Abstract
Modern cells embody metabolic networks containing thousands of elements and form autocatalytic sets of molecules that produce copies of themselves. How the first self-sustaining metabolic networks arose at life's origin is a major open question. Autocatalytic sets smaller than metabolic networks were proposed as transitory intermediates at the origin of life, but evidence for their role in prebiotic evolution is lacking. Here, we identify reflexively autocatalytic food-generated networks (RAFs)—self-sustaining networks that collectively catalyse all their reactions—embedded within microbial metabolism. RAFs in the metabolism of ancient anaerobic autotrophs that live from H2 and CO2 provided with small-molecule catalysts generate acetyl-CoA as well as amino acids and bases, the monomeric components of protein and RNA, but amino acids and bases without organic catalysts do not generate metabolic RAFs. This suggests that RAFs identify attributes of biochemical origins conserved in metabolic networks. RAFs are consistent with an autotrophic origin of metabolism and furthermore indicate that autocatalytic chemical networks preceded proteins and RNA in evolution. RAFs uncover intermediate stages in the emergence of metabolic networks, narrowing the gaps between early Earth chemistry and life.
Keywords: autocatalytic networks, origin of metabolism, biochemical evolution, origin of life, methanogens, acetogens
1. Introduction
Cells are autocatalytic in that they require themselves for reproduction. The origin of the first cells from the elements on the early Earth roughly 4 billion years ago [1–4] must have been stepwise. The nature of autocatalytic systems as intermediate states in that process is of interest. Autocatalytic sets are simpler than cellular metabolism and produce copies of themselves if growth substrates for food and a source of chemical energy for thermodynamic thrust are provided [5–7]. In theory, sets of organic molecules should be able to form autocatalytic systems [8–12], which, if provided with a supply of starting ‘food' molecules, can emerge spontaneously and proliferate via constraints imposed by substrates, catalysts, or thermodynamics [13]. Autocatalytic sets have attracted considerable interest as transitory intermediates between chemical systems and genetically encoded proteins at the origin of life [13–17]. Preliminary studies have shown that coenzymes are often required for their own synthesis and are therefore replicators with autocatalytic properties [18]. However, autocatalytic networks have not been identified in non-enzymatic metabolic networks so far, and evidence for their existence during prebiotic evolution is lacking.
Of special interest for metabolic evolution are a class of mathematical objects called reflexively autocatalytic food-generated networks—RAFs—in which each reaction is catalysed by a molecule from within the network and all molecules can be produced from a set of food molecules by the network itself [19]. RAFs are a precisely defined type of autocatalytic set, for their emergence from this ambient food set. Other autocatalytic networks not food-generated (sometimes called pseudo-RAFs) have the property of persistence but not of emergence from a food supply. In a related class of objects named constructively autocatalytic food-generated networks—CAFs [20], catalysts must either be present in the food set or produced before their first requirement. By contrast, RAFs impose that all necessary catalysts need to be produced by the network at some point, but not necessarily at the first time they are required. This feature models the emergence of specificity, speed, and efficiency in autocatalysis. Depending on the order of non-catalysed reactions, different routes can exist for the formation of an RAF. By depending on food, and representing the emergence of catalytic specificity, RAFs are an appropriate model for the origin of metabolism. Small chemical systems resembling RAFs have been constructed in the laboratory [7,21–23], although still far from the scale of cellular metabolism, which is composed of thousands of reactions. Modern cellular metabolism is enzyme–based, but greater than 60% of enzyme mechanisms described to date involve one or more cofactors [24] and 40% of all proteins crystallised have a bound metal relevant to their function [25]. RAFs can thus be identified in modern metabolism [17] by attributing the catalysis of enzymes to their metals and cofactors in prebiotic evolution [26–31], generalizing the well-known observation that native metals [3,32–36], flavins [37], pyridoxal 5′-phosphate [38,39], S-adenosyl methionine (SAM) [40], NAD [41], CoA [42], thiamine diphosphate [43], folates [44], and other cofactors [26,30,45] can themselves perform catalysis in the absence of enzymes. Also relevant to the inference of RAFs within metabolic networks are the numerous non-enzymatic, spontaneous reactions that are known to occur in metabolism [31,46]. If autocatalytic chemical networks antedate genetically encoded proteins, cofactor-dependent RAFs might have been involved and, if so, should have left evidence for their existence in modern metabolic networks.
In search of RAFs, we investigated different levels of ancient metabolism preserved in modern cells. Starting with the biosphere level of the KEGG database, we first removed all eukaryote-specific reactions, and then peeled back one more layer of time by examining anaerobic metabolism. The detection of a large RAF in anaerobic prokaryotic metabolism prompted us to ask whether RAFs are also preserved in the metabolism of ancient anaerobic autotrophs that trace to the last universal common ancestor, LUCA [47]. As far back as we could look in metabolic evolution, RAFs were found. They were found in the metabolism of the acetogenic bacterium Moorella thermoacetica and the methanogenic archaeon Methanococcus maripaludis, lineages thought to be primitive as they live on the simplest source of carbon and energy known, the H2-CO2 redox couple [1,5,29,47–49], they assimilate geochemically generated carbon species [50,51], they generate ATP from CO2 fixation [5], their core bioenergetic reactions occur abiotically in hydrothermal vents [48,52], and under laboratory conditions [3], their ecology and gene trees link them to LUCA [53], and they still inhabit primordial habitats within the crust today [54]. The RAFs of the methanogen and the acetogen furthermore intersect in a primordial network that generates amino acids, nucleosides, and acetyl-CoA from a starting set of simple food molecules, shedding light on the nature of autocatalytic networks that existed before the first cells arose from the elements on the early Earth.
2. Results
(a). Two-thirds of global prokaryotic metabolism can be annotated with small-molecule catalysis
In search of RAFs in 4-billion-year-old metabolism, we started from all 10 828 KEGG reactions and purged the set of non-primordial reactions in two pruning steps. First, looking at the 8352 reactions assigned to enzymes, we removed reactions that occur only in eukaryotes. Such reactions are unlikely to be primordial, because eukaryotes arose less than 2 billion years ago [2]. Second, from the resulting set we excluded O2-dependent reactions, because O2 is a product of cyanobacterial photosynthesis, which arose about 2.4 billion years ago [55]. These pruning steps left 5847 enzyme-associated reactions, 66% of which involve at least one cofactor, and the remaining 33% were assigned an operational catalyst named ‘peptide' which is attributed to reactions catalysed by enzymes to which no cofactor or metal is currently associated in the databases. From the initial set of 10 828 reactions, we identified, retrieved, and added to the previous set 147 spontaneous reactions, generating a global network comprising 5994 reactions and 5723 metabolites (electronic supplementary material, figure S1 and dataset S1A). The cofactors involved in this ancient anaerobic network are distributed among the six different enzyme commission (EC) classes as shown in figure 1. Cofactors can be both catalysts and substrates or products in reactions (for example, NAD) or just catalysts (metals). Metal catalysis is widespread across all classes of metabolism, and NADs dominate the oxido-reductase reactions. The resulting prokaryotic O2-independent network contains 70% of all 8352 reactions associated with enzymes before removal of O2-dependent and eukaryote-specific reactions, indicating that a great part of metabolism was invented in the anaerobic world [56].
