Abstract
Several structures of complexes between viral attachment proteins and their cellular receptors have been determined recently, enhancing our understanding of the molecular recognition processes that guide formation of virus–receptor complexes. Moreover, these structures also highlight strategies by which highly similar viral proteins within a single virus family can adapt to engage different receptors. Consequences of such differences are altered tropism and pathogenicity. An improved understanding of the molecular details of this specificity switching in receptor binding will help to establish links between receptor tropism, spread, and disease. Moreover, it also has relevance for the design and use of viruses as gene delivery vehicles with altered properties as well as for the identification of target viral epitopes of new vaccines.
Introduction
The interaction between a virus and a host cell receptor is the first step in a complex process that eventually leads to cell infection. Viruses must not only be able to specifically attach to cells in order to gain entry into the cell, but their newly formed progeny must also be able to release themselves from the cell membrane after an infection. As a result, attachment and release processes depend on accurately regulated contacts and affinities between viral proteins and their cognate receptor molecules on the cell surface. However, these interactions are constantly subject to changes because of evolutionary pressure on viruses to increase their infection efficiency [1]. Subtle modifications in the virus coat proteins can have drastic consequences, leading to the emergence of a new pathogen with altered infectivity, tissue tropism, or host range. Well-established examples of such modifications include strains of several picornaviruses such as foot-and-mouth disease virus, human rhinoviruses, and coxsackieviruses, all of which can be adapted to use an alternate receptor for cell entry in tissue culture [2, 3, 4]. Another example is the canine parvovirus, which has emerged as a new pathogen of dogs by gaining the ability to recognize the canine transferrin receptor [5]. Finally, measles virus and the SARS coronavirus are also established models for receptor specificity switching as a result of subtle amino acid changes in their viral attachment proteins [6, 7]. A number of well-resolved structures of viruses or viral proteins in complex with cellular receptors are available, and several of these have already contributed to our understanding of parameters that can alter ligand specificities [7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23•, 24, 25••, 26••, 27••, 28••, 29••, 30••, 31•, 32, 33••, 34••]. In this review, we will focus on three examples: adenoviruses (Ads), paramyxoviruses, and polyomaviruses. In each case, recent structural information has allowed for an improved appreciation of strategies used by these viruses to engage different receptors.
Interactions of adenoviruses with their cellular receptors CAR and CD46
Ads are nonenveloped, icosahedral particles that have a trimeric attachment protein, or fiber, protruding from each of the 12 vertices of the capsid [35]. The fiber consists of a globular head (the knob), a fibrous shaft, and a tail. Fifty-one Ad serotypes, belonging to subgroups A–F, are currently known, and they cause a wide range of diseases in humans [35]. While many Ads attach to cells by engaging the coxsackievirus and adenovirus receptor (CAR) [16, 36] or sialic acid [14, 37], most group B serotypes use the ubiquitously expressed membrane cofactor protein (MCP) CD46 as their receptor [38, 39, 40]. The extracellular region of CD46 contains four ∼60 amino acid units known as short consensus repeats (SCR1–SCR4). The crystal structure of the SCR1–SCR2 region of CD46 in complex with the Ad11 knob revealed that the Ad11 knob profoundly realigns the overall conformation of CD46, reshaping its bent surface structure into an elongated rod [25••]. This conformational change is in part mediated by the extrusion of a single hydrophobic residue from the interface between two CD46 domains. A centrally positioned arginine side chain serves as the primary determinant of binding [41•].
