Abstract
Phylosymbiosis was recently formulated to support a hypothesis-driven framework for the characterization of a new, cross-system trend in host-associated microbiomes. Defining phylosymbiosis as ‘microbial community relationships that recapitulate the phylogeny of their host’, we review the relevant literature and data in the last decade, emphasizing frequently used methods and regular patterns observed in analyses. Quantitative support for phylosymbiosis is provided by statistical methods evaluating higher microbiome variation between host species than within host species, topological similarities between the host phylogeny and microbiome dendrogram, and a positive association between host genetic relationships and microbiome beta diversity. Significant degrees of phylosymbiosis are prevalent, but not universal, in microbiomes of plants and animals from terrestrial and aquatic habitats. Consistent with natural selection shaping phylosymbiosis, microbiome transplant experiments demonstrate reduced host performance and/or fitness upon host–microbiome mismatches. Hybridization can also disrupt phylosymbiotic microbiomes and cause hybrid pathologies. The pervasiveness of phylosymbiosis carries several important implications for advancing knowledge of eco-evolutionary processes that impact host–microbiome interactions and future applications of precision microbiology. Important future steps will be to examine phylosymbiosis beyond bacterial communities, apply evolutionary modelling for an increasingly sophisticated understanding of phylosymbiosis, and unravel the host and microbial mechanisms that contribute to the pattern. This review serves as a gateway to experimental, conceptual and quantitative themes of phylosymbiosis and outlines opportunities ripe for investigation from a diversity of disciplines.
Keywords: symbiosis, phylosymbiosis, microbiome, host–microbe interactions
1. Introduction
The last decade has brought renewed interest in the complexity of microorganisms living in association with hosts, yielding a number of new empirical results, philosophical concepts and research opportunities [1,2]. Any discussion on the study of host–microbiome interactions must begin with clear definitions. Here, we use the term symbiosis (sym—‘together’, bios—‘life’ in Greek) to encompass associations between two or more organisms of different species and without restriction to the length of time of the association or phenotypes produced by the interacting species. Since temporal and functional variation in symbiosis is context-dependent, symbiotic interactions can include a range of obligatory, facultative, transient and permanent associations with varying degrees of specificity and functional costs and benefits.
The last two decades of research and technological advances have placed microbial symbiosis as a nexus of many subdisciplines within and beyond biology. Scholars now have a suite of tools and increased awareness of the major questions to be answered. These include holistic approaches for the identification of ecological [3] and host [4–7] drivers of microbial taxonomic and functional diversity, as well as reductionist approaches that provide evolutionary and mechanistic insights into transmission processes [8] and phenotypic outcomes of symbiosis [1]. The abundance of empirical and theoretical investigations on the ecology and evolution of simple symbioses also comprise fertile ground to build a foundation for the microbiome field that studies frequently complex associations between hosts and their multiple microbial associates. One rapidly growing research area across diverse systems is the recently defined pattern of phylosymbiosis [9]. This review aims to synthesize the topic to provide: (i) a long-lasting definition of the term; (ii) a practical guide to test phylosymbiosis; (iii) an overview of the prevalence of phylosymbiosis; (iv) a discourse on the biological significance of phylosymbiosis; and (v) future directions in phylosymbiosis research.
2. What is and what is not phylosymbiosis?
We use the following quote to describe our initial and basic definition of phylosymbiosis, namely ‘microbial community relationships that recapitulate the phylogeny of their host’ [9]. Phylosymbiosis is first and foremost a significant association between host phylogenetic relationships and host-associated microbial community relationships wherein ‘phylo’ refers to the host clade and ‘symbiosis’ refers to the microbial community in or on the host.
Prior to the introduction of the term phylosymbiosis in a study of Nasonia parasitoid wasp species [9], early investigations specified relationships between host phylogenies or genetic distances with microbial beta diversity in maize [10], insects [5,11] and mammals [4,12]. These studies used bacterial 16S rRNA gene sequencing across multiple host species to demonstrate that closely related species harbour more similar microbiomes than distantly related species. For example, the sister species N. giraulti and N. longicornis diverged approximately 0.4 Ma and harbour more similar 2nd instar larval, pupal and adult microbiomes compared with the microbiome in their outgroup species N. vitripennis [9,11], which diverged approximately 1.0 Ma from the two sister species [13].
Phylosymbiosis may arise from stochastic and/or deterministic evolutionary and ecological forces. For example, stochastic effects include dispersal fluctuations in microbial communities (ecological drift) or shifts in host geographical ranges [14]. Phylosymbiosis can also be shaped by ecological [15–17] and dietary [4] niche variation across host lineages. Deterministic effects include microbial colonization preferences for certain host backgrounds or host regulation in which microbial community composition is influenced by host trait(s) [18]. The first study linking phylosymbiotic patterns to the function of specific host genes found that knockdown of the Hydra armenin antimicrobial peptide disrupted phylosymbiosis [6] commonly observed in several freshwater and laboratory Hydra species [19]. Although phylosymbiosis can potentially arise from long-term, intimate host–microbe associations over evolutionary time, such as through host–microbe coevolution, codiversification [20] and cospeciation [21], importantly it may also be driven by relatively short-term changes in microbiome composition. Indeed, a recent Drosophila melanogaster study revealed the effects of gut microbiome changes on host genomic divergence in as little as five generations [22]. This suggests that rather than being passive agents of phylosymbiosis, microbial communities have the potential to induce host genomic changes that could, in turn, impact the establishment, maintenance or breakdown of phylosymbiosis.
While phylosymbiosis distinguishes itself from non-phylosymbiosis by a significant degree of association between host phylogenetic and microbiome community relationships, it is not universal (§5) and therefore provides a testable hypothesis. Determining the presence of phylosymbiosis is a first step preceding further investigations into eco-evolutionary mechanisms, such as the nature of species–species associations, selective or neutral forces driving phylosymbiosis, and the (in)consequences of the pattern on the host and microbial phenotypes. If phylosymbiosis results from an evolutionary selective pressure, then decreases in host or microbial fitness are expected upon host exposure to microbiomes from different host lineages in an evolutionarily informed manner. Evolutionary selective pressures that result in phylosymbiosis could drive the spread of host traits that regulate microbiome composition or microbial traits that enhance host colonization. In this general light, we refer to ‘functional phylosymbiosis' when the host and/or microbial phenotypes impact or are impacted by phylosymbiotic associations.