Figure 1.
Catalysts in global oxygen-independent prokaryotic metabolism. The catalysis-annotated network separated by enzyme commission (EC) classes with the corresponding cofactors for each. Cofactors are grouped (legend, top right) according to their function in catalysis. NADs, Cobalamins, Folates, Flavins, and Quinones are each a group of equivalent catalysts with common properties and common biosynthetic pathways (for example, NADs stands for NAD(P)(H); all pooling reactions are detailed in electronic supplementary material, dataset S1A).
(b). Autocatalysis in global metabolism expands with a small set of cofactors
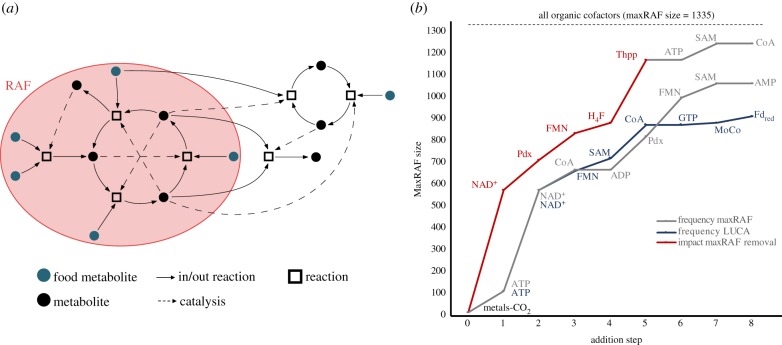
The largest possible RAFs (maxRAFs) in a network are of interest because they represent its largest component of autocatalytic complexity. Figure 2a shows a schematic of an RAF within a metabolic network. The RAF algorithm, first introduced in [19] and refined in [57] and [58], starts with the full reaction network, and iteratively removes reactions that fail to have all their reactants and at least one catalyst able to be produced from the food set via the current reaction set. For this, the ‘closure of the food set' needs to be computed at each iteration (electronic supplementary material, Methods). The algorithm ends when no more reactions can be removed. If the remaining set of reactions is non-empty, it comprises the maxRAF, i.e. the union of all possible RAFs in the original reaction network. If the remaining set is empty, there was no RAF in the original network.
Figure 2.
Autocatalysis in global metabolism expands with a small set of cofactors. (a) Schematic depiction of a reflexively autocatalytic food-generated network (RAF) highlighted (red ellipse) in a metabolic network. Food metabolites (green circles) may enter the RAF allowing subsequent reactions (squares) to occur and other metabolites (black circles) to be produced. Each reaction is catalysed by a metabolite in the network (catalysis shown in dashed arrows). (b) Increasing maxRAF sizes with the sequential addition to the initial food set (with only inorganic compounds) of the organic cofactors (i) with the highest impact on maxRAF size upon removal—NAD, Pdx, FMN H4F, and Thpp (red)—followed by the three most frequent in the largest maxRAF with all organic cofactors added–ATP, SAM, and CoA (grey), (ii) most frequent in the maxRAF with all organic cofactors added (grey), and (iii) most frequent in enzymes predicted to be in LUCA [47] (blue) (Pdx, pyridoxal 5-phosphate; H4F, tetrahydrofolate, Thpp, thiamine diphosphate, MoCo, molybdopterin; Fdred, reduced ferredoxin). Top dashed line shows the maxRAF size obtained when all organic cofactors are added to the food set.
The maxRAFs in the global prokaryotic O2-independent network were identified for different food sets, that is, molecules provided by the environment (electronic supplementary material, table S1). An inorganic food set containing H2O, H2, H+, CO2, CO, , , , , S, H2S, NH3, N2, all metals, FeS clusters and other metal clusters, a generalist acceptor, donor, and metal produced a minute maxRAF with eight reactions linking ammonia, carbon, and sulfide transformations. The addition of formate, methanol, acetate, and pyruvate, which are central metabolites with experimental evidence for synthesis from CO2 and metals [3], doubles the maxRAF size to 16 reactions. In principle, the addition of organic cofactors (electronic supplementary material, table S1) to the food set should generate larger maxRAFs. Sequential addition of the eight most frequent cofactors identified in the LUCA's proteins [47] to the metal–CO2 food set expanded the maxRAF from 16 to 914 reactions (figure 2b). The addition of all cofactors germane to the anaerobic network generates a maxRAF with 1335 reactions spanning 25% of the starting anaerobic network. Sequential addition of the eight compounds that were most frequent in that maxRAF, to the metal–CO2 food set, expands the maxRAF from 16 to 1066 reactions, whereas sequential addition of the five compounds with the greatest impact (upon removal from the food set) on anaerobic maxRAF size followed by the three most frequent in the largest maxRAF yields a final maxRAF of 1248 reactions (figure 2b). These results indicate that RAFs can grow in size through sequential incorporation of organic cofactors (figure 2b). RAFs can thus provide structure, contingency, increasing complexity, and direction to interactions among molecule food sets, given a sustained geochemical source of carbon, energy, and electrons.
(c). Autocatalytic networks point to an early autotrophic metabolism
If autocatalytic sets were instrumental at the origin of metabolism [13], lineages with a physiology very similar to that of the first cells should harbour the most ancient RAFs.
Several lines of evidence indicate that methanogens and acetogens reflect the ancestral state of microbial physiology in the bacteria and archaea, respectively [1,3,5,29,47–52,54]. By investigating the metabolic networks of one archaeon and one bacterium that each satisfy both their carbon and energy needs via H2-dependent CO2 reduction, we can identify their conserved common features. From comparative physiology [59] and from the standpoint of genes that trace to the LUCA [47,53], their shared features should reflect a state predating the divergence of the two prokaryotic domains. Subsets of the global prokaryotic O2-independent network were obtained by parsing the genomes of the acetogen Moorella thermoacetica (Ace) and the methanogen Methanococcus maripaludis (Met). These were completed with reactions from corresponding manually curated genome-scale metabolic models [60,61], resulting in 1193 reactions for Ace and 920 for Met (electronic supplementary material, dataset S1B and S1C). Both the acetogen and the methanogen metabolic networks contain RAFs. When all organic cofactors are added to the food set, the maxRAFs contain 394 and 209 reactions for Ace and Met, respectively, spanning major KEGG functional categories (figure 3; electronic supplementary material, table S1 and figure S2 and S3 and dataset S2).
