Highlights
-
•
Optical tweezers provide a sensitive tool to probe structural dynamics.
-
•
Native and non-native folding can be characterized in detail.
-
•
Effects of intermolecular interactions on dynamics can be probed.
-
•
Mechanisms of complex molecular processes can be determined.
-
•
Allows direct connection between dynamics and function to be observed.
Abstract
Conformational changes are an essential feature of most molecular processes in biology. Optical tweezers have emerged as a powerful tool for probing conformational dynamics at the single-molecule level because of their high resolution and sensitivity, opening new windows on phenomena ranging from folding and ligand binding to enzyme function, molecular machines, and protein aggregation. By measuring conformational changes induced in a molecule by forces applied by optical tweezers, new insight has been gained into the relationship between dynamics and function. We discuss recent advances from studies of how structure forms in proteins and RNA, including non-native structures, fluctuations in disordered proteins, and interactions with chaperones assisting native folding. We also review the development of assays probing the dynamics of complex protein–nucleic acid and protein–protein assemblies that reveal the dynamic interactions between biomolecular machines and their substrates.
Current Opinion in Structural Biology 2015, 34:43–51
This review comes from a themed issue on Biophysical and molecular biological methods
Edited by Ben Schuler and Janet L Smith
For a complete overview see the Issue and the Editorial
Available online 17th July 2015
http://dx.doi.org/10.1016/j.sbi.2015.06.006
0959-440X/© 2015 Elsevier Ltd. All rights reserved.
Introduction
The structural dynamics of macromolecules like protein and RNA are critically tied to their biological function. Numerous processes, from folding to ligand binding and enzymatic catalysis, depend on conformational changes, whether they involve large-scale rearrangements or subtle fluctuations. Studies aimed at elucidating the details of protein and RNA conformational dynamics are thus essential for a full understanding of biological mechanisms. Single-molecule force spectroscopy (SMFS), whereby mechanical forces are applied to individual molecules by a force probe and the length of the molecule is measured to capture the resulting conformational changes [1, 2], is a powerful tool for studying the relationship between conformational dynamics and function, owing to the very high resolution it can achieve and its high sensitivity to rare and transient events [2, 3, 4•]. Here we discuss recent advances made using optical tweezers to study topics such as intra-molecular and inter-molecular interactions in the folding and misfolding of proteins and RNAs and the dynamics underlying the function of molecular motors and complex macromolecular assemblies.
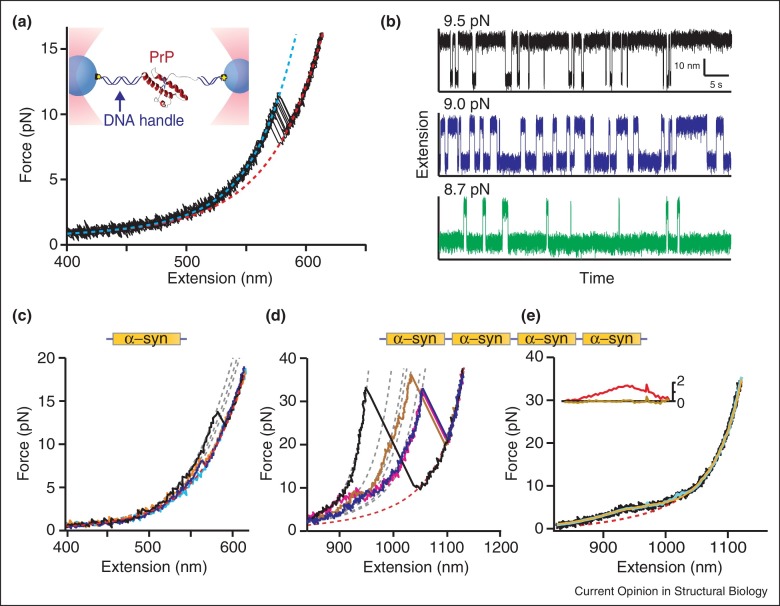
The principles and construction of optical tweezers are reviewed in detail elsewhere; with sufficient care, atomic-scale resolution can be achieved, allowing the discrimination of subtle conformational changes [3, 5, 6]. In a typical apparatus (Figure 1a inset), the molecule of interest is attached (often via DNA handles) to polystyrene beads, which are in turn trapped by laser beams that apply tension to the molecule [7]. Two main measurement modalities are used [8]: first, non-equilibrium measurements, such as force-ramps, where the traps are moved continuously to ramp up/down the force on the molecule (Figure 1a), or force-jumps, where the force is changed discontinuously, and second, equilibrium measurements, where the molecule is held under constant force and/or the traps are kept at a constant position and the extension is measured as the structure fluctuates at equilibrium (Figure 1b).
Figure 1.
Optical tweezers measurements of protein folding/unfolding. (a) DNA handles attached to each end of a protein molecule are bound to beads held under tension in optical traps (inset). Ramping up the force by moving the traps apart, the handles stretch until the protein unfolds abruptly, generating a sawtooth-like rip in the force-extension curve (FEC). Representative FECs (black) for unfolding the protein PrP are fit to wormlike chain (WLC) models for the folded (cyan) and unfolded (red) states. (b) Constant–force trajectories of PrP folding measured at three different forces show abrupt jumps as PrP unfolds/refolds in a two-state process. (c) FECs of α-synuclein monomers usually display no rips (cyan) and behave as if unfolded (red: WLC for unfolded state). Some reveal discrete transitions (black, orange, blue), with different contour lengths (gray: WLC fits). (d,e) FECs of α-synuclein tetramers connected in tandem by peptide linkers. (d) Some FECs show discrete transitions revealing many different structures with different sizes and unfolding forces (dashed lines: WLC fits; gray: folded states, red: unfolded state). (e) FECs without discrete rips were averaged (cyan) and compared to polymer models. Data did not fit a non-interacting WLC model (red; residuals in inset), but did fit a model incorporating rapid structural fluctuations (yellow; residuals in inset).
