different bird species. Vet. Microbiol. 2000;74:3–13. doi: 10.1016/s0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- Alexander D.J. Proceedings of the International Symposium on Emergence and Control of Zoonotic Ortho—An Paramyxovirus Diseases. 2001. Ecology of avian influenza in domestic birds; pp. 25–34. [Google Scholar]
- Alexander D.J. Avian influenza viruses and human health. Dev. Biol. 2006;124:77–84. [PubMed] [Google Scholar]
- Bean W.J., Kawaoka Y., Wood J.M., Pearson J.E., Webster R.G. Characterization of virulent and avirulent A/Chicken/Pennsylvania/83 influenza A viruses: potential role of defective interfering RNAs in nature. J. Virol. 1985;54:151–160. doi: 10.1128/jvi.54.1.151-160.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C., Hall H., Huang J., Klimov A., Cox N., Hay A., Gregory V., Cameron K., Lim W., Subbarao K. Characterization of the surface proteins of influenza A (H5N1) viruses isolated from humans in 1997–1998. Virology. 1999;254:115–123. doi: 10.1006/viro.1998.9529. [DOI] [PubMed] [Google Scholar]
- Bragstad K., Jorgensen P.H., Handberg K.J., Mellergaard S., Corbet S., Fomsgaard A. New avian influenza A virus subtype combination H5N7 identified in Danish mallard ducks. Virus Res. 2005;109:181–190. doi: 10.1016/j.virusres.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Capua I., Marangon S., dalla Pozza M., Terregino C., Cattoli G. Avian Influenza in Italy 1997–2001. Avian Dis. 2003;47:839–843. doi: 10.1637/0005-2086-47.s3.839. [DOI] [PubMed] [Google Scholar]
- Capua I., Alexander D.J. Animal and human health implications of avian influenza infections. Biosci. Rep. 2007;27:359–372. doi: 10.1007/s10540-007-9057-9. [DOI] [PubMed] [Google Scholar]
- Capua I., Marangon S. The use of vaccination to combat multiple introductions of notifiable avian influenza viruses of the H5 and H7 subtypes between 2000 and 2006 in Italy. Vaccine. 2007;25:4987–4995. doi: 10.1016/j.vaccine.2007.01.113. [DOI] [PubMed] [Google Scholar]
- Chen J., Lee K.H., Steinhauer D.A., Stevens D.J., Skehel J.J., Wiley D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- Chen G.W., Chang S.C., Mok C.K., Lo Y.L., Kung Y.N., Huang J.H., Shih Y.H., Wang J.Y., Chang C., Chen C.J., Shih S.R. Genomic signatures of human versus avian influenza A viruses. Emerg. Infect. Dis. 2006;12:1353–1360. doi: 10.3201/eid1209.060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.C., Lee M.S., Wang C.H. Molecular characterisation of H5N2 low pathogenic avian influenza viruses from Taiwan poultry in 2004. Proceedings of the APEC Conference on Avian Influenza; National Chung Hsing University, Taichung, Taiwan; 2005. p. 30. [Google Scholar]
- Cheung C.Y., Poon L.L., Lau A.S., Luk W., Lau Y.L., Shortridge K.F., Gordon S., Guan S., Peiris J.S. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1801–1802. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- Chow A., Ma S., Ling A.E., Chew S.K. Influenza-associated deaths in tropical Singapore. Emerg. Dis. Infect. 2006;12:114–121. doi: 10.3201/eid1201.050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande K., Fried V.A., Ando M., Webster R.G. Glycosylation affects cleavage of an H5N2 influenza virus hemagglutinin and regulates virulence. Proc. Natl. Acad. Sci. U.S.A. 1987;84:36–40. doi: 10.1073/pnas.84.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donatelli I., Campitelli L., Di Trani L., Puzelli S., Selli L., Fioretti A., Alexander D.J., Tollis M., Krauss S., Webster R.G. Characterization of H5N2 influenza viruses from Italian poultry. J. Gen. Virol. 2001;82:623–630. doi: 10.1099/0022-1317-82-3-623. [DOI] [PubMed] [Google Scholar]