Interspecific microbiome transplant experiments are useful in elucidating functional phylosymbiosis. A large-scale phylosymbiosis investigation spanning 24 species across four laboratory-reared host clades (Nasonia wasps, Drosophila flies, mosquitoes and Peromyscus deer mice) demonstrated that interspecific transplants of gut microbial communities between Peromyscus species decreased dry matter digestibility and increased food intake, while transplants between Nasonia species markedly lowered survival to adulthood by nearly half [23]. In addition, interspecific microbiomes are more costly to Nasonia larval growth and pupation than intraspecific microbiomes [24]. Similarly, reciprocal maternal symbiont transplants between two wild, sympatric Ontophagus dung beetle species caused developmental delay and elevated mortality in non-native hosts that persisted to the next generation [25]. Collectively, phylosymbiotic associations that impact host fitness support the premise that hosts are adapted to their native microbiomes rather than non-native microbiomes, although more studies are needed to confirm these associations and effects in captive and wild host populations.
Hybridization between host species causes host–microbiome mismatches since combining independently evolved host genotypes in a hybrid may cause a breakdown in either microbial colonization preferences for certain hosts or host control of the microbiome. As demonstrated in Nasonia [9], house mice [26] and whitefish [27], hybrids have an altered microbiome relative to the parental microbiome, suggesting a reduced capacity for hosts to regulate their microbiomes and an increased capacity for pathogenic microbes to bloom. These breakdowns in host–microbiome interactions can associate with maladaptive phenotypes in hybrids including immune dysfunction, pathology, inviability and sterility [9,26] that can reduce interbreeding between species or populations. In Nasonia, the lethality of hybrids between the older species pair was rescued by germ-free rearing and restored by feeding an inoculum of select, resident gut bacterial species from parents to germ-free hybrids [9]. By contrast, hybrids between a younger Nasonia species pair did not have an altered microbiome nor suffer functional costs. Collectively, the results from interspecific microbiome transplant experiments and host hybridization studies illustrate that host–microbiome interactions across host species can have important functional consequences that impact evolutionary events within and between species, including wedging host populations into species.
Having now summarized phylosymbiosis, we briefly accentuate what phylosymbiosis is not, for clarity. Phylosymbiosis does not necessarily imply vertical transmission, mutualistic interactions or evolutionary splitting from a common ancestor via coevolution, cospeciation, co-diversification or cocladogenesis. Although these processes may lead to phylosymbiosis, the pattern may alternatively arise by antagonistic interactions and/or horizontal microbial transmission whereby interactions between hosts and environmental microbes establish phylosymbiosis anew each generation. As such, phylosymbiosis has varied underpinnings subject to empirical investigation, and it may appear at certain points of time and space rather than be stable throughout a host's entire lifespan.
3. A practical guide to studying phylosymbiosis
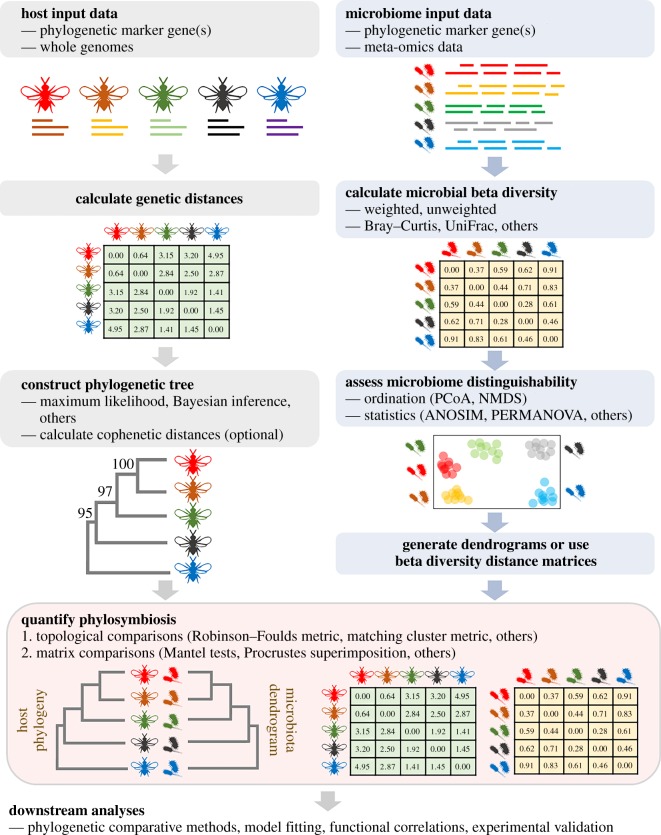
Investigations of phylosymbiosis vary in approach (qualitative versus quantitative), methodology and statistical power [18]. Thus, a clear, consistent and robust workflow to detect phylosymbiosis is desirable for newcomers and experts alike. Here, we suggest a comprehensive workflow for examining phylosymbiosis (figure 1).
Figure 1.
Sequential overview of bioinformatic methods commonly used for phylosymbiosis analyses. (Online version in colour.)
(a). Host taxa and input data
Because phylosymbiosis detection involves the collection of replicated samples across multiple taxa, both optimization of statistical sensitivity [28] and specificity [18], as well as minimization of sequencing batch effects, are crucial for differentiating between noise and signal. Although our 2016 study showed that rooted trees with four Nasonia species are sufficient to detect phylosymbiosis within the clade [23], we suggest the use of appropriate power and effect size analyses (reviewed in [29] for microbiome data) to determine sufficient replicates and taxa for the optimization of statistical power [28]. Sampling multiple individuals per species will help resolve noise from signal in microbial community relationships, but further study is required on how replicates of inter- and intraspecies samples are best used in studying phylosymbiosis across host clades that can vary in divergence times. If available, experimental designs of successful phylosymbiosis studies with similar sample types can also be adapted accordingly [30]. Previous studies have successfully detected phylosymbiosis in host taxa spanning approximately 0.3–100 Myr of evolutionary history [21,23], and whether longer times since a last common ancestor impacts phylosymbiosis detection requires further study. Nucleotide or amino acid sequence(s) from host species can be used to generate a phylogenetic or phylogenomic tree that is confidently supported at branching nodes with bootstrap [31] or other measures [32] and across several phylogenetic inference methods (e.g. maximum likelihood [33] and Bayesian inference [34]). Because an accurate host phylogenetic topology is essential for evaluating phylosymbiosis, the tree should be free from systematic artefacts such as long-branch attraction and polytomies should be resolved in the host phylogeny when possible. As methods used to reconstruct a host phylogeny from a sequence alignment have been extensively reviewed [35], we will not discuss them further here. With a host evolutionary tree, pairwise host distances can also be represented as cophenetic distances, computed as the sum of branch lengths connecting a pair of terminal nodes on a phylogenetic tree [36].
