Abstract
The 3D jury system has predicted the methyltransferase fold for the nsp13 protein of the SARS coronavirus. Based on the conservation of a characteristic tetrad of residues, the mRNA cap-1 methyltransferase function has been assigned to this protein, which has potential implications for antiviral therapy.
Main Text
The latest outbreak of the severe acute respiratory syndrome (SARS) epidemic has led to thousands of potentially lethally infected patients and hundreds of deaths. These numbers are likely to rise, and the spreading disease is already causing major medical and economical concerns. Meanwhile, the SARS coronavirus identified as the pathogen responsible for the disaster has been isolated, and its genome sequenced Marra et al. 2003, Rota et al. 2003.
We have applied the 3D jury meta predictor (Ginalski et al., 2003) to annotate the structure and function of proteins encoded by the viral positive-strand ssRNA. Novel fold recognition methods utilize the global network of independent structure prediction servers. Detection of patterns of structural similarity between diverse models is used to consistently select the correct fold from a set of borderline predictions. Such methods made a dramatic impact on the last critical assessment of protein structure prediction (CASP-5 experiment) conducted in the summer of 2002. One of the most interesting findings obtained during the SARS genome annotation process is a surprisingly reliable (3D jury score >100) assignment of the methyltransferase fold to the nsp13 (GI:30133975) domain located in the C-terminal part of the almost 7000 amino acid large pp1ab viral polyprotein (Figure 1) . Standard sequence comparison tools such as PSI-BLAST or RPS-BLAST applied using the conserved domain database (Marchler-Bauer et al., 2003) failed to assign any function to this domain. The domain belongs to the ancient family of AdoMet-dependent ribose 2′-O-methyltransferases, which has been adapted by numerous viruses before the three domains of life evolved form the last universal common ancestor (LUCA) (Feder et al., 2003). The enzymatic role of the protein was confirmed by the presence of the conserved tetrad of residues K-D-K-E essential for mRNA cap-1 (mGpppNm) formation.
Figure 1.
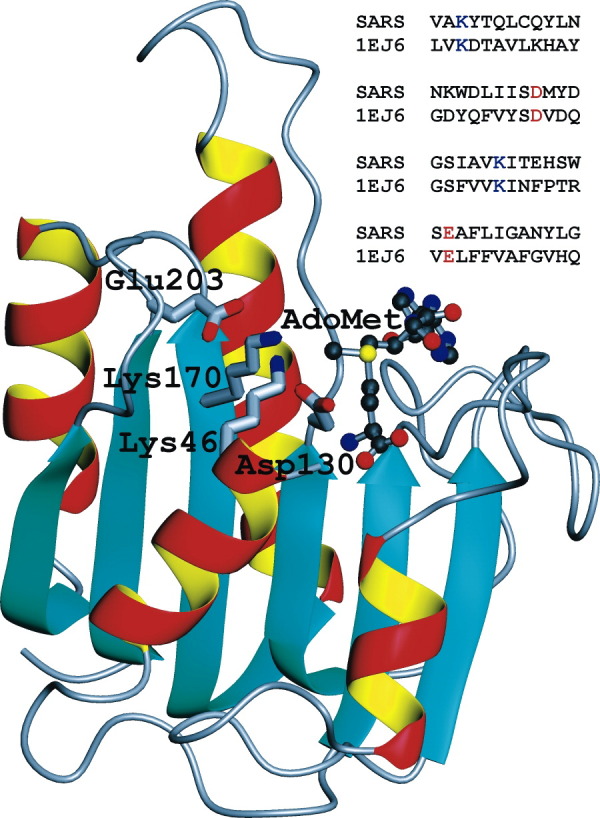
3D Model of the nsp13 Domain of the SARS Coronavirus pp1ab Polyprotein
This model is based on the reassigned (Bujnicki and Rychlewski, 2001) cap-1 methyltransferase of the reovirus λ2 protein (1ej6 [Reinisch et al., 2000]). While other templates (1eiz or 1ej0) obtained marginally higher 3D jury scores, the selected template had the lowest number of insertions and deletions. Side chains of the conserved tetrad of residues (K-D-K-E) essential for cap-1 methylation and the docked AdoMet cofactor are shown. Four blocks of aligned motifs containing the conserved, function-specific residues are shown in upper right corner.
The mRNA cap methylation is found indispensable for efficient replication of many viruses Bach et al. 1995, Woyciniuk et al. 1995, Vlot et al. 2002 and represents an active area for drug development. Nevertheless, direct inhibitors of the nsp13 enzyme may fail to suppress viral replication, as the cap-1 formation seems to be less critical than the preceding cap-0 (mGpppN) formation Latner et al. 2002, Wu and Guarino 2003. The existence of the cap-1-forming enzyme in the genome would suggest that the virus also requires the AdoMet-dependent cap-0 methyltransferase. Both functions can be inhibited by carbocyclic analogs of adenosine, such as Neplanocin A or 3-deazaneplanocin A, which interfere with the AdoMet-AdoHcy metabolism of the host cell De Clercq 1998, Bray et al. 2002. Those compounds could complement other therapeutic strategies aimed at blocking enzymatic functions such as the RNA-dependent RNA polymerase, the protease, or the helicase encoded by the SARS virus.
References
- Bach C., Cramer A., Scholtissek C. J. Gen. Virol. 1995;76:1025–1032. doi: 10.1099/0022-1317-76-4-1025. [DOI] [PubMed] [Google Scholar]
- Bray M., Raymond J.L., Geisbert T., Baker R.O. Antiviral Res. 2002;55:151–159. doi: 10.1016/s0166-3542(02)00018-9. [DOI] [PubMed] [Google Scholar]
- Bujnicki, J.M., and Rychlewski, L. (2001). Genome Biol. 2, RESEARCH0038. [DOI] [PMC free article] [PubMed]
- De Clercq E. Nucleosides Nucleotides. 1998;17:625–634. doi: 10.1080/07328319808005205. [DOI] [PubMed] [Google Scholar]
- Feder M., Pas J., Wyrwicz L.S., Bujnicki J.M. Gene. 2003;302:129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- Ginalski K., Elofsson A., Fischer D., Rychlewski L. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Latner D.R., Thompson J.M., Gershon P.D., Storrs C., Condit R.C. Virology. 2002;301:64–80. doi: 10.1006/viro.2002.1538. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., DeWeese-Scott C., Fedorova N.D., Geer L.Y., He S., Hurwitz D.I., Jackson J.D., Jacobs A.R., Lanczycki C.J., et al. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Jones, S.J., Astell, C.R., Holt, R.A., Brooks-Wilson, A., Butterfield, Y.S., Khattra, J., Asano, J.K., Barber, S.A., Chan, S.Y., et al. (2003). Science, in press. Published online May 1, 2003. 10.11261/science.1085953.
- Reinisch K.M., Nibert M.L., Harrison S.C. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., et al. Science, in press. Published online. 2003;May 1:2003. 10.11261science1055952. [Google Scholar]
- Vlot A.C., Menard A., Bol J.F. J. Virol. 2002;76:11321–11328. doi: 10.1128/JVI.76.22.11321-11328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyciniuk P., Linder M., Scholtissek C. Virus Res. 1995;35:91–99. doi: 10.1016/0168-1702(94)00085-q. [DOI] [PubMed] [Google Scholar]
- Wu X., Guarino L.A. J. Virol. 2003;77:3430–3440. doi: 10.1128/JVI.77.6.3430-3440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