Adapted from Ref. [26] ((a) and (b), copyright (2012) National Academy of Sciences, USA) and Refs. [40, 42•] ((c)–(e)).
Conformational changes generate characteristic features in these measurements owing to the sudden change in molecular extension during cooperative transitions: sawtooth-shaped ‘rips’ in force-extension curves (FECs) (Figure 1a), and steps in equilibrium trajectories (Figure 1b). These measurements yield information such as the molecule's contour length change (yielding the number of amino acids or nucleotides involved in structural transitions), the presence and number of intermediates on a given pathway, the existence of alternative pathways, and how each state is connected [9]. Furthermore, kinetic and thermodynamic information can be obtained including the force-dependent microscopic rates for transitions [10, 11, 12, 13•], the free energy of each state [14, 15, 16], the position and height of energy barriers between states [12, 17, 18, 19], the full profile of the energy landscape governing the conformational dynamics [8, 18, 20, 21, 22, 23, 24], and the diffusion coefficient setting the timescale for microscopic dynamics [8, 25, 26, 27••]. Equilibrium and non-equilibrium measurements yield similar information but from different approaches, which can be exploited for specific purposes (e.g. to select kinetically for a specific pathway [26]) or to enhance analysis reliability via independent measures of the desired information.
Probing folding/unfolding transitions and conformational dynamics in ordered and disordered proteins
Optical tweezers are providing significant insight into the structure formation process in proteins. Stigler et al. deciphered the complex network of states involved in the folding of calmodulin [10] and Yu et al. did the same for the prion protein [26, 28], in each case characterizing the thermodynamic and kinetic properties of native and non-native intermediates in detail. Neupane et al. showed how the statistics of the transition paths can prove that the reaction coordinate used in SMFS is good and that 1D diffusive models of the folding are justified [29•]. Marqusee and colleagues probed the properties of different stages of folding, showing that molten globules can be distinguished from fully native states through their increased compliance [30] and extending classic phi analysis of transition states into the single-molecule regime [31]. Measurements of the unfolding and refolding of monomeric HIV-1-protease validated simulations suggesting the existence of multiple pathways [32], revealing not only two-state unfolding but also unfolding through an intermediate and an ensemble of partially folded states en route to the native state, which themselves unfolded via multiple pathways [33]. Studies of SNARE complex assembly, which involves a stable four-helix bundle, helped clarify how it drives membrane fusion to allow transport of molecules between different membranes [34, 35] and identified rare misfolded states of coiled-coils [36] that may be involved in diseases related to SNARE misfolding [37].
A notable recent advance has been the extension of optical tweezers SMFS to study intrinsically disordered proteins (IDPs), which play many important roles [38, 39] but present a special challenge for conformational analysis because of their lack of stable structure. Optical tweezers are well-suited, because of their high force stability and low stiffness [3], to probing the low-energy fluctuations and marginally stable structures expected in IDPs. Studying α-synuclein, an IDP whose aggregation is associated with Parkinson's disease, Neupane et al. [40] captured infrequent fluctuations into a diverse set of transient metastable structures (Figure 1c). Linking copies of α-synuclein to create tandem oligomers, the complexity and diversity of the metastable structures increased with the oligomer size (Figure 1d). Not surprisingly for an IDP, however, most FECs displayed no discrete rips; nevertheless, these curves still contained significant information about the conformational dynamics. Indeed, FECs without rips deviated from the pure wormlike chain behavior expected for a non-interacting polymer [41], exhibiting a shoulder-like feature at low force (Figure 1e). This feature suggests rapid quasi-equilibrium fluctuations into structures that are only marginally stable [42•], consistent with the picture of a collapsed, molten-globule-like state for α-synuclein held together by long-range contacts that emerged from structural and computational work [43, 44]. Similar shoulder-like features were seen previously in FECs of the protein villin, owing to its ultrafast dynamics which prevented resolution of discrete transitions [4•]. Through analysis of the fast kinetics of α-synuclein, the energy landscape was reconstructed and found to be flat with low barriers, but featuring slow diffusion owing to landscape roughness. Such a flat but rough landscape was expected to be a hallmark of IDPs, but had not been quantified experimentally.
Probing interactions that influence protein misfolding and aggregation
Another area of particular interest is protein misfolding and its relation to disease [45], which is well-suited for study by single-molecule methods because of their ability to distinguish and characterize even transient components of heterogeneous mixtures [46], as shown by recent work with optical tweezers. For example, dimeric prion protein was found to misfold into a stable aggregated state via multiple intermediates, with much slower diffusion than for native folding indicating a rougher energy landscape for misfolding [27••]. The slow refolding of the enzyme luciferase and its propensity to aggregate were linked to the formation of a misfolded state [47]. The effects of calcium concentration on misfolding of a calcium sensor, neuronal calcium sensor-1 (NCS-1) [48•] suggested the missing link between Ca2+ dysregulation, misfolding, and a NCS protein involved in neurodegenerative disorders [49, 50].