(b). Microbiome input data
Phylosymbiosis analysis requires microbial diversity data from each host lineage. Short-read sequencing of microbial phylogenetic marker genes (e.g. 16S rRNA gene) is common and economical for microbial profiling. Processed sequenced reads can be analysed by one of two current methods. First, they can be clustered into operational taxonomic units (OTUs) at different sequence cutoffs (e.g. 97% and 99%) with and/or without reference sequence database [37,38]. OTU clustering cutoffs reflect genetic distances between taxa over evolutionary time and may affect phylosymbiosis detection [39]; such variability has also been observed in practice (reviewed in [18]). Second, reads can be resolved into amplicon sequence variants (ASVs) without clustering, which may offer single-nucleotide resolution, though sequencing error rates should be accounted for [40]. For the greatest sensitivity in phylosymbiosis assessment, meta-omics datasets are advantageous because finer-scale taxonomic and functional profiling can be achieved [41]. Metagenomic sequence data were used to demonstrate viral phylosymbiosis in Nasonia [42] as well as the varying effects of host phylogeny and ecology on the composition and functions of non-human, primate gut microbiomes [43,44].
(c). Microbial beta diversity measures
Microbial beta diversity, which measures dissimilarities in microbial composition and structure across host samples, is conventionally used to measure phylosymbiosis. Binary measures, such as Jaccard distance and Sørensen–Dice distance [45,46], are calculated with OTU presence/absence data. Quantitative descriptors of OTU abundances can also compute beta diversity, including the Bray–Curtis dissimilarity [47] derived from Motyka et al.'s coefficient [48]. Phylogeny-based metrics, such as weighted and unweighted unique fraction (UniFrac), use phylogenetic distances between communities (samples) to calculate microbial community differences, necessitating the use of a microbial phylogenetic tree as input to calculate the total community distance [49].
Because beta diversity metrics reflect different aspects of dissimilarity, the choice of metric is study specific and depends partly on the microbial composition and evolutionary history of the lineages studied. Binary metrics based on presence/absence are more sensitive to variations in rare taxa and were implemented to study host specificity of sponge microbiomes, where rare taxa comprised more than 90% of distinct OTUs [50]. Binary metrics may also be sensitive to recent microbial diversification because recently diverged OTUs/ASVs will exert the same effect as OTUs/ASVs with a longer divergence history [39]. By contrast, quantitative metrics are more sensitive to variations in abundant taxa. Besides taxonomy-based phylosymbiosis studies [23,51–53], quantitative metrics have also been applied to metagenomics data [42,43]. Metrics that consider phylogenetic relationships between OTUs, such as UniFrac distances, [54] are applied in many other phylosymbiosis studies, including bats [55], corals [20] and mammals [4,43].
Microbiome distinguishability, or the characteristic of being able to significantly differentiate microbial communities of host lineages under evaluation, is a prerequisite for phylosymbiosis and should be tested before evaluating the phylosymbiosis prediction that more similar host species harbour more similar microbiomes [20,23,51–53]. Microbiome distinguishability can be visualized from beta diversity data and categorical sample grouping data using ordination plots, such as principal coordinate analysis (PCoA) and non-metric multidimensional scaling (NMDS) plots [56]. In addition, microbiome distinguishability can be further evaluated using typically non-parametric multivariable analyses, such as analysis of similarities (ANOSIM) [57] and variants of permutational multivariate analysis of variance (PERMANOVA) [58]. Specific pairwise comparisons of intra- and interspecific microbial beta diversity distances can also be performed with an appropriate non-parametric two-sample test [23].
(d). Quantifying phylosymbiosis
The determination of phylosymbiosis relies on evaluating a significant association between host phylogenetic relationships and host-associated microbial community distances. To this end, topological congruency tests directly compare topologies of a host phylogenetic tree and a microbiome dendrogram [23,42,51–53,59]. To generate a hierarchical dendrogram, several agglomerative hierarchical clustering methods (reviewed in [56]) can cluster microbial beta diversity distances. The most commonly used method, unweighted pair group method with arithmetic mean (UPGMA), performs pairwise sample clustering from their average dissimilarity values and gives all samples equal weights [60]. Compared with linkage clustering approaches, UPGMA prioritizes relationships among groups over individual samples [56]. By assigning equal weights to all samples, UPGMA assumes that samples in each group are representative of groups in the larger reference population [56]. As such, it may be sensitive to sample sizes and may generate unstable topologies with imbalanced data where some groups are oversampled while some are undersampled. Newer clustering methods, such as the phylogenetically aware squash clustering method, directly compute distances between samples (rather than differences between beta diversity distances) based on their positions on a microbial phylogenetic tree [61]. In general, the effects of clustering methods on phylosymbiosis detection require further study.
Topological comparison metrics, such as the Robinson–Foulds metric and the more robust and sensitive matching cluster metric, are frequently used to detect phylosymbiosis [23,42,51,52,59,62]. Robinson–Foulds analyses the distance between two trees as the smallest number of operations required to convert one topology to the other [63], while matching cluster considers congruency at the subtree level and is, therefore, a more refined evaluation of small topological changes that affect incongruence [64]. Statistical significance (p-values) has been evaluated by determining the probability of 100 000 randomized bifurcating dendrogram topologies yielding equivalent or more congruent phylosymbiotic patterns than the microbiome dendrogram [23]. Moving forward, improved randomization techniques that preserve conspecific relationships will be useful in reducing false positives. Normalized Robinson–Foulds and matching cluster scores can be calculated as the number of differences between the two topologies divided by the total possible congruency scores for the two trees, with normalized distances ranging from 0 (complete congruence) to 1 (complete incongruence) [23].