The location of the CD46-binding surface is completely distinct from that used by other Ads to bind to CAR (Figure 1a and b). The structure of the complex between Ad12 knob and CAR [16] showed that the Ad12 knob AB-loop serves as the most important determinant of CAR binding as it contributes over 50% to the protein–protein interaction, including three hydrogen bonds involving residues that are conserved in CAR binding Ads serotypes. The elongated AB-loop forms a platform that spans the width of CAR. By comparison, CD46 engages a long and narrow surface formed by the DG-loop, HI-loop, and IJ-loop from two protomers in the Ad11 knob [25••]. The CAR-binding AB-loop is located on a different face of the knob, almost exactly opposite from the site of CD46 attachment. While the overall structures of the Ad11 and Ad12 knobs are very similar, closer inspection of the binding regions for CD46 and CAR reveals that each knob has subtly altered its loop structure in order to gain specificity for its receptor. Non-CAR-binding Ad knobs typically carry an insertion in the AB-loop, which, in the case of Ad11, protrudes from the core β-sheet and likely helps to prevent binding to CAR. Superposition of the Ad12 knob–CAR complex with the Ad11 knob brings the most protruding Cα-atom of the Ad11 AB-loop into very close proximity to the nearest Cα atom of CAR (Figure 1c and e). The distance between the Cα atoms is only about 4 Å, which effectively would prevent CAR from forming a complex with the Ad11 knob in a manner similar to the interaction seen in the Ad12 knob–CAR complex. Conversely, CAR-binding Ad knobs have very short DG- and HI-loops, lacking most of the CD46-binding surface (Figure 1d and f). Interestingly, the modes of CAR and CD46 binding are quite different. The CAR-binding surface of Ad12 features several discrete, smaller contact points separated by large solvent-filled cavities [16], whereas the CD46-binding surface in the Ad11 knob is extensive, continuous, and devoid of solvent molecules [25••]. Despite these differences, both adenovirus types feature large buried surface areas and bind to their respective receptors CAR and CD46 with high affinity [25••, 42]. Ads therefore have evolved to bind different receptors not by adjusting an initial binding surface but by creating an alternative second one while deconstructing the first.
Figure 1.
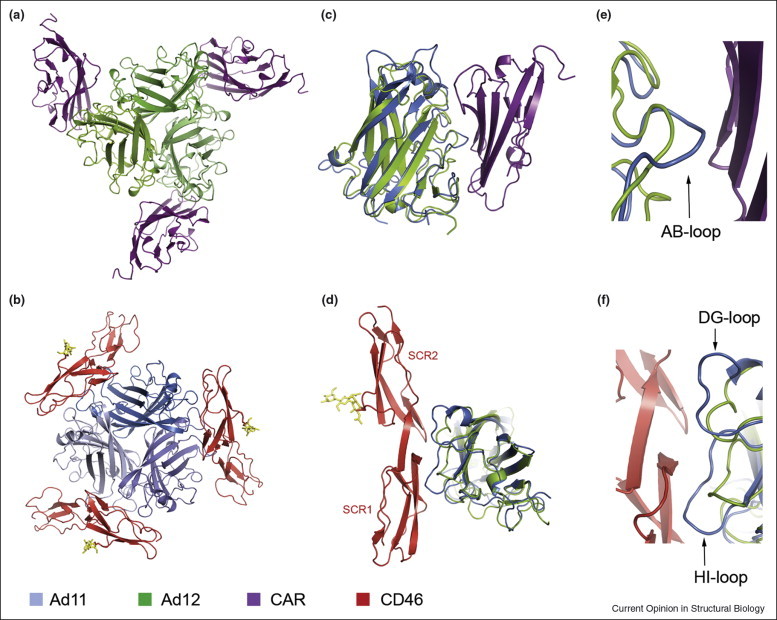
Interactions of the Adenovirus knob with receptors CAR and CD46. (a) Complex between the Ad12 knob and the N-terminal domain of CAR [16]. (b) Complex between the Ad11 knob and the SCR1–SCR2 portion of CD46 [25••]. The orientation of the Ad knobs in (a) and (b) is similar. (c) and (e) Views into the Ad12 knob–CAR interface, with the Ad11 knob superimposed onto the Ad12 knob. CAR binds to a surface that is mostly generated by the AB-loop of the Ad12 knob. The corresponding loop in Ad11 carries a protruding insertion that would prevent binding to CAR in a similar manner. (d) and (f) Views into the Ad11 knob–CD46 interface, with the Ad12 knob superimposed onto the Ad11 knob. CD46 binds to a platform formed by the DG-loop and HI-loop of the Ad11 knob. These loops are much shorter in Ad12. For (c) and (d), complexes were superimposed using all corresponding Ad knob residues. Only one Ad knob monomer is shown in each case, although a second monomer also contributes some contacts in both complexes. Panels (e) and (f) are close-up views of the interfaces shown in (c) and (d), respectively. In all panels, the color code is as follows: CAR, magenta; Ad12 knob, green; Ad11 knob blue; CD46 SCR1–SCR2, red.