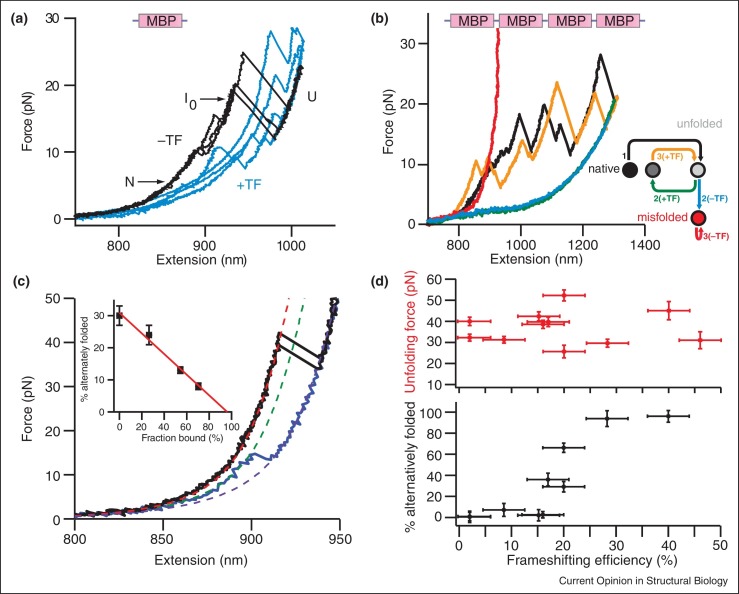
Of course optical tweezers’ utility extends beyond studying proteins in isolation to probing the relationship between dynamics and function under conditions that more closely recapitulate key aspects of the cellular environment. Kim et al. studied how the binding of von Willebrand factor (VWF) to platelets is regulated by hydrodynamic forces in the vasculature, revealing that force activates platelet binding and increases the effects of disease-related mutations [51]. Especially fascinating is recent work probing the activity of molecular chaperones, which are required for the folding of many proteins but whose mechanisms remain poorly understood at the molecular level [52]. Full understanding of chaperone activity must also be tied to insights into protein misfolding in the absence of chaperones. Tans and colleagues [53••] investigated how the general bacterial chaperone trigger factor (TF) influences the folding of maltose-binding protein (MBP), which folds efficiently in isolation, and a MBP tandem-repeat oligomer (MBP4), which has a high tendency to misfold and aggregate. Unfolding curves of MBP typically showed a single intermediate state without TF present, whereas they contained additional intermediate states with TF present (Figure 2a). The MBP4 oligomer tended to form a stable aggregate that could not be unfolded in the absence of TF, but adding TF dramatically changed the picture: although misfolding interactions generating non-native structures were still common, they were typically much weaker, and most of the protein chain was now natively folded (Figure 2b). Taken together, these results suggest that TF promotes native folding by protecting partially folded states from long-range interactions driving stable misfolded states [53••].
Figure 2.
Effects of chaperones and small-molecule ligands on protein and RNA folding. (a) In the absence of TF, MBP monomers (black) unfold via an intermediate state (I0) between the native (N) and unfolded (U) states. In the presence of 1 μM TF (blue), MBP frequently occupies a variety of intermediate states. (b) FECs of a tandem MBP tetramer show more complex behavior. Initial unfolding (black) shows features corresponding to four natively folded MBP monomers. When refolded without TF (blue), subsequent pulling (red) reveals no unfolding, indicating the tetramer misfolded with tight inter-domain interactions. When refolded with 1 μM TF (green), subsequent pulling (orange) showed extension changes characteristic of native interactions (e.g. at ∼1230 nm) and weak non-native interactions (at ∼1100 nm), but no tight ones. Inset: sequence of pulling curves and states observed. (c) Most FECs for the SARS frameshifting stimulatory pseudoknot showed a length change, found from WLC fits (dashed lines), consistent with the native structure (black), but some revealed a smaller, alternative structure (blue). Inset: the extent of alternative structure formation decreased linearly with the fraction of ligand-bound pseudoknots and extrapolated to zero at 96 ± 8% binding, indicating that ligand binding effectively eliminates the formation of alternative structures, mirroring the suppression of −1 PRF to near-background levels caused by the ligand [67]. (d) The average unfolding force of pseudoknots is uncorrelated with efficiency of −1 PRF stimulation (upper panel), indicating that mechanical stability is not a primary determinant of −1 PRF efficiency. Higher frameshifting efficiency is instead correlated with increased tendency to form alternate structures (lower panel).
Adapted from Ref. [53••] ((a) and (b), by permission from Macmillan publishers Ltd: Nature, copyright (2013)), Ref. [68] ((c), copyright (2014) American Chemical Society) and Ref. [65•] ((d), copyright (2012) National Academy of Sciences, USA).
Conformational dynamics in RNA
SMFS studies of RNA have helped to connect dynamics in complex folding pathways — manifested for example by the tendency of some RNAs to form alternative structures — to function. Measurements of the E. coli rpsO gene operator transcript, which folds into either a pseudoknot or double-hairpin conformation but binds the gene product only in the pseudoknot conformation, showed that the two transcript structures can interchange spontaneously [54], which is unusual because conformational switching in RNA typically requires regulatory factors such as metabolites, non-coding RNAs, or proteins [55, 56]. Studies of RNA kissing-loops have led to better understanding of the structural determinants of these complexes, including roles for flanking nucleotides [57]. Riboswitches are an especially interesting class of RNAs that undergo large-scale, functional conformational changes upon ligand binding [58]. Anthony et al. deciphered the ligand-dependent and ligand-independent steps for the metabolite-sensing domain of the TPP riboswitch [59], and Neupane et al. showed how structural changes in the metabolite-sensing domain of the add riboswitch are communicated to alter the structure of the neighboring domain controlling gene expression [60]. By studying pbuE riboswitch dynamics while the RNA was being transcribed, Frieda et al. were able to demonstrate kinetically controlled folding directly and predict the regulatory outcome — transcription termination or run-through — from the initial RNA dynamics [61•].