Matrix correlation methods identify phylosymbiosis by comparing the similarities between host-derived and microbial-derived distance matrices. Methods implemented in phylosymbiosis studies [20,21,39,50,65–72] include variations of the Mantel test, which statistically evaluates the linear correlation between all corresponding elements from two independent matrices by permutation [73] and the more powerful Procrustean superimposition approach, which rotates and fits two matrices to minimize their differences association [74]. Partial Mantel tests [75] measuring correlations between two matrices while controlling for the effects of a third variable described in another matrix are also used to evaluate associations between microbial communities and multiple aspects of host characteristics, such as phylogeny, identity, genetic distances and geographical distances [39,66,67,69].
Although both topology-based and matrix-based tests are specific and sensitive enough to detect phylosymbiosis in a variety of empirical cases, there are several differences between them. Topological comparison metrics do not use branch length information as there is no a priori reason to assume rates of host evolution in each lineage should equal rates of ecological community change in the microbiome. Indeed, rates of microbiome change may be expected to be far more rapid than the gradual evolution of host genetic changes. As such, tests of topology without relative branch lengths are conservative relative to matrix correlation methods that directly rely on comparisons of host genetic divergence with microbial community dissimilarity. A simulation analysis suggested that the Mantel test has higher sensitivity and power than the Robinson–Foulds metric when phylosymbiosis is based on the assumption of microbial preferences for a host trait [19]. The practical relevance of this conclusion is not clear because phylosymbiosis will arise from reasons other than microbial colonization preferences, such as host preferences, neutral processes and microbe–microbe interactions. Moreover, the performance between the Mantel test and the more sensitive topology-based matching cluster distance was not evaluated in this simulation, and such comparisons are likely to yield different insights. Systematic benchmarking of type I and II error rates of phylosymbiosis measurement methods across various possible scenarios will aid experimental design and result interpretation. As such, research opportunities for the development and implementation of improved phylosymbiosis detection methods are ample.
(e). Parameter selection
Phylosymbiosis detection involves the selection of various parameters, such as OTU identity cutoff, beta diversity metric, clustering method and congruency test, each with their strengths and limitations that will vary with study design and questions. Although various parameter combinations can be tested and compared simultaneously [39], in the case when only a few of all possible parameter combinations detect phylosymbiosis, we recommend cautious interpretation of results with respect to the chosen parameters. If available, results should also be compared to those from previous phylosymbiosis studies with similar sample types using the same parameter combinations. Experimental replication is also necessary to confirm phylosymbiosis, especially when it is not consistently detected.
(f). Phylogenetic comparative methods
The effects of phylogenetic signal, defined as ‘a tendency for related species to resemble each other more than they resemble species drawn at random from the tree’ [76], on univariate traits (e.g. microbial alpha diversity) have been examined in parallel with phylosymbiosis studies [66,67]. Phylogenetic signal indices like Pagel's λ [77] and Blomberg's K [78] are based on a random Brownian model of trait evolution [79], but can also be used with and compared to more complex models that take into account natural selection. Although these methods are less commonly used on multivariable data and have not yet been applied to evaluate phylosymbiosis explicitly, they are promising alternatives for not only examining host phylogenetic signal on microbial beta diversity, but also testing evolutionary models relevant to phylosymbiosis.
Phylogenetic comparative methods, such as phylogenetic independent contrasts [79] and phylogenetic generalized linear mixed models (pGLMMs) [80], predict the evolutionary correlation between two or more discrete or continuous traits given a known phylogeny and an evolutionary model. These can also be integrated into phylosymbiosis studies. pGLMMs were recently implemented in coral microbiome [20] and passerine feather microbiome studies [71] to examine the effects of latitude and colony size on coral alpha diversity, cophylogenetic coral–bacteria relationships, and relationships between alpha diversity and relative abundances of bacteriocin-producing bacteria and keratinolytic feather damaging bacteria. Because phylosymbiosis may arise from ecological (among other) forces, these methods can be useful in understanding the various ecological interactions that possibly underlie phylosymbiosis.
Overall, as meta-omics and trait evolution analyses become more widely applicable to phylosymbiosis, one compelling direction of future phylosymbiosis investigations in silico is to venture beyond host phylogenetic effects on microbial diversity to resolve linkages between host phylogeny, host functions, microbial diversity, microbial functions, selective forces and environmental factors.
4. The prevalence of phylosymbiosis
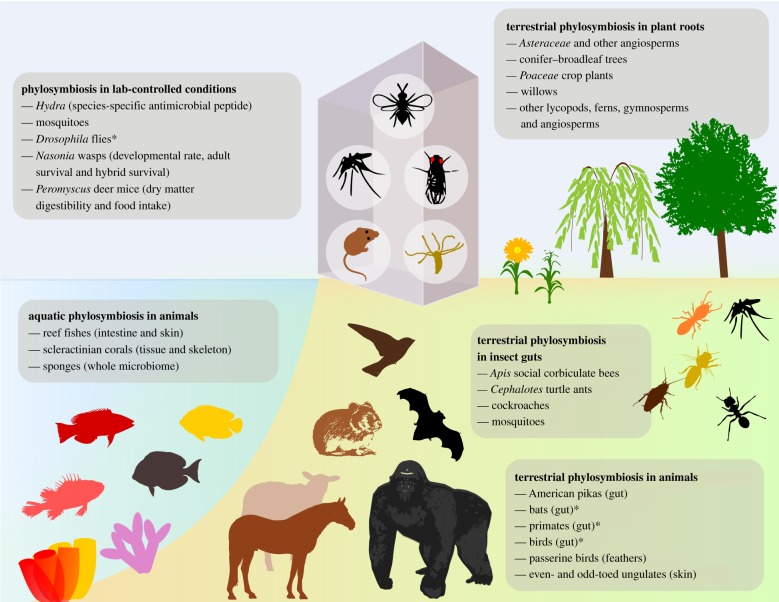
A major goal of microbiome science is to find general paradigms and rules, if any, that are comparable across varied systems. In this light, phylosymbiosis is emerging as a bona fide trend because of its frequent recurrence across eukaryotic host systems (figure 2). Phylosymbiosis in insects include viromes of Nasonia parasitoid jewel wasps [42] and gut microbiomes of cockroaches, termites [81], lab-reared [23] and wild mosquitoes [59], Cephalotes turtle ants [39] and Apis social corbiculate bees [69]. In Drosophila flies, phylosymbiosis patterns are either weakly supported [23] or not detected [82] in laboratory strains and wild populations.