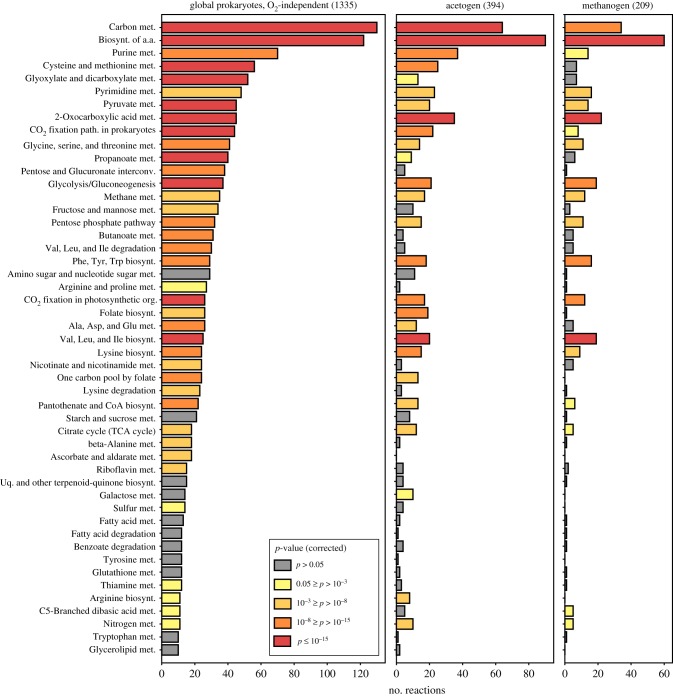
Figure 3.
Autocatalytic networks point to an early autotrophic metabolism. The number of reactions in each functional category for three maxRAFs and functional enrichment relative to the global O2-independent prokaryotic network. Colours represent bins of corrected p-values (Fisher's exact test with Benjamini–Hochberg false discovery rate (FDR) multiple-testing correction). From left to right, maxRAF obtained for (sizes in brackets): global O2-independent prokaryotic network, acetogen (Ace) and methanogen (Met). Categories are sorted according to the number of reactions in the first maxRAF, from largest to smallest; only categories where this maxRAF had more than 10 reactions are shown.
Carbon fixation and biosynthetic pathways are represented and amino acid biosynthesis is highly enriched in all maxRAFs, recovering autotrophic components of early autocatalytic metabolism. Note that, none of the food sets described so far in these analyses contained ‘peptide'. Addition of the generic ‘peptide' catalyst to the food set increases the maxRAF sizes obtained with the global anaerobic network, Met, and Ace by 93%, 47%, and 25%, respectively (electronic supplementary material, table S1). This indicates that adding protein catalysis expands cofactor-supported autocatalytic sets, but it does so to a much lesser degree in the metabolism of Met and Ace than it does in the global O2-independent prokaryotic network.
(d). Metabolism at the origin of LUCA was autocatalytic and autotrophic
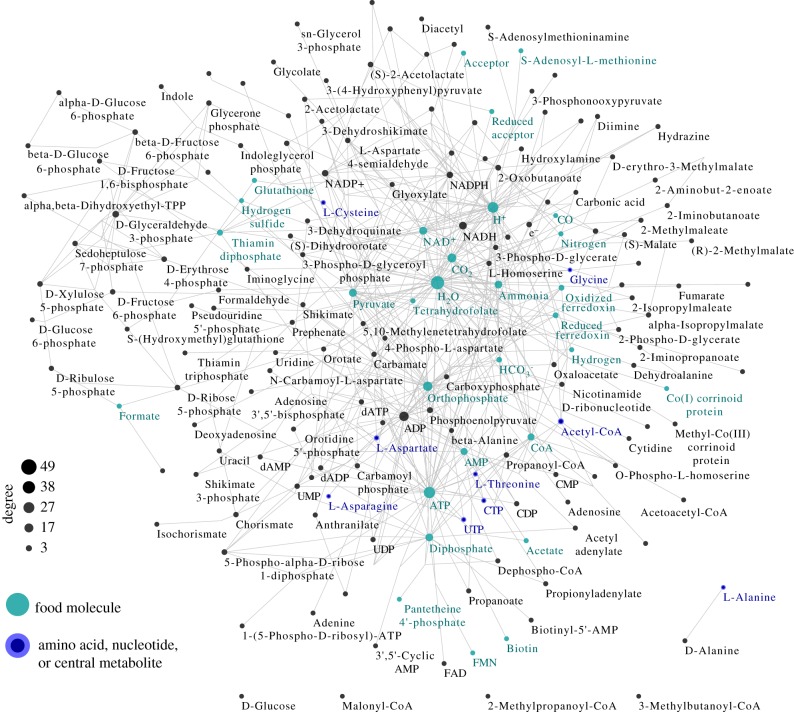
The intersection of the Ace and Met maxRAFs should be more ancient than each of them individually. Three-quarters of the (smaller) Met maxRAF overlap with the (larger) Ace maxRAF in a connected network harbouring 172 reactions and 175 metabolites (figure 4; electronic supplementary material, figure S4; individual maxRAFs from Ace and Met in electronic supplementary material, figure S2 and S3). Six metabolites are disconnected, meaning the species interconvert them using different pathways; one example is that of glucose, catabolism of which arose after LUCA [62]. Highly connected food metabolites in the primordial network (more than 13 edges) include H2O, ATP, protons, phosphate, CO2, NAD+, pyruvate, ammonia, diphosphate, coenzyme A, and AMP; highly connected non-food metabolites (more than eight edges) include ADP, NADH, and other pyridine dinucleotides, glyceraldehyde-3-phosphate, and acetyl-CoA (electronic supplementary material, dataset S3). The network is able to produce six amino acids—asparagine, aspartate, alanine, glycine, cysteine, and threonine—plus the two nucleosides UTP and CTP. Cytochromes and quinones do not figure into the network.
Figure 4.
Core metabolism at the origin of the last universal common ancestor (LUCA). Intersection of the maxRAFs obtained with the networks of Moorella thermoacetica and Methanococcus maripaludis with a food set with organic cofactors (only metabolic interconversions are depicted; catalysis arcs are omitted for clarity). Six metabolites, including D-glucose and L-alanine (bottom), are in the intersection but disconnected from the remaining network. The node size is scaled according to the degree, with food molecules highlighted in green and relevant products in dark blue. ‘Acceptor' and ‘Reduced Acceptor’ are abstract redox molecules as represented in KEGG metabolism.
A different look at the primordial network reveals a hierarchical and highly connected organization (electronic supplementary material, figure S4) with a half-moon core structure with node degree varying from 49 to 4 (electronic supplementary material, dataset S3). Food molecules cluster in the most connected area, suggesting that autocatalytic metabolism is initiated by a handful of central substrate molecules with degree higher than 10.