Recent work with optical tweezers also highlighted the unexpected importance of conformational dynamics for the function of RNA pseudoknots stimulating −1 programmed ribosomal frameshifting (−1 PRF) [62]. Early work identifying a correlation between the pseudoknot unfolding force and −1 PRF stimulation efficiency [63, 64] was shown not to extend to a larger panel of viral pseudoknots; instead, −1 PRF efficiency was correlated with pseudoknot conformational plasticity [65•], reflected in the tendency of the RNA to refold into alternative structures (Figure 2c,d). The importance of pseudoknot dynamics in stimulating frameshifting was corroborated by studies of the frameshift signal from human CCR5 mRNA, showing that it manifests several distinct unfolding pathways when mechanically destabilized [66], and by measurements of the effect of a ligand that abolishes −1 PRF upon binding specifically to the pseudoknot from the SARS coronavirus [67], showing that the ligand reduced the conformational plasticity of the pseudoknot proportional to the amount of ligand bound (Figure 2c inset) [68]. While such studies provide important insights, they investigate only one part of the −1 PRF mechanism, which involves interactions between many different elements. A more complete picture emerges from experiments probing the full translation complex, like those described below.
Complex assemblies in motion: dynamics tied to function
In recent years, optical tweezers have increasingly been applied to reveal the functional dynamics of complex macromolecular assemblies, especially those of protein–nucleic acid complexes. Recent work on nucleosomes, the basic unit of DNA compaction consisting of DNA wrapped around a histone core, elegantly showed how local conformational transitions in single nucleosomes govern DNA unwrapping [69••], demonstrated alternate pathways for nucleosome unwinding [70], and showed through the use of torque-wrench tweezers [71] that torque can modulate nucleosome stability in a way that may regulate histone exchange during transcription and replication [72]. These studies have important implications for how DNA accessibility to enzymes might be regulated at the level of DNA sequence and modifications. In a similar vein, Ma et al. investigated the effects of torsion from DNA supercoiling on transcription by RNA polymerase, finding that torsion modulates the transcription rate but RNA polymerase can generate sufficient torque to melt DNA of arbitrary sequence [73]. Turning to translation, the mechanics of translation were studied by Bustamante, Tinoco, and colleagues: after demonstrating that individual codon steps could be discerned and showing that ribosomes apply force actively to unfold structures in the mRNA during translation [74, 75], they investigated how the ribosome affects nascent polypeptide folding [76] and showed that nascent chain folding near the surface of the exit tunnel exerts a force that can rescue sequence-induced ribosome stalling [77•].
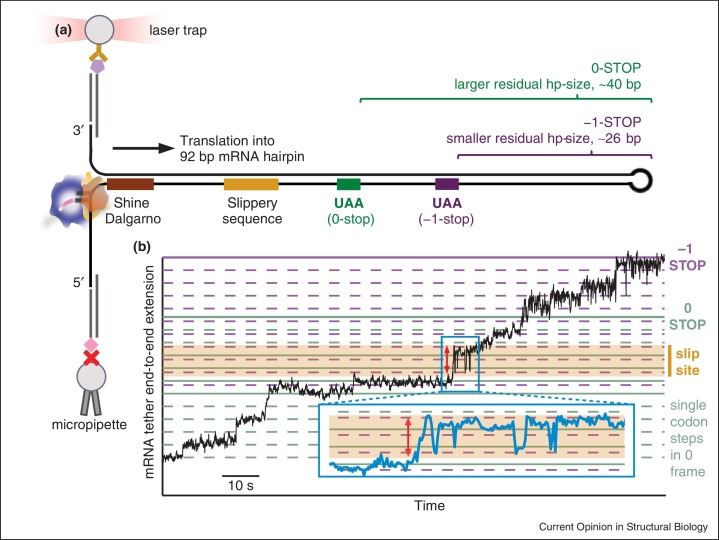
Tinoco and colleagues also applied a single-ribosome translocation assay to probe −1 PRF stimulated by a structured RNA hairpin from the E. coli dnaX gene [78••]. Tight regulation of −1 PRF efficiency is achieved by three elements in the mRNA: a 7-nt slippery sequence where −1 PRF occurs, and an internal Shine–Dalgarno sequence and downstream hairpin flanking the slippery sequence. Codon-by-codon translation was monitored along a mRNA template embedding this frameshift signal within a 92-bp hairpin held under tension by optical tweezers (Figure 3a). Back-and-forth motions of the ribosome around the slippery sequence (Figure 3b), previously detected by single-molecule fluorescence measurements [79, 80], were corroborated and shown to occur regardless of the presence of a frameshift, suggesting that they are a property of the mechanics of the frameshift signal rather than a feature of −1 PRF itself. Notably, the fluctuation timescale was similar to that for the dynamics of the ribosome 30S body and head during regular translation [81], suggesting that these fluctuations reflect conformational excursions of the 30S head during multiple forward translocation attempts [78••].
Figure 3.
Translocation dynamics in programmed ribosomal frameshifting. (a) An mRNA hairpin molecule containing the frameshift signal from dnaX (internal Shine–Dalgarno sequence, slippery site, stimulatory hairpin) and bound with a single ribosome is held under tension. The hairpin unzips by 3 basepairs/codon as the ribosome translocates, increasing the tether length. When the first 0-frame codon in the slippery site resides in the ribosome P site, a 55-basepair hairpin remains downstream. To determine the ribosome location on the mRNA at the end of each trajectory, hairpin portions not unwound by the ribosome are mechanically unfolded. Ribosome termination at the −1 stop codon leaves a smaller residual hairpin than at the 0 stop. (b) A single-ribosome translation trajectory along the mRNA construct shows individual translocation steps (green and purple lines: single-codon steps in 0 and −1 frames, respectively). The ribosome continually translocates against a hairpin, but characteristic fluctuations in mRNA extension (red arrow) occur around the slippery sequence region (orange-shaded area), reflecting dynamics in the ribosome induced by the frameshift signal.