Figure 2.
Representative diversity of phylosymbiosis across host species, tissues, habitats and functions. Asterisks denote taxa with mixed evidence of phylosymbiosis. (Online version in colour.)
The first phylosymbiosis study on mammalian gut microbiomes [4] demonstrated the effects of animal phylogeny and diet on gut microbial community dissimilarity [12,21,23,39,70,83]. Studies focusing on gut microbiomes of specific animal groups detected phylosymbiosis in American pikas [51] and Peromyscus deer mice [23,52], no phylosymbiosis in western chipmunks [84], and mixed evidence of phylosymbiosis in primates [17,43,44,70], bats [55,85] and birds [62,68,86,87]. A recent large-scale study revealed much stronger effects of host phylogeny and diet on the gut microbiomes of non-flying mammals than those of bats and birds [72]. Besides gut or faecal microbiomes, animal surface microbiomes have also been analysed for phylosymbiotic associations [88], which for example occur on mammalian skin [53] and passerine feathers [71], but not on amphibian skin [3]. A meta-analysis of phylosymbiosis literature highlighted an increased prevalence of the trend in microbiomes inhabiting internal host compartments in relation to those inhabiting external host compartments [18]. However, the finding may be inherently biased due to the larger number of studies investigating phylosymbiosis in the gut in relation to other external host compartments.
Beyond terrestrial and associated habitats, research interest in phylosymbiotic associations in aquatic habitats is steadily growing (figure 2), spanning global sponge microbiome surveys [67,89,90] and taxon-specific sponge surveys [50,65,66] with mixed results. Two previous studies in sponges showed significant correlations between host phylogeny and microbial beta diversity [66,67]. In Australian scleractinian corals, phylosymbiosis was generally observed in tissue and skeleton compartments, but not mucus specimens that are predominantly influenced by the environment [20], suggesting different anatomical impacts on the pattern. Phylosymbiosis and host dietary impacts also occur on the skin microbiomes of 44 fish species from the western Indian Ocean [91], but do not exist on the surface microbiomes of sympatric kelp species [92].
Phylosymbiosis has been assessed in plants, mainly to distinguish the effects of host phylogeny and soil determinants on microbial beta diversity. A comparative analysis of lycopods, ferns, gymnosperms and angiosperms across a coastal tropical soil chronosequence indicated host phylogeny is a secondary but statistically significant factor shaping root-associated bacterial community structure, after soil age [15]. More taxonomically and/or spatially restricted surveys have also revealed phylosymbiosis between rhizobacterial communities and Poaceae crop plants [93], endosphere bacterial communities and 30 plant species [94], rhizosphere-associated fungal communities and willows from hydrocarbon-contaminated soils [95], root-associated eumycotan fungal communities and Asteraceae flowering plants in a dry grassland [96], ectomycorrhizal fungal communities and conifer–broadleaf forest trees [97], and ectomycorrhizal fungal communities and Estonian Salicaceae willows [98]. Contrarily, qualitative incongruency between Brassicaceae host phylogeny and their root microbiomes has been observed [99], whereas non-statistically significant phylosymbiotic correlations have been reported in other plant microbiome studies [16,100].
5. Significance and future directions of phylosymbiosis
Microbiome research will continue to be revolutionized by the multi-omics era, where a deluge of data has enabled unprecedented insights into the extensive taxonomic, genetic and functional composition of microbial communities and their associated hosts. Such large-scale accumulation of empirical and theoretical findings can potentiate the development of new hypotheses, unifying concepts and frameworks across diverse host–microbiome systems. Indeed, the recurrence of phylosymbiosis across host systems lends itself to large comparative surveys across kingdoms of life that may uncover taxonomic range restrictions of phylosymbiosis as well as the environmental parameters (e.g. soil and water properties) and ecological interactions (e.g. diet and predator–prey relationships) that determine the boundaries of where and when phylosymbiosis occurs. If the microbiome field will have general trends to test in new systems, phylosymbiosis is well poised for this circumstance.
Phylosymbiosis distinguishes itself from non-phylosymbiosis by characterizing a significant degree of association between host phylogenetic and microbiome community relationships. It provides a testable hypothesis, reflects the variation likely to be seen in nature and is amenable to explanation by mechanisms that require further investigation. The determination of whether phylosymbiosis is present or not is a first step preceding further investigations into mechanistic details, such as the nature of species–species associations and the type(s) of ecological and evolutionary genetic processes underpinning phylosymbiosis.
Phylosymbiosis also engenders a holistic view of ecology and evolution in which hosts are communities or holobionts whose microbial members can contribute to genetic and phenotypic variation subject to natural selection. Several questions have been conventionally overlooked. For example, what are the microbial effects on host allele frequencies? Does host gene flow in natural populations impact microbiome variation and phylosymbiosis? Is phylosymbiosis associated with the acceleration or deceleration of host speciation? What are the genetic and mechanistic factors that regulate phylosymbiosis and how do these factors vary across populations or species? Collectively, studies determining the magnitude of ecological, evolutionary and genetic forces in structuring phylosymbiosis represent an important area of future research.
6. Conclusion
Phylosymbiosis defines a link between host evolutionary relationships and microbial diversity that is quantifiable and applicable across living systems. As research in this area proliferates, a definition, conceptual framework and workflow for assessing phylosymbiosis will facilitate identification of phylosymbiotic host–microbe interactions. Future cause-and-effect studies of phylosymbiosis will bring a further mechanistic understanding of the evolutionary, genetic and molecular bases. Just as no mature theory of evolutionary genetics was possible until we understood the mode of inheritance, no mature principle of evolutionary ecology for host-associated microbiomes seems possible until we understand the general mechanisms establishing host–microbiome associations.
Supplementary Material
Acknowledgements
We thank Andrew Brooks, Brittany Leigh, Edward van Opstal and three anonymous reviewers for helpful reviews of previous versions of the manuscript.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the National Science Foundation (grant no. DEB 1046149) and the Vanderbilt Microbiome Initiative.