(e). Primordial metabolism is enriched in metal catalysis, ancient genes, and autotrophic functions
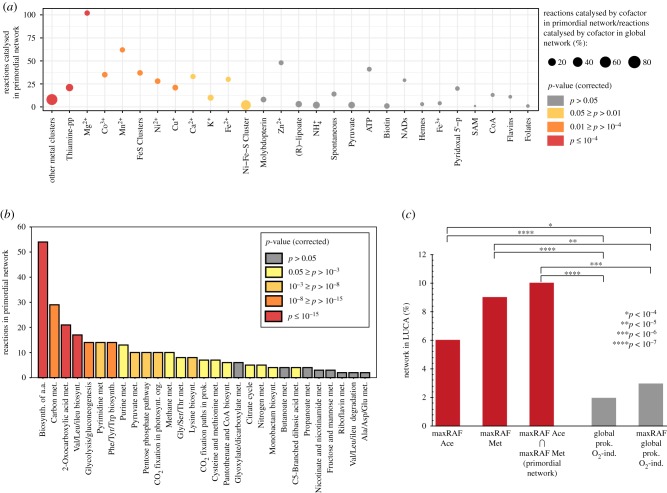
In search of the distinct contributions for autocatalysis, we tested for enrichment in individual catalysts, functions, and ancient genes encoding for reactions in the primordial network. There is a significant enrichment for metal and metal–sulfur cluster catalysis (figure 5a), whereas thiamine diphosphate (a carrier of C2 units in metabolism) is the only organic cofactor that is significantly enriched in catalysing the primordial network when compared with the global network, even though several others are present and essential for the network to grow (figure 5b). The primordial network is also enriched in reactions for amino acid biosynthesis, carbon metabolism, and 2-oxocarboxylic acid metabolism when compared with the global network (figure 5b). Comparing reactions in the primordial network to those catalysed by genes that can be traced to LUCA by independent phylogenetic criteria [47] uncovers highly significant enrichment relative to both the global network and its maxRAF (figure 5c). The maxRAF obtained within the primordial network contains 120 reactions and is enriched in amino acid and carbon metabolism but produces cysteine as the sole amino acid, which is noteworthy because cysteine is the hub of sulfur metabolism and also is the sole ligand for incorporating Fe–S and Fe–Ni–S clusters in proteins (electronic supplementary material, figure S5).
Figure 5.
Properties of the core metabolism at the origin of the last universal common ancestor (LUCA). (a) The number of reactions catalysed by each cofactor in the primordial network (overlapping network between the acetogen and methanogen maxRAFs) and enrichment for each cofactor when compared with the number of reactions catalysed by that cofactor in the global O2-independent prokaryotic network. Circle size indicates the ratio between the number of reactions in the primordial network and number of reactions in the global network catalysed by each cofactor; colour indicates the corrected p-value (Fisher's exact test with Benjamini–Hochberg FDR correction). (b) The number of reactions in each functional category in the primordial network and functional enrichment relative to the global O2-independent prokaryotic network. Colour indicates bins of corrected p-values (Fisher's exact test with Benjamini–Hochberg FDR correction). (c) The proportion of metabolic networks predicted to be in LUCA [47] and enrichment of the individual maxRAFs and the primordial network (red) compared with the global network and the maxRAF obtained with it (grey) (Fisher's exact test).
(f). Autocatalysis, ATP, NAD, and monomers
Crucial catalysts can be identified by removing them from the food set. NAD+ is strongly embedded in the RAF and its removal reduces the size of the maxRAF by approximately 50% (electronic supplementary material, figure S6). The role of NADs, which we use to collectively designate NAD(P)+ and NAD(P)H, in the food set of the largest maxRAFs is striking. It exhibits the strongest effect we observed for any cofactor. NAD donates and accepts hydride for redox-dependent reaction catalysis. The strict dependence of the largest maxRAFs upon NAD reflects the circumstance that microbial metabolism, without exception, always involves redox reactions [63]. Though Fe–S clusters are more ancient redox cofactors than NAD(P)H [64], they perform one-electron transfer reactions and have been replaced in evolution by NAD and other two-electron carriers [65,66]. Fe–S clusters also heavily impact maxRAF sizes, together with pyridoxal-5-phosphate and divalent metals.
Surprisingly, when we remove ATP from the food set of organic cofactors, this has no impact on the size of the maxRAF (the number of reactions in the maxRAF remains unchanged), both for the individual networks (electronic supplementary material, figure S6) and LUCA's network (electronic supplementary material, figure S4). Why does ATP removal from the food set with organic cofactors have no effect on RAFs? ATP is an essential intermediate in the maxRAF, but it is not required to kick-start it when other organics are present. This reflects the increasingly evident role of alternative energy currencies in primordial metabolism [6], such as acyl phosphates [29], thioesters [7], and reduced ferredoxin [49,67]. Alternative energy currencies are particularly common in anaerobes [49].
RAFs provided with a food set containing catalysts can generate amino acids and bases (figure 4), but the converse is not true: adding amino acids and bases to the simplest food set, which includes inorganic catalysts and CO2 (electronic supplementary material, table S1), produces a miniscule 33-reaction maxRAF (electronic supplementary material, figure S7). The maxRAF contains 47 metabolites, 27 of which are food molecules. This indicates that primordial autocatalytic networks embedded in microbial metabolism generated amino acids and bases using small-molecule catalysis.
3. Discussion
Autocatalytic networks are objects of molecular self-organization [8–12]. Their salient property in the study of early biochemical evolution is the capacity to grow in size and complexity. Compounds generated from the food set become part of the network, hence autocatalytic networks can start small and grow, in principle to a size approaching the complexity of metabolic networks of modern cells [15], and very little catalysis by individual elements is required for autocatalytic networks to emerge [19,20]. Reflexively autocatalytic and food-generated networks—RAFs—are a particularly interesting formalization of collectively autocatalytic sets, as they capture a property germane to life: they require a constant supply of an environmentally provided food source in order to grow [19]. In that sense, RAFs reflect metabolic networks in real cells, in that growth substrates are converted to end products, a proportion of which comprises the substance of cells. But RAFs are far simpler than metabolism because they can start very small.
RAFs have not been applied in the study of the evolution of chemical networks that led to the metabolism of modern cells, themselves large natural autocatalytic networks. By embracing the simple and robust premise that reactions catalysed by simple molecules and inorganic compounds preceded metabolic reactions catalysed by enzymes [17,26,30], we have retooled RAFs into an analytical instrument to investigate the nature of metabolic evolution.
Our analyses started with the enzymatic and spontaneous reactions charted in modern metabolism and used RAFs as a filter to uncover elements with self-organizational properties, to address the nature of processes in the earliest phases of evolution, before the origin of eukaryotes and before the appearance of oxygen. We found evidence for a role of autocatalytic networks at the onset of metabolism. The largest RAF that we identified in the whole prokaryotic anaerobic biochemical space has 1335 reactions and points to early autotrophy. This RAF is larger than the genome size of the smallest free-living archaeon, Methanothermus fervidus [68]. With a genome coding for 1311 proteins and 50 RNA genes, M. fervidus lives from H2 and CO2 as carbon and energy sources (the food set) and requires only inorganic, geochemical nutrients, no other cells for survival [69]. H2 and CO2 were present in abundance on the early Earth and may have given rise to the first metabolic pathways that brought forth the first archaeal and bacteria cells [3,6,47]. Our anaerobic RAF is, however, smaller than the reaction network in the smallest genome of bacteria that live from H2 and CO2, which is found in the acetogen Thermoanaerobacter kivui, encoding 2378 proteins [70].