Adapted from Ref. [78••], copyright (2015), with permission from Elsevier.
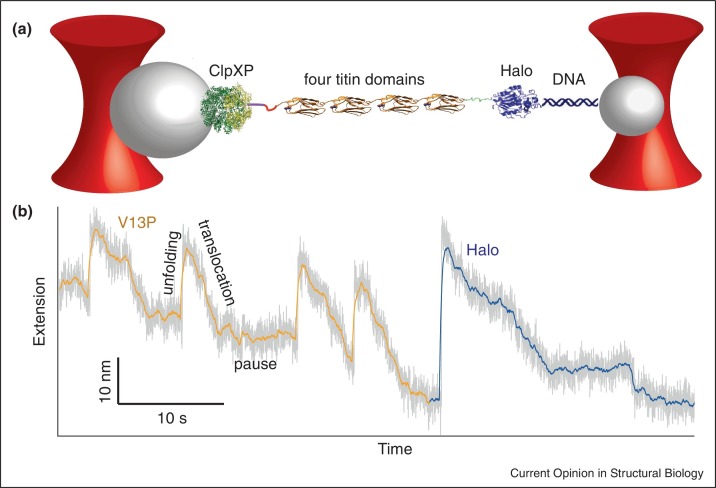
The ability of optical tweezers to track substrate movement through molecular machines has been applied to other systems, too, such as elegant work investigating how the bacterial protease ClpXP unfolds and translocates a wide variety of substrates [82, 83••]. To probe the interactions between protease and substrate, Cordova et al. [83••] engaged ClpXP with a substrate consisting of four titin domains (Figure 4a). Measurements under constant force displayed three signatures of ClpXP mechanical function: first, abrupt extension increases due to protein unfolding; second, gradual extension decreases due to translocation of the unfolded polypeptide; and third, unchanged extension representing a pre-unfolding dwell between completed translocation of an unfolded domain and denaturation of the next native domain (Figure 4b). As expected, titin mutants with lower stability were less resistant to degradation. These measurements led to a model of how ClpXP uses ATP to move along its substrate: ClpXP takes steps of variable sizes (1–4 nm), in no defined order but with steps of similar sizes clustering together, indicating interplay between stochastic and deterministic behavior of the subunits. Incorporating hydrolysis-incompetent mutant subunits into ClpXP showed that the largest steps required no more than two subunits. This study also illustrates nicely how a complete mechanistic understanding of complex biological processes requires fitting single-molecule data into a wider framework provided by other techniques: here, the degradation rate measured by optical tweezers was consistently higher than in ensemble experiments, suggesting that the rate-limiting step in solution is the threading of the substrate into the protease, a step done before the tweezers measurements started.
Figure 4.
Unfolding and translocation of single proteins by the protease ClpXP. (a) ClpXP is attached to one laser-trapped bead and engaged with a multidomain substrate consisting of four titin domains separated by short peptide linkers and a Halo-tag domain, which is attached to a second laser-trapped bead via a DNA linker. (b) A trajectory for ClpXP unfolding and translocation of the V13P titin mutant (gray, unaveraged; gold, averaged data for V13P domains; purple, averaged data for Halo domain), in which the titin domain is destabilized to increase ClpXP degradation, shows three main features. Unfolding of individual domains by ClpXP causes abrupt increases in the construct extension under tension (upward movement), whereas translocation decreases the extension gradually as the protease reels in the unfolded polypeptide chain (downward movement). After each domain is translocated, ClpXP pauses for a variable dwell time before unfolding the next domain.
Figure adapted from Ref. [83••], copyright (2014), with permission from Elsevier.
Outlook
Models describing the activity of proteins and RNAs have not always included the role of conformational dynamics in determining function. Optical tweezers provide a powerful tool for probing the dynamics of biomolecules at the single-molecule level, providing new information about the relationship between dynamics and function. Studying the dynamics of molecules in isolation continues to yield valuable new insights, but it is the application of optical tweezers to more complex systems that is particularly exciting. Sensitive and high-resolution measurements with optical tweezers hold promise for elucidating the conformational dynamics underlying the complicated mechanisms of phenomena such as ribosomal translocation and frameshifting, co-transcriptional and co-translational folding, protein misfolding and chaperone action, and protein degradation by proteases. With a constant flow of new information from such experiments to complement results from other single-molecule and ensemble techniques, it will be exciting to see how mechanistic models evolve in the coming years.
Conflict of interest statement
Nothing to declare.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgement
This work was supported by Alberta Innovates Technology Futures.
References
- 1.Bustamante C., Chemla Y.R., Forde N.R., Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 2.Woodside M.T., García-García C., Block S.M. Folding and unfolding single RNA molecules under tension. Curr Opin Chem Biol. 2008;12:640–646. doi: 10.1016/j.cbpa.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenleaf W.J., Woodside M.T., Block S.M. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•.Žoldák G., Stigler J., Pelz B., Li H., Rief M. Ultrafast folding kinetics and cooperativity of villin headpiece in single-molecule force spectroscopy. Proc Natl Acad Sci U S A. 2013;110:18156–18161. doi: 10.1073/pnas.1311495110. [DOI] [PMC free article] [PubMed] [Google Scholar]; Expands the accessible dynamic range of single-molecule force spectroscopy by optical tweezers to the microsecond range, showing that fast-folding proteins near the folding ‘speed limit’ can be studied in the single-molecule regime.