- Duan L., Campitelli L., Fan X.H., Leung Y.H.A., Vijaykrishna D., Zhang J.X., Donatelli I., Delogu M., Li K.S., Foni E., Chiapponi C., Wu W.L., Kai H., Webster R.G., Shortridge K.F., Peiris J.S.M., Dmith G.J.D., Chen H., Guan Y. Characterisation of low-pathogenic H5 subtype influenza viruses from Eurasia: implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 2007;81:7529–7539. doi: 10.1128/JVI.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Schneeberger P.M., Rozendaal F.W., Broekman J.M., Kemink S.A., Munster V., Kuiken T., Rimmelzwaan G.F., Schutten M., Van Doornum G.J., Koch G., Bosman A., Koopmans M., Osterhaus A.D. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Munster V., Wallensten A., Bestebroer T.M., Herfst S., Smith D., Rimmelzwaan G.F., Olsen B., Osterhaus A.D. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from blackheaded gulls. J. Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G., Herwig A., Klenk H.D. Interaction of polymerase subunit PB2 and NP with importin alpha 1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4:e11. doi: 10.1371/journal.ppat.0040011. doi:10.371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidet N., Cattoli G., Hammoumi S., Newman S.H., Hagemeijer W., Takekawa J.Y., Cappelle J., Dodman T., Joannis T., Gil P., Monne I., Fusaro A., Capua I., Manu S., Micheloni P., Ottosson U., Mshelbwala J.H., Lubroth J., Domenech J., Monicat F. Evidence of infection by H5N2 highly pathogenic avian influenza viruses in healthy wild waterfowl. PLoS Pathog. 2008;4:e1000127. doi: 10.1371/journal.ppat.1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M., Suarez D.L., Crawford J.M., Latimer J.W., Slemons R.D., Swayne D.E., Perdue M.L. Evolution of H5 subtype avian influenza A viruses in North America. Virus Res. 1997;2:115–124. doi: 10.1016/s0168-1702(97)00087-7. [DOI] [PubMed] [Google Scholar]
- Gorman O.T., Bean W.J., Kawaoka Y., Webster R.G. Evolution of the nucleoprotein gene of influenza A virus. J. Virol. 1990;64:1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y., Stevens D.J., Skehel J.J., Wiley D.C. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale B.G., Randall R.E., Ortín J., Jackson D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Hay A.J., Wolstenholme A.J., Skehel J.J., Smith M.H. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4(11):3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindshaw V.S., Webster R.G., Rodriguez R.J. Influenza A viruses: combinations of hemagglutinin and neuraminidase subtypes isolated from animals and other sources. Arch. Virol. 1981;67:191–201. doi: 10.1007/BF01318130. [DOI] [PubMed] [Google Scholar]
- Hirst M., Astell C.R., Griffith M., Coughlin S.M., Moksa M., Zeng T., Smailus D.E., Holt R.A., Jones S., Marra M.A., Petric M., Krajden M., Lawrence D., Mak A., Chow R., Skowronski D.M., Tweed S.A., Goh S.H., Brunham R.C., Robinson J., Bowes V., Sojonky K., Byrne S.K., Li Yan., Kobassa D., Booth T., Mark P. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg. Infect. Dis. 2004;10:2192–2195. doi: 10.3201/eid1012.040743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Horimoto T., Rivera E., Pearson J., Senne D., Krauss S., Kawaoka Y., Webster R.G. Origin and molecular changes associated with emergence of a highly pathogenic H5N influenza virus in Mexico. Virology. 1995;213:223–230. doi: 10.1006/viro.1995.1562. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Naeve C.W., Webster R.G. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology. 1984;139:303–316. doi: 10.1016/0042-6822(84)90376-3. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Webster R.G. Evolution of the A/Chicken/Pennsylvania/83 (H5N2) influenza virus. Virology. 1985;146:130–137. doi: 10.1016/0042-6822(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Krauss S., Webster R.G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Senne D.A., Linares J.A., Woolcock P.R., Stallknecht D.E., Spackman E., Swayne D.E., Suarez D.L. Characterisation of recent H5 subtype avian influenza viruses from US poultry. Avian Pathol. 2004;33:288–297. doi: 10.1080/0307945042000203407. [DOI] [PubMed] [Google Scholar]