Methanothermus fervidus and T. kivui harbour primitive forms of methanogenesis and acetogenesis in that they both lack cytochromes and quinones, suggesting that they represent energy metabolic relics from the earliest phases of biochemical evolution on the primordial Earth, before anaerobic respiratory chains had evolved [29]. To investigate this aspect further, we examined the best annotated metabolic networks existing for H2-CO2-dependent archaea and bacteria, the methanogen Methanococcus maripaludis and the acetogen Moorella thermoacetica. Remarkably, a food set containing only small abiogenic molecules and a handful of organic cofactors generates sizeable RAFs in each of the networks, with 209 and 394 reactions, respectively. The inclusion of organic molecules as catalysts in our food set is in line with a premise common to all scientific theories for the origin of life, namely that the environment provided starting material from which metabolism and life evolved. The small sizes of maxRAFs compared to full metabolic networks in the genome-scale metabolic models are contingent upon two aspects. First, all reactions in the maxRAFs presented here require small-molecule catalysis. This excludes important edges in the network catalysed by peptides only, that allow for significant expansion, when the generic catalyst ‘peptide' is added to the food set (electronic supplementary material, table S1). Moreover, in genome-scale models, a method known as gap-filling allows genome-scale metabolic models to be connected to the necessary degree for the production of all biomass components [71]. To avoid the introduction of noise and over-fitting of our model for autocatalysis, we have not performed gap-filling at any stage in this work. Note that the genome-scale models used here also require organic cofactors to produce biomass [60,61].
RAFs uncover elements of metabolic evolution that predate the divergence of archaea and bacteria from the LUCA. The intersection of the RAFs of M. maripaludis and M. thermoacetica uncovers commonalities—a core, conserved network with 172 reactions that is enriched in metal catalysis and carbon-metal bonds [72] that points both to early autotrophy [5] and to the origin of the genetic repertoire of LUCA [47]. This conserved network does not produce all 20 amino acids, even though LUCA probably used all of them, given the universality of the genetic code. This apparent contradiction is reconciled by the virtual certainty that LUCA, before it became a free-living cell, was auxotrophic for some amino acids (and other components) that were provided by the environment [73]. Our model does not include catalysed transport reactions, and these would be an interesting addition in future formulations. Our results indicate that some enzyme catalysis had to be invented to allow for a sustained production of several amino acids. Our results also show that the kick-start of autocatalysis in anaerobic metabolism does not require ATP in the food set, even though ATP is an essential intermediate in all maxRAFs. This relates to the use of alternative energetic currencies in anaerobic prokaryotes [49] and recent findings that suggest that complexity in early metabolic reaction systems could have emerged without phosphate [6]. More importantly, NAD plays an essential role in kick-starting all sizeable maxRAFs obtained here. This underscores the special role of redox chemistry in primordial catalysis [49,74].
An important insight uncovered by RAFs is the observation that although a food set with organic cofactors sparks a large autocatalytic metabolic network that generates amino acids and bases, the opposite does not occur: adding amino acids and bases to the simplest food set (which includes inorganic catalysts and CO2) only produces a minute RAF with 33 reactions. The result that autocatalytic networks detectable in microbial metabolism as RAFs generate amino acids and bases using small-molecule catalysts is in accord with the recent report of amino acid synthesis catalysed by native metals [35], and with the physiology of extant anaerobic autotrophs: amino acids and bases are sequestered end-products of H2- and CO2-dependent metabolism, they are polymerised to make the substance of cells.
RAFs can serve as a guide for the identification and construction of larger, biologically relevant autocatalytic reaction networks. The synthesis of compounds characteristic of the metabolism of acetogens and methanogens, intermediates and end products of the acetyl-CoA pathway, and of the incomplete citric acid cycle from CO2 using only the catalysis of native metals [3,32–36], as well as the demonstrated catalytic power of organic cofactors without their enzymes including flavins [37], pyridoxal 5′-phosphate [38,39], SAM [40], NAD [41], and others [42–45] encourages the investigation of more complex autocatalytic networks in laboratory reactors. Individual reactions in the maxRAFs presented here can be tested in independent experiments with the catalysts assigned (electronic supplementary material, dataset S2). Though we have not explored subRAFs [75] embedded within the (larger) maxRAFs in the present work, this may be an interesting route for further investigation, amenable to laboratory experiments and/or the incorporation of experimental data [58]. The increasing availability of large-scale kinetic, thermodynamic, and inhibition data for metabolism will allow further exploration of RAFs [58]. The plausibility and sustainability of RAFs under various scenarios has been explored in extensive simulations [76].
Our results are directly relevant to two deeply divided schools of thought concerning the nature of chemical reactions at the origin of life: genetics first and metabolism first. The genetics first school, or RNA world, holds that the origin of RNA molecules marked the origin of life-like processes, and that RNA both self-replicated and possessed catalytic abilities that led to the emergence of biochemical reactions [77,78]. In that view, the origin of the bases that drove that process forward is decoupled from biochemical processes that are germane to modern cellular metabolism. Proponents of the RNA world [77,78] might criticize that we did not investigate the possible catalytic role of RNA in the metabolic networks presented here. However, no reactions in KEGG have RNA catalysts assigned in Uniprot, an observation concerning both the nature of catalysis in microbial metabolism and the limitations inherent to current databases. If we count RNA-related cofactors as representing the RNA world [26], the participation of RNA-like bases in RAFs is ubiquitous, but as monomers, with polymers playing no role. Hence, addition of a generic polymer ‘RNA' has no impact on RAFs while the generic catalyst ‘peptide' does, whereby we note that KEGG does not include the full processes of transcription, splicing, and translation, which require the genetic code, an innovation that arose subsequent to the phase of biochemical evolution probed here by RAFs.
The metabolism-first school holds that spontaneous (exergonic) chemical reactions preceded reactions catalysed by genetic material, and that those exergonic reactions continuously gave rise to substrate-product relationships [29,79]. From such reactions, more complex interaction networks with autocatalytic properties arose [16,80], in which elements of the set intervened in reactions of the set, providing structure and direction to product accumulation. Our results indicate that RNA monomers can readily arise from autocatalytic networks—though the converse is not true—and the nature of the products accumulated in RAFs will include nucleic acids. In other words, RAFs applied to ancient autotrophic metabolism reveal a vector of autopoietic genesis that detects RNA as emergent from metabolism.