References
- 1.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Theis KR, et al. 2016. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems 1, e00028-16 ( 10.1128/mSystems.00028-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bletz MC, Archer H, Harris RN, McKenzie VJ, Rabemananjara FCE, Rakotoarison A, Vences M. 2017. Host ecology rather than host phylogeny drives amphibian skin microbial community structure in the biodiversity hotspot of Madagascar. Front. Microbiol 8, 1530 ( 10.3389/fmicb.2017.01530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320, 1647–1651. ( 10.1126/science.1155725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colman DR, Toolson EC, Takacs-Vesbach C. 2012. Do diet and taxonomy influence insect gut bacterial communities? Mol. Ecol. 21, 5124–5137. ( 10.1111/j.1365-294X.2012.05752.x) [DOI] [PubMed] [Google Scholar]
- 6.Franzenburg S, Walter J, Kunzel S, Wang J, Baines JF, Bosch TCG, Fraune S. 2013. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl Acad. Sci. USA 110, E3730 ( 10.1073/pnas.1304960110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bost A, Martinson VG, Franzenburg S, Adair KL, Albasi A, Wells MT, Douglas AE. 2018. Functional variation in the gut microbiome of wild Drosophila populations. Mol. Ecol. 27, 2834–2845. ( 10.1111/mec.14728) [DOI] [PubMed] [Google Scholar]
- 8.Funkhouser LJ, Bordenstein SR. 2013. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 11, e1001631 ( 10.1371/journal.pbio.1001631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brucker RM, Bordenstein SR. 2013. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667–669. ( 10.1126/science.1240659) [DOI] [PubMed] [Google Scholar]
- 10.Bouffaud M, Kyselkova M, Gouesnard B, Grundmann G, Muller D, Moenne-Loccoz Y. 2012. Is diversification history of maize influencing selection of soil bacteria by roots? Mol. Ecol. 21, 195–206. ( 10.1111/j.1365-294X.2011.05359.x) [DOI] [PubMed] [Google Scholar]
- 11.Brucker RM, Bordenstein SR. 2012. The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution 66, 349–362. ( 10.1111/j.1558-5646.2011.01454.x) [DOI] [PubMed] [Google Scholar]
- 12.Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, Hugenholtz P, Achtman M. 2010. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 8, e1000546 ( 10.1371/journal.pbio.1000546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werren JH, et al. 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327, 343–348. ( 10.1126/science.1178028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, Nachman MW. 2017. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc. Natl Acad. Sci. USA 114, 13 768–13 773. ( 10.1073/pnas.1700122114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeoh YK, Dennis PG, Paungfoo-Lonhienne C, Weber L, Brackin R, Ragan MA, Schmidt S, Hugenholtz P. 2017. Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat. Commun. 8, 1–9. ( 10.1038/s41467-016-0009-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erlandson S, Wei X, Savage J, Cavender-Bares J, Peay K. 2018. Soil abiotic variables are more important than Salicaceae phylogeny or habitat specialization in determining soil microbial community structure. Mol. Ecol. 27, 2007–2024. ( 10.1111/mec.14576) [DOI] [PubMed] [Google Scholar]
- 17.Grieneisen LE, Charpentier MJ, Alberts SC, Blekhman R, Bradburd G, Tung J, Archie EA. 2019. Genes, geology and germs: gut microbiota across a primate hybrid zone are explained by site soil properties, not host species. Proc. R. Soc. B 286, 20190431 ( 10.1098/rspb.2019.0431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazel F, Davis KM, Loudon A, Kwong WK, Groussin M, Parfrey LW. 2018. Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems 3, e00097-18 ( 10.1128/mSystems.00097-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraune S, Bosch TCG. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl Acad. Sci. USA 104, 13 146–13 151. ( 10.1073/pnas.0703375104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, Thurber RV, Zaneveld JR. 2018. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9, 4921 ( 10.1038/s41467-018-07275-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, Alm EJ. 2017. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 8, 14319 ( 10.1038/ncomms14319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudman SM, et al. 2019. Microbiome composition shapes rapid genomic adaptation of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 116, 20 025–20 032. ( 10.1073/pnas.1907787116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR. 2016. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 ( 10.1371/journal.pbio.2000225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Opstal EJ, Bordenstein SR. 2019. Phylosymbiosis impacts adaptive traits in Nasonia wasps. MBio 10, e00887-19 ( 10.1128/mBio.00887-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker ES, Dury GJ, Moczek AP. 2019. Transgenerational developmental effects of species-specific, maternally transmitted microbiota in Onthophagus dung beetles. Ecol. Entomol. 44, 274–282. ( 10.1111/een.12703) [DOI] [Google Scholar]
- 26.Wang J, et al. 2015. Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat. Commun. 6, 6440 ( 10.1038/ncomms7440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sevellec M, Laporte M, Bernatchez A, Derome N, Bernatchez L. 2019. Evidence for host effect on the intestinal microbiota of whitefish (Coregonus sp.) species pairs and their hybrids. Ecol. Evol. 9, 11 762–11 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen J. 1988. Statistical power analysis for the behavioral sciences, 2nd edn Hillsdale, NJ: Lawrence Erlbaum Associates. [Google Scholar]
- 29.Debelius J, Song SJ, Vazquez-Baeza Y, Xu ZZ, Gonzalez A, Knight R. 2016. Tiny microbes, enormous impacts: what matters in gut microbiome studies? Genome Biol. 17, 217 ( 10.1186/s13059-016-1086-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conesa A, et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 ( 10.1186/s13059-016-0881-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. ( 10.1111/j.1558-5646.1985.tb00420.x) [DOI] [PubMed] [Google Scholar]
- 32.Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. ( 10.1080/10635150600755453) [DOI] [PubMed] [Google Scholar]
- 33.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 34.Mau B, Newton MA. 1997. Phylogenetic inference for binary data on dendograms using Markov chain Monte Carlo. J. Comput. Graph. Stat. 6, 122–131. ( 10.1080/10618600.1997.10474731) [DOI] [Google Scholar]