- 5.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustamante C., Chemla Y.R., Moffitt J.R. High-resolution dual-trap optical tweezers with differential detection: an introduction. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.top60. [DOI] [PubMed] [Google Scholar]
- 7.Woodside M.T., Valentine M.T. Single-molecule manipulation using optical traps. In: Hinterdorfer P., Oijen A., editors. Handbook of Single-Molecule Biophysics. Springer US; 2009. pp. 341–370. [Google Scholar]
- 8.Woodside M.T., Block S.M. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu Rev Biophys. 2014;43:19–39. doi: 10.1146/annurev-biophys-051013-022754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X., Ma L., Zhang Y. High-resolution optical tweezers for single-molecule manipulation. Yale J Biol Med. 2013;86:367–383. [PMC free article] [PubMed] [Google Scholar]
- 10.Stigler J., Ziegler F., Gieseke A., Gebhardt J.C.M., Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann A., Woodside M.T. Signal-pair correlation analysis of single-molecule trajectories. Angew Chem Int Ed Engl. 2011;50:12643–12646. doi: 10.1002/anie.201104033. [DOI] [PubMed] [Google Scholar]
- 12.Dudko O.K., Hummer G., Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc Natl Acad Sci U S A. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Zhang Y., Dudko O.K. A transformation for the mechanical fingerprints of complex biomolecular interactions. Proc Natl Acad Sci U S A. 2013;110:16432–16437. doi: 10.1073/pnas.1309101110. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides a theoretical framework for analyzing force spectroscopy data from multi-state systems to extract the microscopic rate constants for all transitions.
- 14.Jarzynski C. Nonequilibrium equality for free energy differences. Phys Rev Lett. 1997;78:2690–2693. [Google Scholar]
- 15.Crooks G.E. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys Rev E. 1999;60:2721–2726. doi: 10.1103/physreve.60.2721. [DOI] [PubMed] [Google Scholar]
- 16.Alemany A., Mossa A., Junier I., Ritort F. Experimental free-energy measurements of kinetic molecular states using fluctuation theorems. Nat Phys. 2012;8:688–694. [Google Scholar]
- 17.Dudko O.K., Hummer G., Szabo A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys Rev Lett. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- 18.Manuel A.P., Lambert J., Woodside M.T. Reconstructing folding energy landscapes from splitting probability analysis of single-molecule trajectories. Proc Natl Acad Sci U S A. 2015;112:7183–7188. doi: 10.1073/pnas.1419490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierse C.A., Dudko O.K. Kinetics and energetics of biomolecular folding and binding. Biophys J. 2013;105:L19–L22. doi: 10.1016/j.bpj.2013.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A.N., Vincent A., Neupane K., Yu H., Wang F., Woodside M.T. Experimental validation of free-energy-landscape reconstruction from non-equilibrium single-molecule force spectroscopy measurements. Nat Phys. 2011;7:631–634. [Google Scholar]
- 21.Engel M.C., Ritchie D.B., Foster D.A.N., Beach K.S.D., Woodside M.T. Reconstructing folding energy landscape profiles from nonequilibrium pulling curves with an inverse Weierstrass integral transform. Phys Rev Lett. 2014;113:238104. doi: 10.1103/PhysRevLett.113.238104. [DOI] [PubMed] [Google Scholar]
- 22.Hummer G., Szabo A. Free energy reconstruction from nonequilibrium single-molecule pulling experiments. Proc Natl Acad Sci U S A. 2001;98:3658–3661. doi: 10.1073/pnas.071034098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodside M.T., Anthony P.C., Behnke-Parks W.M., Larizadeh K., Herschlag D., Block S.M. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lannon H., Haghpanah J.S., Montclare J.K., Vanden-Eijnden E., Brujic J. Force-clamp experiments reveal the free-energy profile and diffusion coefficient of the collapse of protein molecules. Phys Rev Lett. 2013;110:128301. doi: 10.1103/PhysRevLett.110.128301. [DOI] [PubMed] [Google Scholar]
- 25.Neupane K., Ritchie D.B., Yu H., Foster D.A.N., Wang F., Woodside M.T. Transition path times for nucleic acid folding determined from energy-landscape analysis of single-molecule trajectories. Phys Rev Lett. 2012;109:068102. doi: 10.1103/PhysRevLett.109.068102. [DOI] [PubMed] [Google Scholar]
- 26.Yu H., Gupta A.N., Liu X., Neupane K., Brigley A.M., Sosova I., Woodside M.T. Energy landscape analysis of native folding of the prion protein yields the diffusion constant, transition path time, and rates. Proc Natl Acad Sci U S A. 2012;109:14452–14457. doi: 10.1073/pnas.1206190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Yu H., Dee D.R., Liu X., Brigley A.M., Sosova I., Woodside M.T. Protein misfolding occurs by slow diffusion across multiple barriers in a rough energy landscape. Proc Natl Acad Sci U S A. 2015;112:8308–8313. doi: 10.1073/pnas.1419197112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that prion protein dimers misfold into a stable aggregated state via multiple intermediates with much slower diffusion than for native folding; includes first direct observations of microscopic transition paths in protein folding.
- 28.Yu H., Liu X., Neupane K., Gupta A.N., Brigley A.M., Solanki A., Sosova I., Woodside M.T. Direct observation of multiple misfolding pathways in a single prion protein molecule. Proc Natl Acad Sci U S A. 2012;109:5283–5288. doi: 10.1073/pnas.1107736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Neupane K., Manuel A.P., Lambert J., Woodside M.T. Transition-path probability as a test of reaction-coordinate quality reveals DNA hairpin folding is a one-dimensional diffusive process. J Phys Chem Lett. 2015;6:1005–1010. doi: 10.1021/acs.jpclett.5b00176. [DOI] [PubMed] [Google Scholar]; Demonstrates how to test the quality of the experimental observable used to monitor the progress of folding reactions in terms of its ability to capture the dynamics of the system.