4. Material and methods
Detailed methods including annotation procedures and algorithms can be found in the electronic supplementary material.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Filipa L. Sousa, Martina Preiner, and anonymous reviewers for comments.
Data accessibility
All biochemical data used here is publicly available in KEGG, Uniprot, and in the publications of the genome-scale metabolic models of Moorella thermoacetica [60] and Methanococcus maripaludis [61]. The curated and annotated networks are provided in electronic supplementary material, dataset S1. A custom-made implementation of the maxRAF algorithm was used for the analysis in this paper and is available at https://www.canterbury.ac.nz/engineering/schools/mathematics-statistics/research/bio/downloads/raf/. An example of an input file (global prokaryotic O2-independent network, food set with all small molecules, abiotic carbon, and organic cofactors) is given in electronic supplementary material, dataset S4. A more general-purpose and interactive RAF analyser can be found online at https://github.com/husonlab/catlynet, including several more examples and explanations.
Authors' contributions
J.C.X. collected and integrated data from databases, curated models, and literature. J.C.X., M.S., S.K., and W.F.M. designed the project. W.H. and M.S. wrote the pseudocode and algorithm for detection of maxRAFs and W.H. performed maxRAF identification. J.C.X. and W.F.M. wrote the first manuscript draft. J.C.X., W.H., S.K., M.S., and W.F.M. contributed in data interpretation and writing the final manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by grants from the European Research Council (666053) and the Volkswagen Foundation (93 046) to W.F.M. W.H. thanks the Institute for Advanced Study, University of Amsterdam, The Netherlands, for partial support in the form of a fellowship.
References
- 1.Baross JA. 2018. The rocky road to biomolecules. Nature 564, 42–43. ( 10.1038/d41586-018-07262-8) [DOI] [PubMed] [Google Scholar]
- 2.Betts HC, Puttick MN, Clark JW, Williams TA, Donoghue PCJJ, Pisani D. 2018. Integrated genomic and fossil evidence illuminates life's early evolution and eukaryote origin. Nat. Ecol. Evol. 2, 1556–1562. ( 10.1038/s41559-018-0644-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varma SJ, Muchowska KB, Chatelain P, Moran J. 2018. Native iron reduces CO2 to intermediates and end-products of the acetyl-CoA pathway. Nat. Ecol. Evol. 2, 1019–1024. ( 10.1038/s41559-018-0542-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tashiro T, Ishida A, Hori M, Igisu M, Koike M, Méjean P, Takahata N, Sano Y, Komiya T. 2017. Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature 549, 516–518. ( 10.1038/nature24019) [DOI] [PubMed] [Google Scholar]
- 5.Fuchs G. 2011. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu. Rev. Microbiol. 65, 631–658. ( 10.1146/annurev-micro-090110-102801) [DOI] [PubMed] [Google Scholar]
- 6.Goldford JE, Hartman H, Smith TF, Segrè D. 2017. Remnants of an ancient metabolism without phosphate. Cell 168, 1126–1134.e9. 10.1016/j.cell.2017.02.001 [DOI] [PubMed] [Google Scholar]
- 7.Semenov SN, Kraft LJ, Ainla A, Zhao M, Baghbanzadeh M, Campbell VE, Kang K, Fox JM, Whitesides GM. 2016. Autocatalytic, bistable, oscillatory networks of biologically relevant organic reactions. Nature 537, 656–660. ( 10.1038/nature19776) [DOI] [PubMed] [Google Scholar]
- 8.Kauffman SA. 1971. Cellular homeostasis, epigenesis and replication in randomly aggregated macromolecular systems. J. Cybern. 1, 71–96. ( 10.1080/01969727108545830) [DOI] [Google Scholar]
- 9.Gánti T. 1971. Az élet princípiuma. Budapest: Gondolat. [Google Scholar]
- 10.Eigen M. 1971. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 58, 465–523. ( 10.1007/BF00623322) [DOI] [PubMed] [Google Scholar]
- 11.King GAM. 1978. Autocatalysis. Chem. Soc. Rev. 7, 297 ( 10.1039/cs9780700297) [DOI] [Google Scholar]
- 12.Dyson FJ. 1982. A model for the origin of life. J. Mol. Evol. 18, 344–350. ( 10.1007/BF01733901) [DOI] [PubMed] [Google Scholar]
- 13.Kauffman SA. 1986. Autocatalytic sets of proteins. J. Theor. Biol. 119, 1–24. ( 10.1016/S0022-5193(86)80047-9) [DOI] [PubMed] [Google Scholar]
- 14.Wächtershäuser G. 1990. Evolution of the first metabolic cycles. Proc. Natl Acad. Sci. USA 87, 200–204. ( 10.1073/pnas.87.1.200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hordijk W, Hein J, Steel M. 2010. Autocatalytic sets and the origin of life. Entropy 12, 1733–1742. ( 10.3390/e12071733) [DOI] [Google Scholar]
- 16.Smith E, Morowitz HJ. 2004. Universality in intermediary metabolism. Proc. Natl Acad. Sci. USA 101, 13 168–13 173. ( 10.1073/pnas.0404922101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sousa FL, Hordijk W, Steel M, Martin WF. 2015. Autocatalytic sets in E. coli metabolism. J. Syst. Chem. 6, 4 ( 10.1186/s13322-015-0009-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kun Á, Papp B, Szathmáry E, Kun A. 2008. Computational identification of obligatorily autocatalytic replicators embedded in metabolic networks. Genome Biol. 9, R51 ( 10.1186/gb-2008-9-3-r51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hordijk W, Steel M. 2004. Detecting autocatalytic, self-sustaining sets in chemical reaction systems. J. Theor. Biol. 227, 451–461. ( 10.1016/j.jtbi.2003.11.020) [DOI] [PubMed] [Google Scholar]
- 20.Mossel E, Steel M. 2005. Random biochemical networks: the probability of self-sustaining autocatalysis. J. Theor. Biol. 233, 327–336. ( 10.1016/j.jtbi.2004.10.011) [DOI] [PubMed] [Google Scholar]
- 21.Ashkenasy G, Jagasia R, Yadav M, Ghadiri MR. 2004. Design of a directed molecular network. Proc. Natl Acad. Sci. USA 101, 10 872–10 877. ( 10.1073/pnas.0402674101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaidya N, Manapat ML, Chen IA, Xulvi-Brunet R, Hayden EJ, Lehman N. 2012. Spontaneous network formation among cooperative RNA replicators. Nature 491, 72–77. ( 10.1038/nature11549) [DOI] [PubMed] [Google Scholar]
- 23.Miras HN, Mathis C, Xuan W, Long D-L, Pow R, Cronin L. 2019. Spontaneous formation of autocatalytic sets with self-replicating inorganic metal oxide clusters ( 10.26434/CHEMRXIV.9598442.V1) [DOI] [PMC free article] [PubMed]