Cellular receptor recognition in the paramyxovirus family
The paramyxoviruses are enveloped, negative-stranded RNA viruses that include relevant human and animal pathogens [43]. In the viruses of the paramyxovirus family, cell attachment and virus–cell membrane fusion are mediated by two distinct membrane glycoproteins. The proteins responsible for cell attachment, which will be the focus of this section, exhibit some diversity at the functional level as well as at the level of the cellular receptor to which they bind. Rubulaviruses (e.g. Mumps virus), avulaviruses (e.g. Newcastle disease virus), and respiroviruses (e.g. Sendai virus) bind to cell surface sialic acid via the hemagglutinin–neuraminidase (HN) attachment glycoprotein, a bifunctional protein engaged in recognition as well as hydrolysis of sialic acid. Both activities are absent in other paramyxoviruses such as the morbilliviruses (e.g. measles virus), henipaviruses (Hendra and Nipah viruses), and pneumoviruses (e.g. respiratory syncytial virus) [43]. These latter viruses recognize protein receptors via the hemagglutinin (H) glycoprotein in morbilliviruses or the attachment glycoprotein G in henipaviruses and pneumoviruses.
The paramyxovirus attachment proteins are type II membrane proteins anchored to the virus envelope by a single transmembrane domain [43]. Their extracellular region can be divided into an N-terminal stalk region that serves as a spacer, and a C-terminal globular domain that has receptor-binding activity. In the virus envelope, the attachment proteins are present as disulphide-linked homodimers, and there are indications of tetramer formation in some cases [44, 45, 46]. The attachment proteins form complexes with the fusion (F) proteins, which are also located in the viral envelope. Receptor binding must trigger rearrangements in these complexes that alter the structure of F and result in fusion of the viral and cellular membranes at neutral pH [47].
The C-terminal, globular domains of paramyxovirus HN glycoproteins from Newcastle disease virus (NDV), human parainfluenza type III (PIV3), and parainfluenza virus 5 (SV5) all fold into highly similar six-bladed β-propeller structures [12, 13, 21]. The high degree of similarity is confirmed by the structural alignment shown in Figure 2 . Recent work by several groups has now resulted in crystal structures of three additional paramyxovirus attachment glycoproteins: measles virus H (MV-H) [26••, 27••] as well as Nipah virus G and Hendra virus G (NiV-G and HeV-G, respectively) [29••, 30••]. As NiV-G and HeV-G are very similar, only NiV-G will be discussed in detail here. The MV-H and NiV-G structures superimpose well with those of the previously determined HN proteins (Figure 2), although, as noted before [29••], the agreement with NiV-G is somewhat better compared to MV-H. Despite these structural similarities, NiV-G and MV-H function very differently from the HN proteins as they lack the conserved residues engaged in sialic acid binding and hydrolysis (red in Figure 2). Instead, NiV-G interacts with the ephrin-B2 (EFNB2) and ephrin-B3 (EFNB3) receptors [48, 49], whereas MV-H can bind either CD46 [50, 51] or Signaling Lymphocytic Activation Molecule (SLAM) [52], depending on the MV strain. Thus, a specificity switch from carbohydrate to protein receptor is seen in NiV-G as well as in MV-H. Interestingly, however, the strategy that made this switch possible is very different in the two cases.
Figure 2.