- 30.Elms P.J., Chodera J.D., Bustamante C., Marqusee S. The molten globule state is unusually deformable under mechanical force. Proc Natl Acad Sci U S A. 2012;109:3796–3801. doi: 10.1073/pnas.1115519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagannathan B., Elms P.J., Bustamante C., Marqusee S. Direct observation of a force-induced switch in the anisotropic mechanical unfolding pathway of a protein. Proc Natl Acad Sci U S A. 2012;109:17820–17825. doi: 10.1073/pnas.1201800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonomi M., Barducci A., Gervasio F.L., Parrinello M. Multiple routes and milestones in the folding of HIV-1 protease monomer. PLoS One. 2010;5:e13208. doi: 10.1371/journal.pone.0013208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caldarini M., Sonar P., Valpapuram I., Tavella D., Volonté C., Pandini V., Vanoni M.A., Aliverti A., Broglia R.A., Tiana G. The complex folding behavior of HIV-1-protease monomer revealed by optical-tweezer single-molecule experiments and molecular dynamics simulations. Biophys Chem. 2014;195:32–42. doi: 10.1016/j.bpc.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Zorman S., Rebane A.A., Ma L., Yang G., Molski M.A., Coleman J., Pincet F., Rothman J.E., Zhang Y. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. eLife. 2014;3:e03348. doi: 10.7554/eLife.03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y., Zorman S., Gundersen G., Xi Z., Ma L., Sirinakis G., Rothman J.E., Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xi Z., Gao Y., Sirinakis G., Guo H., Zhang Y. Single-molecule observation of helix staggering, sliding, and coiled coil misfolding. Proc Natl Acad Sci U S A. 2012;109:5711–5716. doi: 10.1073/pnas.1116784109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burré J., Sharma M., Tsetsenis T., Buchman V., Etherton M.R., Südhof T.C. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uversky V.N. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci Landmark Ed. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 40.Neupane K., Solanki A., Sosova I., Belov M., Woodside M.T. Diverse metastable structures formed by small oligomers of α-synuclein probed by force spectroscopy. PLoS One. 2014;9:e86495. doi: 10.1371/journal.pone.0086495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M.D., Yin H., Landick R., Gelles J., Block S.M. Stretching DNA with optical tweezers. Biophys J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Solanki A., Neupane K., Woodside M.T. Single-molecule force spectroscopy of rapidly fluctuating, marginally stable structures in the intrinsically disordered protein α-synuclein. Phys Rev Lett. 2014;112:158103. doi: 10.1103/PhysRevLett.112.158103. [DOI] [PubMed] [Google Scholar]; Demonstrates that optical tweezers are well-suited for probing the low-energy fluctuations and marginally stable structures present in IDPs, reconstructing the landscape for structural fluctuations in α-synuclein.
- 43.Dedmon M.M., Lindorff-Larsen K., Christodoulou J., Vendruscolo M., Dobson C.M. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 44.Bertoncini C.W., Jung Y.-S., Fernandez C.O., Hoyer W., Griesinger C., Jovin T.M., Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci U S A. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann A., Neupane K., Woodside M.T. Single-molecule assays for investigating protein misfolding and aggregation. Phys Chem Chem Phys. 2013;15:7934–7948. doi: 10.1039/c3cp44564j. [DOI] [PubMed] [Google Scholar]
- 47.Mashaghi A., Mashaghi S., Tans S.J. Misfolding of luciferase at the single-molecule level. Angew Chem Int Ed. 2014;53:10390–10393. doi: 10.1002/anie.201405566. [DOI] [PubMed] [Google Scholar]
- 48•.Heidarsson P.O., Naqvi M.M., Otazo M.R., Mossa A., Kragelund B.B., Cecconi C. Direct single-molecule observation of calcium-dependent misfolding in human neuronal calcium sensor-1. Proc Natl Acad Sci U S A. 2014;111:13069–13074. doi: 10.1073/pnas.1401065111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides a basis for better understanding the link between protein misfolding and calcium dysregulation, and how they are related to neurodegeneration, by monitoring calcium-dependent misfolding reactions of the human neuronal calcium sensor-1.
- 49.Zündorf G., Reiser G. Calcium dysregulation and homeostasis of neural calcium in the molecular mechanisms of neurodegenerative diseases provide multiple targets for neuroprotection. Antioxid Redox Signal. 2011;14:1275–1288. doi: 10.1089/ars.2010.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berridge M.J. Calcium signalling remodelling and disease. Biochem Soc Trans. 2012;40:297–309. doi: 10.1042/BST20110766. [DOI] [PubMed] [Google Scholar]
- 51.Kim J., Hudson N.E., Springer T.A. Force-induced on-rate switching and modulation by mutations in gain-of-function von Willebrand diseases. Proc Natl Acad Sci U S A. 2015;112:4648–4653. doi: 10.1073/pnas.1501689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 53••.Mashaghi A., Kramer G., Bechtluft P., Zachmann-Brand B., Driessen A.J.M., Bukau B., Tans S.J. Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature. 2013;500:98–101. doi: 10.1038/nature12293. [DOI] [PubMed] [Google Scholar]; Provides the first direct evidence that the molecular basis of chaperone activity includes reshaping of protein folding landscapes, demonstrating that trigger factor binds partially folded structures that ultimately convert to the native state and promotes correct folding by protecting partially folded states from distant interactions leading to stable aggregates.