- 24.Ribeiro AJM, Holliday GL, Furnham N, Tyzack JD, Ferris K, Thornton JM. 2018. Mechanism and Catalytic Site Atlas (M-CSA): a database of enzyme reaction mechanisms and active sites. Nucleic Acids Res. 46, D618–D623. ( 10.1093/nar/gkx1012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guengerich FP. 2016. Metals in biology 2016: molecular basis of selection of metals by enzymes. J. Biol. Chem. 291, 20 838–20 839. ( 10.1074/jbc.R116.749259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White HB. 1976. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 7, 101–104. ( 10.1007/BF01732468) [DOI] [PubMed] [Google Scholar]
- 27.Penny D. 2005. An interpretive review of the origin of life research. Biol. Phil. 20, 633–671. ( 10.1007/s10539-004-7342-6) [DOI] [Google Scholar]
- 28.Shapiro R. 2006. Small molecule interactions were central to the origin of life. Q Rev. Biol. 81, 105–126. ( 10.1086/506024) [DOI] [PubMed] [Google Scholar]
- 29.Martin W, Russell MJ. 2007. On the origin of biochemistry at an alkaline hydrothermal vent. Phil. Trans. R. Soc. B 362, 1887–1926. ( 10.1098/rstb.2006.1881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stockbridge RB, Lewis CA, Yuan Y, Wolfenden R. 2010. Impact of temperature on the time required for the establishment of primordial biochemistry, and for the evolution of enzymes. Proc. Natl Acad. Sci. USA 107, 22 102–22 105. ( 10.1073/pnas.1013647107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller MA, Piedrafita G, Ralser M. 2015. The widespread role of non-enzymatic reactions in cellular metabolism. Curr. Opin Biotechnol. 34, 153–161. ( 10.1016/j.copbio.2014.12.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitani A, Tsunetsugu S, Suzuki A, Ito S, Sasaki K. 1995. Fe(III)-ion-catalysed non-enzymatic transformation of adenosine diphosphate into adenosine triphosphate part II. Evidence of catalytic nature of Fe ions. Bioelectrochem. Bioenerg. 36, 47–51. ( 10.1016/0302-4598(94)01751-L) [DOI] [Google Scholar]
- 33.Huber C, Wächtershäuser G. 1997. Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science 276, 245–247. ( 10.1126/science.276.5310.245) [DOI] [PubMed] [Google Scholar]
- 34.Keller MA, Turchyn AV, Ralser M. 2014. Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Mol. Syst. Biol. 10, 1–12. ( 10.1002/msb.20145228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muchowska KB, Varma SJ, Moran J. 2019. Synthesis and breakdown of universal metabolic precursors promoted by iron. Nature 569, 104–107. ( 10.1038/s41586-019-1151-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preiner M, et al. 2020. A hydrogen-dependent geochemical analogue of primordial carbon and energy metabolism. Nat. Ecol. Evo. ( 10.1038/s41559-020-1125-6) [DOI] [PubMed]
- 37.Argueta EA, Amoh AN, Kafle P, Schneider TL. 2015. Unusual non-enzymatic flavin catalysis enhances understanding of flavoenzymes. FEBS Lett. 589, 880–884. ( 10.1016/j.febslet.2015.02.034) [DOI] [PubMed] [Google Scholar]
- 38.Metzler DE, Snell EE. 1952. Deamination of serine. I. Catalytic deamination of serine and cysteine by pyridoxal and metal salts. J. Biol. Chem. 198, 353–361. [PubMed] [Google Scholar]
- 39.Zabinski RF, Toney MD. 2001. Metal ion inhibition of nonenzymatic pyridoxal phosphate catalyzed decarboxylation and transamination. J. Am. Chem. Soc. 123, 193–198. ( 10.1021/ja0026354) [DOI] [PubMed] [Google Scholar]
- 40.Barrows LR, Magee PN. 1982. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis 3, 349–351. ( 10.1093/carcin/3.3.349) [DOI] [PubMed] [Google Scholar]
- 41.Betanzos-Lara S, Liu Z, Habtemariam A, Pizarro AM, Qamar B, Sadler PJ. 2012. Organometallic ruthenium and iridium transfer-hydrogenation catalysts using coenzyme NADH as a cofactor. Angew Chemie Int. Ed. 51, 3897–3900. ( 10.1002/anie.201108175) [DOI] [PubMed] [Google Scholar]
- 42.Wagner GR, Payne RM. 2013. Widespread and enzyme-independent Nɛ-acetylation and Nɛ-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29 036–29 045. ( 10.1074/jbc.M113.486753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizuhara S, Handler P. 1954. Mechanism of thiamine-catalyzed reactions. J. Am. Chem. Soc. 76, 571–573. ( 10.1021/ja01631a071) [DOI] [Google Scholar]
- 44.Benkovic SJ. 1978. On the mechanisms of folate cofactors. Acc. Chem. Res. 11, 314–320. ( 10.1021/ar50128a005) [DOI] [Google Scholar]
- 45.Bazhenova TA, Bazhenova MA, Petrova GN, Mironova SA, Strelets VV. 2000. Catalytic behavior of the nitrogenase iron-molybdenum cofactor extracted from the enzyme in the reduction of C2H2 under nonenzymatic conditions. Kinet. Catal. 41, 499–510. ( 10.1007/BF02756066) [DOI] [Google Scholar]
- 46.Nakada HI, Weinhouse S. 1953. Non-enzymatic transamination with glyoxylic acid and various amino acids. J. Biol. Chem. 204, 831–836. [PubMed] [Google Scholar]
- 47.Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF. 2016. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116 ( 10.1038/nmicrobiol.2016.116) [DOI] [PubMed] [Google Scholar]
- 48.McCollom TM, Seewald JS. 2007. Abiotic synthesis of organic compounds in deep-sea hydrothermal environments. Chem. Rev. 107, 382–401. ( 10.1021/cr0503660) [DOI] [PubMed] [Google Scholar]
- 49.Müller V, Chowdhury NP, Basen M. 2018. Electron bifurcation: a long-hidden energy-coupling mechanism. Annu. Rev. Microbiol. 72, 331–353. ( 10.1146/annurev-micro-090816-093440) [DOI] [PubMed] [Google Scholar]
- 50.Stupperich E, Fuchs G. 1984. Autotrophic synthesis of activated acetic acid from two CO2 in methanobacterium thermoautotrophicum. Arch. Microbiol. 139, 8–13. ( 10.1007/BF00692704) [DOI] [Google Scholar]
- 51.Lang SQ, Butterfield DA, Schulte M, Kelley DS, Lilley MD. 2010. Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim. Cosmochim. Acta 74, 941–952. ( 10.1016/j.gca.2009.10.045) [DOI] [Google Scholar]
- 52.McDermott JM, Seewald JS, German CR, Sylva SP. 2015. Pathways for abiotic organic synthesis at submarine hydrothermal fields. Proc. Natl Acad. Sci. USA 112, 7668–7672. ( 10.1073/pnas.1506295112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss MC, Preiner M, Xavier JC, Zimorski V, Martin F. 2018. The last universal common ancestor between ancient Earth chemistry and the onset of genetics. PLoS Genet. 14, e1007518 ( 10.1371/journal.pgen.1007518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ijiri A, et al. 2018. Deep-biosphere methane production stimulated by geofluids in the Nankai accretionary complex. Sci. Adv. 4, eaao4631 ( 10.1126/sciadv.aao4631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer WW, Hemp J, Johnson JE. 2016. Evolution of oxygenic photosynthesis. Annu. Rev. Earth Planet Sci. 44, 647–683. ( 10.1146/annurev-earth-060313-054810) [DOI] [Google Scholar]