- Lee C.W., Senne D.A., Suarez D.L. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J. Virol. 2004;78:8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Swayne D.E., Linares J.A., Senne D.A., Suarez D.L. H5N2 Avian influenza outbreak in Texas in 2004: the first highly pathogenic strain in the United States in 20 years? J. Virol. 2005;79:11412–11421. doi: 10.1128/JVI.79.17.11412-11421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V.J., Chen M.I., Chan S.P., Wong C.S., Cutter J., Goh K.T., Tambyah P.A. Influenza pandemics in Singapore, a tropical, globally connected city. Emerg. Dis. Infect. 2007;13:1052–1056. doi: 10.3201/eid1307.061313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.S., Guan Y., Wang J., Smith G.J., Xu K.M., Duan L., Rahardjo A.P., Puthavathana P., Buranathai C., Nguyen T.D., Estoepangestie A.T., Chaisingh A., Auewarakul P., Long H.T., Hanh N.T., Webby R.J., Poon L.L., Chen H., Shirtridge K.F., Yuen K.Y., Webster R.G., Peiris J.S. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in Eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- Matrosovich M., Zhou N., Kawaoka Y., Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 1999;73:1145–1146. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah D., Krug R.M. Influenza virus virulence and its molecular determinants. Adv. Virus Res. 2005;65:121–145. doi: 10.1016/S0065-3527(05)65004-X. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Compans R.W. Effects of glucosamine, 2-deoxyglucose, and tunicamycin on glycosylation, sulfation, and assembly of influenza viral proteins. Virology. 1978;84:303–319. doi: 10.1016/0042-6822(78)90250-7. [DOI] [PubMed] [Google Scholar]
- Obenauer J.C., Denson J., Mehta P.K., Su X., Mukatira S., Finkelstein D.B., Xu X., Wang J., Ma J., Fan Y., Rakestraw K.M., Webster R.G., Hoffmann E., Krauss S., Zheng J., Zhang Z., Naeve C.W. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1562–1563. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- Okamatsu M., Saito T., Mase M., Tsukamoto K., Yamaguchi S. Characterisation of influenza A viruses isolated from chickens in Japan. Avian Dis. 2007;50:474–475. doi: 10.1637/7573-033106R1.1. [DOI] [PubMed] [Google Scholar]
- Okazaki K., Kawaoka Y., Webster R.G. Evolutionary pathways of the PA genes of influenza A viruses. Virology. 1989;172:601–608. doi: 10.1016/0042-6822(89)90202-x. [DOI] [PubMed] [Google Scholar]
- Pasick J., Handel K., Robinson J., Copps J., Ridd D., Hills K., Kehler J., Cottam-Birt C., Neufeld J., Berhance Y., Czub S. Intersegmental recombination between the haeagglutinin and the matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. J. Gen. Virol. 2005:727–731. doi: 10.1099/vir.0.80478-0. [DOI] [PubMed] [Google Scholar]
- Perdue M.L., Garcia M., Senne D., Fraire M. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 1997;49(2):173–186. doi: 10.1016/s0168-1702(97)01468-8. [DOI] [PubMed] [Google Scholar]
- Pillai S.P., Suarez D.L., Pantin-Jackwood M., Lee C.W. Pathogenicity and transmission studies of H5N2 parrot avian influenza virus of Mexican lineage in different poultry species. Vet. Microbiol. 2008;129:48–57. doi: 10.1016/j.vetmic.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Pinto L.H., Holsinger L.J., Lamb R.A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- Poddar S.K. Influenza virus types and subtypes detection by single step tube multiplex reverse transcription-polymerase chain reaction (RT-PCR) and agarose gel electrophoresis. J. Virol. Methods. 2002;99:63–70. doi: 10.1016/s0166-0934(01)00380-9. [DOI] [PubMed] [Google Scholar]