- 35.Wiley EO, Lieberman BS. 2012. Phylogenetics: the theory and practice of phylogenetics, 2nd edn Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 36.Sokal RR, Rohlf FJ. 1962. The comparison of dendrograms by objective methods. Taxon 11, 33–40. ( 10.2307/1217208) [DOI] [Google Scholar]
- 37.Rideout JR, et al. 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2, e545 ( 10.7287/peerj.545v0.2/reviews/1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopylova E, et al. 2016. Open-source sequence clustering methods improve the state of the art. mSystems 1, e00003-15 ( 10.1128/mSystems.00003-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders JG, Powell S, Kronauer DJC, Vasconcelos HL, Frederickson ME, Pierce NE. 2014. Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol. Ecol. 23, 1268–1283. ( 10.1111/mec.12611) [DOI] [PubMed] [Google Scholar]
- 40.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. ( 10.1038/ismej.2017.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medina M, Sachs JL. 2010. Symbiont genomics, our new tangled bank. Genomics 95, 129–137. ( 10.1016/j.ygeno.2009.12.004) [DOI] [PubMed] [Google Scholar]
- 42.Leigh BA, Bordenstein SR, Brooks AW, Mikaelyan A, Bordenstein SR. 2018. Finer-scale phylosymbiosis: insights from insect viromes. mSystems 3, e00131-18 ( 10.1128/mSystems.00131-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amato KR, et al. 2019. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 13, 576–587. ( 10.1038/s41396-018-0175-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amato KR, et al. 2019. Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol. 20, 201 ( 10.1186/s13059-019-1807-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dice LR. 1945. Measures of the amount of ecologic association between species. Ecology 26, 297–302. [Google Scholar]
- 46.Sørensen TJ. 1948. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Kongelige Danske Videnskabernes Selskab 5, 1–34. [Google Scholar]
- 47.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 326–349. ( 10.2307/1942268) [DOI] [Google Scholar]
- 48.Motyka J, Dobrzanski B, Zawadzki S. 1950. Wstepne badania nad lagami potudniowowschodniej Lubelszezyzny [Preliminary studies on meadows in the southeast of the provinee Lublin]. Univ. Mariae Curie-Sktodowska Ann. Sect E 5, 367–447. [Google Scholar]
- 49.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. ( 10.1128/AEM.71.12.8228-8235.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reveillaud J, Maignien L, Murat Eren A, Huber JA, Apprill A, Sogin ML, Vanreusel A. 2014. Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 8, 1198–1209. ( 10.1038/ismej.2013.227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohl KD, Varner J, Wilkening JL, Dearing MD. 2018. Gut microbial communities of American pikas (Ochotona princeps): evidence for phylosymbiosis and adaptations to novel diets. J. Anim. Ecol. 87, 323–330. ( 10.1111/1365-2656.12692) [DOI] [PubMed] [Google Scholar]
- 52.Kohl KD, Dearing MD, Bordenstein SR. 2018. Microbial communities exhibit host species distinguishability and phylosymbiosis along the length of the gastrointestinal tract. Mol. Ecol. 27, 1874–1883. ( 10.1111/mec.14460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross AA, Muller KM, Weese JS, Neufeld JD. 2018. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class Mammalia. Proc. Natl Acad. Sci. USA 115, E5786–E5795. ( 10.1073/pnas.1801302115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, Ley RE. 2014. Conducting a microbiome study. Cell 158, 250–262. ( 10.1016/j.cell.2014.06.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phillips CD, et al. 2012. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol. Ecol. 21, 2617–2627. ( 10.1111/j.1365-294X.2012.05568.x) [DOI] [PubMed] [Google Scholar]
- 56.Legendre P, Legendre L. 1998. Numerical ecology. Amsterdam, the Netherlands: Elsevier Science B.V. [Google Scholar]
- 57.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. ( 10.1111/j.1442-9993.1993.tb00438.x) [DOI] [Google Scholar]
- 58.McArdle BH, Anderson MJ. 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. ( 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2) [DOI] [Google Scholar]
- 59.Novakova E, Woodhams DC, Rodriguez-Ruano SM, Brucker RM, Leff JW, Maharaj A, Amir A, Knight R, Scott J. 2017. Mosquito microbiome dynamics, a background for prevalence and seasonality of West Nile Virus. Front. Microbiol. 8, 526 ( 10.3389/fmicb.2017.00526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michener CD, Sokal RR. 1957. A quantitative approach to a problem in classification. Evolution 11, 130–162. ( 10.1111/j.1558-5646.1957.tb02884.x) [DOI] [Google Scholar]
- 61.Matsen IV FA, Evans SN. 2013. Edge principal components and squash clustering: using the special structure of phylogenetic placement data for sample comparison. PLoS ONE 8, e56859 ( 10.1371/journal.pone.0056859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laviad-Shitrit S, Izhaki I, Lalzar M, Halpern M. 2019. Comparative analysis of intestine microbiota of four wild waterbird species. Front. Microbiol. 10, 1911 ( 10.3389/fmicb.2019.01911) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson DF, Foulds LR. 1981. Comparison of phylogenetic trees. Math. Biosci. 53, 131–147. ( 10.1016/0025-5564(81)90043-2) [DOI] [Google Scholar]
- 64.Bogdanowicz D, Giaro K. 2013. On a matching distance between rooted phylogenetic trees. Int. J. Appl. Math. Comput. Sci. 23, 669–684. ( 10.2478/amcs-2013-0050) [DOI] [Google Scholar]
- 65.Schottner S, Hoffmann F, Cardenas P, Rapp HT, Boetius A, Ramette A. 2013. Relationships between host phylogeny, host type and bacterial community diversity in cold-water coral reef sponges. PLoS ONE 8, e55505 ( 10.1371/journal.pone.0055505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Easson CG, Thacker RW. 2014. Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 5, 532 ( 10.3389/fmicb.2014.00532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas T, et al. 2016. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7, 11870 ( 10.1038/ncomms11870) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kropáčková L, et al. 2017. Codiversification of gastrointestinal microbiota and phylogeny in passerines is not explained by ecological divergence. Mol. Ecol. 26, 5292–5304. ( 10.1111/mec.14144) [DOI] [PubMed] [Google Scholar]