- 54.Wu Y.-J., Wu C.-H., Yeh A.Y.-C., Wen J.-D. Folding a stable RNA pseudoknot through rearrangement of two hairpin structures. Nucleic Acids Res. 2014;42:4505–4515. doi: 10.1093/nar/gkt1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serganov A., Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkler W.C., Breaker R.R. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 57.Stephenson W., Asare-Okai P.N., Chen A.A., Keller S., Santiago R., Tenenbaum S.A., Garcia A.E., Fabris D., Li P.T.X. The essential role of stacking adenines in a two-base-pair RNA kissing complex. J Am Chem Soc. 2013;135:5602–5611. doi: 10.1021/ja310820h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Savinov A., Perez C.F., Block S.M. Single-molecule studies of riboswitch folding. Biochim Biophys Acta. 2014;1839:1030–1045. doi: 10.1016/j.bbagrm.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anthony P.C., Perez C.F., García-García C., Block S.M. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci U S A. 2012;109:1485–1489. doi: 10.1073/pnas.1115045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neupane K., Yu H., Foster D.A.N., Wang F., Woodside M.T. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011;39:7677–7687. doi: 10.1093/nar/gkr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Frieda K.L., Block S.M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides a direct view of how folding of mRNA and binding of regulatory ligands couple with transcription to regulate gene expression in a dynamic process.
- 62.Brierley I., Gilbert R.J.C., Pennell S. Pseudoknot-dependent programmed — 1 ribosomal frameshifting: structures, mechanisms and models. In: Atkins J.F., Gesteland R.F., editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; New York: 2010. pp. 149–174. [Google Scholar]
- 63.Chen G., Chang K.-Y., Chou M.-Y., Bustamante C., Tinoco I. Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of −1 ribosomal frameshifting. Proc Natl Acad Sci U S A. 2009;106:12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansen T.M., Reihani S.N.S., Oddershede L.B., Sørensen M.A. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc Natl Acad Sci U S A. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Ritchie D.B., Foster D.A.N., Woodside M.T. Programmed −1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc Natl Acad Sci U S A. 2012;109:16167–16172. doi: 10.1073/pnas.1204114109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Revisits the role of RNA pseudoknots in determining programmed frameshifting efficiency. Provides evidence that RNA conformational plasticity, rather than mechanical stability as was widely believed, is a primary determinant of frameshifting levels.
- 66.De Messieres M., Chang J.-C., Belew A.T., Meskauskas A., Dinman J.D., La Porta A. Single-molecule measurements of the CCR5 mRNA unfolding pathways. Biophys J. 2014;106:244–252. doi: 10.1016/j.bpj.2013.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park S.-J., Kim Y.-G., Park H.-J. Identification of RNA pseudoknot-binding ligand that inhibits the −1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J Am Chem Soc. 2011;133:10094–10100. doi: 10.1021/ja1098325. [DOI] [PubMed] [Google Scholar]
- 68.Ritchie D.B., Soong J., Sikkema W.K.A., Woodside M.T. Anti-frameshifting ligand reduces the conformational plasticity of the SARS virus pseudoknot. J Am Chem Soc. 2014;136:2196–2199. doi: 10.1021/ja410344b. [DOI] [PubMed] [Google Scholar]
- 69••.Ngo T.T.M., Zhang Q., Zhou R., Yodh J.G., Ha T. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell. 2015;160:1135–1144. doi: 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Finds that nucleosomes unravel asymmetrically under tension and that the direction of unwrapping is controlled by DNA flexibility, demonstrating that the two sides of a nucleosome are orchestrated such that the opening of one end helps to stabilize the other end and revealing a novel mechanism for regulation of gene accessibility by DNA sequence and modifications.
- 70.Schlingman D.J., Mack A.H., Kamenetska M., Mochrie S.G.J., Regan L. Routes to DNA accessibility: alternative pathways for nucleosome unwinding. Biophys J. 2014;107:384–392. doi: 10.1016/j.bpj.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.La Porta A., Wang M.D. Optical torque wrench: angular trapping, rotation, and torque detection of quartz microparticles. Phys Rev Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 72.Sheinin M.Y., Li M., Soltani M., Luger K., Wang M.D. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun. 2013;4:2579. doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma J., Bai L., Wang M.D. Transcription under torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wen J.-D., Lancaster L., Hodges C., Zeri A.-C., Yoshimura S.H., Noller H.F., Bustamante C., Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qu X., Wen J.-D., Lancaster L., Noller H.F., Bustamante C., Tinoco I. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaiser C.M., Goldman D.H., Chodera J.D., Tinoco I., Bustamante C. The ribosome modulates nascent protein folding. Science. 2011;334:1723–1727. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77•.Goldman D.H., Kaiser C.M., Milin A., Righini M., Tinoco I., Bustamante C. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science. 2015;348:457–460. doi: 10.1126/science.1261909. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that force generated by nascent chain folding near the ribosome exit tunnel is sufficient to rescue stalling caused by a sequence still in the tunnel.
- 78••.Yan S., Wen J.-D., Bustamante C., Tinoco I. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; First direct measurement of ribosome-dependent mRNA dynamics during programmed frameshifting, demonstrating multiple ribosomal translocation attempts while in register with a slippery sequence.
- 79.Chen J., Petrov A., Johansson M., Tsai A., O’Leary S.E., Puglisi J.D. Dynamic pathways of −1 translational frameshifting. Nature. 2014;512:328–332. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim H.-K., Liu F., Fei J., Bustamante C., Gonzalez R.L., Tinoco I. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc Natl Acad Sci U S A. 2014;111:5538–5543. doi: 10.1073/pnas.1403457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo Z., Noller H.F. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sen M., Maillard R.A., Nyquist K., Rodriguez-Aliaga P., Pressé S., Martin A., Bustamante C. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155:636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83••.Cordova J.C., Olivares A.O., Shin Y., Stinson B.M., Calmat S., Schmitz K.R., Aubin-Tam M.-E., Baker T.A., Lang M.J., Sauer R.T. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158:647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]; Measures single-molecule degradation using different multidomain substrates and ClpXP variants to reveal how ATP hydrolysis and translocation steps of different sizes relate to solution degradation and the physical properties of substrate proteins.