- Reading P.C., Tate M.D., Pickett D.L., Brooks A.G. Glycosylation as a target for recognition of influenza viruses by the innate immune system. Adv. Exp. Med. Biol. 2007;598:279–292. doi: 10.1007/978-0-387-71767-8_20. [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Rohde W., Von Hoyningen V., Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- Seo S.H., Hoffmann E., Webster R.G. The NS1 gene of H5N1 influenza viruses circumvents the host anti-viral cytokine responses. Virus Res. 2004;103:107–113. doi: 10.1016/j.virusres.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Shaw M., Cooper L., Xu X., Thompson W., Krauss S., Guan Y., Zhou N., Klimov A., Cox N., Webster R., Lim W., Shortridge K., Subbarao K. Molecular changes associated with the transmission of avian influenza A H5N1 and H9N2 to humans. J. Med. Virol. 2002;66:107–114. doi: 10.1002/jmv.2118. [DOI] [PubMed] [Google Scholar]
- Sheng M., Sala C. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Shinya K., Hamm S., Hatta M., Ito H., Ito T., Kawaoka Y. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology. 2004;320:258–266. doi: 10.1016/j.virol.2003.11.030. [DOI] [PubMed] [Google Scholar]
- Spackman E., Senne D.A., Myers T.J., Bulaga L.L., Garber L.P., Perdue M., Lohman K., Daum L.T., Suarez D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez D.L., Senne D.A., Banks J., Brown I.H., Essen S.C., Lee C.W., Manvell R.J., Mathieu-Benson C., Moreno V., Pedersen J.C., Panigrahy B., Rojas H., Spackman E., Alexander D.J. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg. Infect. Dis. 2004;10:693–699. doi: 10.3201/eid1004.030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao E.K., London W., Murphy B.R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue R.J., Hay A.J. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B.H., SARS Investigation Team from DMERI, SGH Strategies adopted and lessons learnt during the sevre acute respiratory syndrome crisis in Singapore. Rev. Med. Virol. 2005;15:57–70. doi: 10.1002/rmv.458. [DOI] [PubMed] [Google Scholar]
- Tarendeau F., Crepin T., Guilligay D., Ruigrok R.W.H., Cusack S., Hart D.J. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 2008;4:e1000136. doi: 10.1371/journal.ppat.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J.K., Reid A.H., Lourens R.M., Wang R., Jin G., Fanning T.G. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- Viseshakul N., Thanawongnuwech R., Amonsin A., Suradhat S., Payungporn S., Keawchareon J., Oraveerakul K., Wongyanin P., Plitkul S., Theamboonlers A., Poovorawan Y. The genome sequence analysis of H5N1 avian influenza A virus isolated from the outbreak among poultry populations in Thailand. Virology. 2004;328:169–176. doi: 10.1016/j.virol.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Webster R.G., Kawaoka Y., Bean W.J. Molecular changes in A/Chicken/ Pennsylvania/83 (H5N2) influenza virus associated with acquisition of virulence. Virology. 1986;149:165–173. doi: 10.1016/0042-6822(86)90118-2. [DOI] [PubMed] [Google Scholar]
- Wiley D.C., Wilson I.A., Skehel J.J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Wilson I.A., Skehek J.J., Wiley D.C. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- World Health Organisation for Animal Health, 2008. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2008. Avian Influenza, pp. 465–481 (Chapter 2.3.4). Accessed at http://www.oie.int/eng/normes/en_mmanual.htm?e1d10 on 5th August 2008.
- Wright P.F., Webster R.G. In: Fields Virology. 5th edition. Knipe D.M., Howley P.M., editors. Lippincott Williams & Wilkins; 2007. (Chapter 47) [Google Scholar]
- Yuen K.Y., Chan P.K., Peiris M., Tsang D.N., Que T.L., Shortridge K.F., Cheung P.T., To W.K., Ho E.T., Sung R., Cheng A.F., Members of the H5N1 Study Group Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]