- 69.Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, Jaffé R, Moran NA. 2017. Dynamic microbiome evolution in social bees. Sci. Adv. 3, e1600513 ( 10.1126/sciadv.1600513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaulke CA, Arnold HK, Humphreys IR, Kembel SW, O'Dwyer JP, Sharpton TJ. 2018. Ecophylogenetics clarifies the evolutionary association between mammals and their gut microbiota. MBio 9, e01348-18 ( 10.1128/mBio.01348-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Javurkova VG, Kreisinger J, Prochazka P, Pozgayova M, Sevcikova K, Brlik V, Adamík P, Heneberg P, Porkert J. 2019. Unveiled feather microcosm: feather microbiota of passerine birds is closely associated with host species identity and bacteriocin-producing bacteria. ISME J. 13, 2363–2376. ( 10.1038/s41396-019-0438-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song SJ, et al. 2020. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. mBio 11, e02901-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220. [PubMed] [Google Scholar]
- 74.Peres-Neto P, Jackson DA. 2001. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 129, 169–178. ( 10.1007/s004420100720) [DOI] [PubMed] [Google Scholar]
- 75.Smouse PE, Long JC, Sokal RR. 1986. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Biol. 35, 627–632. ( 10.2307/2413122) [DOI] [Google Scholar]
- 76.Blomberg SP, Garland T Jr. 2002. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 15, 899–910. ( 10.1046/j.1420-9101.2002.00472.x) [DOI] [Google Scholar]
- 77.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 78.Blomberg SP, Garland JRT, Ives AR. 2003. Testing for phylogenetic signal in comparative date: behavioral traits are more labile. Evolution 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 79.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15. ( 10.1086/284325) [DOI] [Google Scholar]
- 80.Ives AR, Helmus MR. 2011. Generalized linear mixed models for phylogenetic analyses of community structure. Ecol. Monogr. 81, 511–525. ( 10.1890/10-1264.1) [DOI] [Google Scholar]
- 81.Dietrich C, Kohler T, Brune A. 2014. The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl. Environ. Microbiol. 80, 2261–2269. ( 10.1128/AEM.04206-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wong AC, Chaston JM, Douglas AE. 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 7, 1922–1932. ( 10.1038/ismej.2013.86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moeller AH, et al. 2016. Cospeciation of gut microbiota with hominids. Science 353, 380–382. ( 10.1126/science.aaf3951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grond K, Bell KC, Demboski JR, Santos M, Sullivan JM, Hird SM. 2019. No evidence for phylosymbiosis in western chipmunk species. FEMS Microbiol. Ecol. 96, fiz182 ( 10.1093/femsec/fiz182) [DOI] [PubMed] [Google Scholar]
- 85.Lutz HL, Jackson EW, Webala PW, Babyesiza WS, Kerbis Peterhans JC, Demos TC, Patterson BD, Gilbert JA. 2019. Ecology and host identity outweigh evolutionary history in shaping the bat microbiome. mSystems 4, e00511-19 ( 10.1128/msystems.00511-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hird SM, Sanchez C, Carstens BC, Brumfield RT. 2015. Comparative gut microbiota of 59 neotropical bird species. Front. Microbiol. 6, 1403 ( 10.3389/fmicb.2015.01403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu H, Chen Z, Gao G, Sun C, Li Y, Zhu Y. 2019. Characterization and comparison of gut microbiomes in nine species of parrots in captivity. Symbiosis 78, 241 ( 10.1007/s13199-019-00613-7) [DOI] [Google Scholar]
- 88.Ross AA, Rodrigues Hoffmann A, Neufeld JD. 2019. The skin microbiome of vertebrates. Microbiome 7, 79 ( 10.1186/s40168-019-0694-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schmitt S, et al. 2012. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 6, 564–576. ( 10.1038/ismej.2011.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lurgi M, Thomas T, Wemheuer B, Webster NS, Montoya JM. 2019. Modularity and predicted functions of the global sponge–microbiome network. Nat. Commun. 10, 992 ( 10.1038/s41467-019-08925-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiarello M, et al. 2018. Skin microbiome of coral reef fish is highly variable and driven by host phylogeny and diet. Microbiome 6, 147 ( 10.1186/s40168-018-0530-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lemay MA, Martone PT, Keeling PJ, Burt JM, Krumhansl KA, Sanders RD, Parfrey LW. 2018. Sympatric kelp species share a large portion of their surface bacterial communities. Environ. Microbiol. 20, 658–670. ( 10.1111/1462-2920.13993) [DOI] [PubMed] [Google Scholar]
- 93.Bouffaud M, Poirier M, Muller D, Moënne-Loccoz Y. 2014. Root microbiome relates to plant host evolution in maize and other Poaceae. Environ. Microbiol. 16, 2804–2814. ( 10.1111/1462-2920.12442) [DOI] [PubMed] [Google Scholar]
- 94.Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MTJ. 2018. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl Acad. Sci. USA 115, E1157–E1165. ( 10.1073/pnas.1717617115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bell TH, El-Din Hassan S, Lauron-Moreau A, Al-Otaibi F, Hijri M, Yergeau E, St-Arnaud M. 2014. Linkage between bacterial and fungal rhizosphere communities in hydrocarbon-contaminated soils is related to plant phylogeny. ISME J. 8, 331–343. ( 10.1038/ismej.2013.149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wehner J, Powell JR, Muller LAH, Caruso T, Veresoglou SD, Hempel S, Rillig MC, van der Heijden M. 2014. Determinants of root-associated fungal communities within Asteraceae in a semi-arid grassland. J. Ecol. 102, 425–436. ( 10.1111/1365-2745.12197) [DOI] [Google Scholar]
- 97.Ishida TA, Nara K, Hogetsu T. 2007. Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer-broadleaf forests. New Phytol. 174, 430–440. ( 10.1111/j.1469-8137.2007.02016.x) [DOI] [PubMed] [Google Scholar]
- 98.Tedersoo L, Mett M, Ishida TA, Bahram M. 2013. Phylogenetic relationships among host plants explain differences in fungal species richness and community composition in ectomycorrhizal symbiosis. New Phytol. 199, 822–831. ( 10.1111/nph.12328) [DOI] [PubMed] [Google Scholar]
- 99.Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. 2014. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl Acad. Sci. USA 111, 585–592. ( 10.1073/pnas.1321597111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vincent JB, Weiblen GD, May G. 2016. Host associations and beta diversity of fungal endophyte communities in New Guinea rainforest trees. Mol. Ecol. 25, 825–841. ( 10.1111/mec.13510) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.