- 56.Raymond J, Segrè D. 2006. The effect of oxygen on biochemical networks and the evolution of complex life. Science 311, 1764–1767. ( 10.1126/science.1118439) [DOI] [PubMed] [Google Scholar]
- 57.Hordijk W, Kauffman SA, Steel M. 2011. Required levels of catalysis for emergence of autocatalytic sets in models of chemical reaction systems. Int. J. Mol. Sci. 12, 3085–3101. ( 10.3390/ijms12053085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steel M, Hordijk W, Xavier JC. 2019. Autocatalytic networks in biology: structural theory and algorithms. J. R. Soc. Interface 16, 20180808 ( 10.1098/rsif.2018.0808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Decker K, Jungermann K, Thauer RK. 1970. Energy production in anaerobic organisms. Angew Chemie Int. Ed. English 9, 138–158. ( 10.1002/anie.197001381) [DOI] [PubMed] [Google Scholar]
- 60.Islam MA, Zengler K, Edwards EA, Mahadevan R, Stephanopoulos G. 2015. Investigating Moorella thermoacetica metabolism with a genome-scale constraint-based metabolic model Integr. Biol. 7, 869–882. ( 10.1039/C5IB00095E) [DOI] [PubMed] [Google Scholar]
- 61.Richards MA, Lie TJ, Zhang J, Ragsdale SW, Leigh JA, Price ND. 2016. Exploring hydrogenotrophic methanogenesis: a genome scale metabolic reconstruction of Methanococcus maripaludis J. Bacteriol. 198, 3379–3390. ( 10.1128/JB.00571-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schönheit P, Buckel W, Martin WF. 2016. On the origin of heterotrophy. Trends Microbiol. 24, 12–25. ( 10.1016/j.tim.2015.10.003) [DOI] [PubMed] [Google Scholar]
- 63.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180. ( 10.1128/MMBR.41.1.100-180.1977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eck RV, Dayhoff MO. 1966. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 152, 363–366. ( 10.1126/science.152.3720.363) [DOI] [PubMed] [Google Scholar]
- 65.Hall DO, Cammack R, Rao KK. 1971. Role for ferredoxins in the origin of life and biological evolution. Nature 233, 136–138. ( 10.1038/233136a0) [DOI] [PubMed] [Google Scholar]
- 66.Daniel RM, Danson MJ. 1995. Did primitive microorganisms use nonhem iron proteins in place of NAD/P? J. Mol. Evol. 40, 559–563. ( 10.1007/BF00160501) [DOI] [Google Scholar]
- 67.Herrmann G, Jayamani E, Mai G, Buckel W. 2008. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190, 784–791. ( 10.1128/JB.01422-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martínez-Cano DJ, et al. 2015. Evolution of small prokaryotic genomes. Front. Microbiol. 5, 742 ( 10.3389/fmicb.2014.00742) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson I, et al. 2010. Complete genome sequence of Methanothermus fervidus type strain (V24ST). Stand. Genomic Sci. 3, 315–324. ( 10.4056/sigs.1283367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hess V, Poehlein A, Weghoff M, Daniel R, Müller V. 2014. A genome-guided analysis of energy conservation in the thermophilic, cytochrome-free acetogenic bacterium Thermoanaerobacter kivui. BMC Genomics 15, 1139 ( 10.1186/1471-2164-15-1139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thiele I, Vlassis N, Fleming RMT. 2014. fastGapFill: efficient gap filling in metabolic networks. Bioinformatics 30, 2529–2531. ( 10.1093/bioinformatics/btu321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin WF. 2019. Carbon–metal bonds: rare and primordial in metabolism. Trends Biochem. Sci. 44, 807–818. ( 10.1016/j.tibs.2019.04.010) [DOI] [PubMed] [Google Scholar]
- 73.Ménez B, Pisapia C, Andreani M, Jamme F, Vanbellingen QP, Brunelle A, Richard L, Dumas P, Réfrégiers M. 2018. Abiotic synthesis of amino acids in the recesses of the oceanic lithosphere. Nature 564, 59–63. ( 10.1038/s41586-018-0684-z) [DOI] [PubMed] [Google Scholar]
- 74.Preiner M, Xavier JC, Vieira A do N, Kleinermanns K, Allen JF, Martin WF. 2019. Catalysts, autocatalysis and the origin of metabolism. Interface Focus 9, 20190072 ( 10.1098/rsfs.2019.0072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hordijk W, Steel M, Kauffman S. 2012. The structure of autocatalytic sets: evolvability, enablement, and emergence. Acta Biotheor. 60, 379–392. ( 10.1007/s10441-012-9165-1) [DOI] [PubMed] [Google Scholar]
- 76.Serra R, Villani M. 2017. Modelling protocells, 1st edn Dordrecht: Springer Netherlands. [Google Scholar]
- 77.Orgel LE. 2008. The implausibility of metabolic cycles on the prebiotic Earth. PLoS Biol. 6, e18 ( 10.1371/journal.pbio.0060018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD. 2015. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 7, 301–307. ( 10.1038/nchem.2202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morowitz HJ, Kostelnik JD, Yang J, Cody GD. 2000. The origin of intermediary metabolism. Proc. Natl Acad. Sci. USA 97, 7704–7708. ( 10.1073/pnas.110153997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kauffman SA. 2011. Approaches to the origin of life on earth. Life 1, 34–48. ( 10.3390/life1010034) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All biochemical data used here is publicly available in KEGG, Uniprot, and in the publications of the genome-scale metabolic models of Moorella thermoacetica [60] and Methanococcus maripaludis [61]. The curated and annotated networks are provided in electronic supplementary material, dataset S1. A custom-made implementation of the maxRAF algorithm was used for the analysis in this paper and is available at https://www.canterbury.ac.nz/engineering/schools/mathematics-statistics/research/bio/downloads/raf/. An example of an input file (global prokaryotic O2-independent network, food set with all small molecules, abiotic carbon, and organic cofactors) is given in electronic supplementary material, dataset S4. A more general-purpose and interactive RAF analyser can be found online at https://github.com/husonlab/catlynet, including several more examples and explanations.