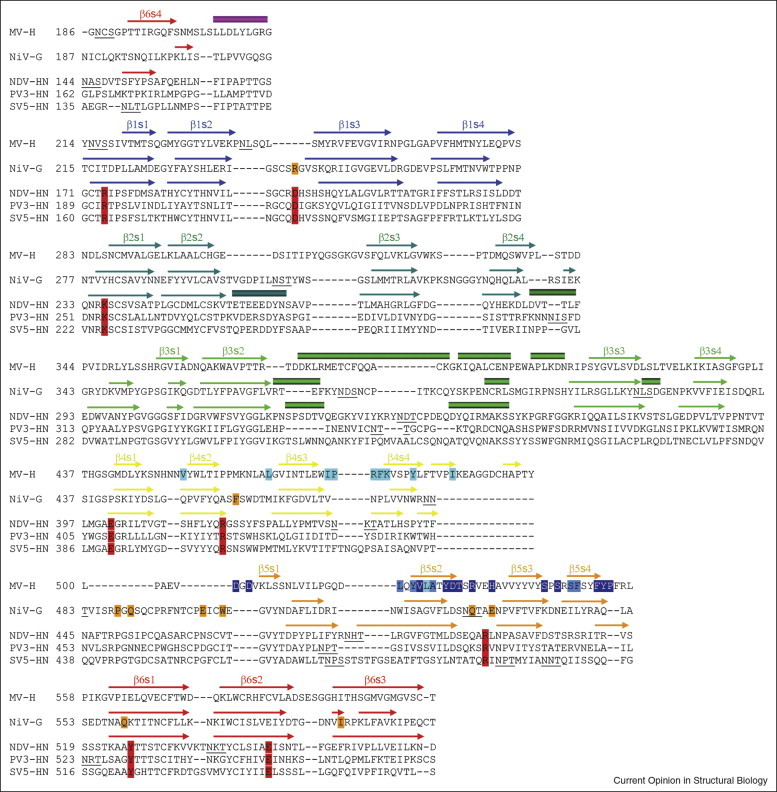
Structural alignment of the paramyxovirus cell attachment proteins. The β-propeller domains of the indicated protein structures were aligned with the program Modeller (program website: http://salilab.org/modeller/modeller.html) with a gap penalty of 1.75. β-Strands and α-helices are represented with arrows and rectangles, respectively. The four β-strands forming each blade are labeled s1–s4, and the six blades in the propeller are labeled β1–β6. Residues engaged in sialic acid binding by the HN proteins are boxed in red [12, 13, 21], residues involved in binding to the EFNB2/EFNB3 receptors by the NiV-G protein are in orange [29••, 30••], and some of the MV-H residues engaged in binding to the CD46 and SLAM receptors are shown in light and dark blue, respectively, whereas residues interacting with both receptor proteins are medium blue [26••, 27••]. Glycosylation sites are underlined.
Structures of complexes between NiV-G in complex with both EFNB2 and EFNB3 receptors are available [29••, 30••], allowing for a detailed comparison of the NiV-G receptor binding mode with that of the sialic acid binding HN proteins (Figure 3 ). The receptor-binding surfaces in HN and NiV-G overlap, and both sialic acid and the EFNB2/EFNB3 receptors bind to sites at the recessed center of the respective β-propeller [29••, 30••]. Protruding hydrophobic residues at the long GH-loop of EFNB2 interact with residues in NiV-G that lie very close to the sialic acid binding site in HN (Figure 3). Thus, NiV-G has an altered contact surface at the center of the propeller. Interestingly, engagement of EFNB2 is accompanied by conformational changes in two NiV-G surface loops that carry EFNB2-contacting residues [53•]. These loops appear to be rather flexible in the unliganded NiV-G structure and only lock into place upon engagement of EFNB2. As only the structure of unliganded MV-H is known so far, we have to rely on receptor-binding data to discuss the putative location of the CD46-binding and SLAM-binding sites on this protein. Both receptor-binding sites map onto the side of the MV-H propeller [26••, 27••] (blue in Figure 3). The putative receptor-binding region in MV-H includes the end of the fourth blade and the beginning of the fifth blade in the β-propeller, a region that exhibits the largest structural difference between MV-H and the other paramyxovirus proteins (Figure 2). This region lacks glycosylation specifically in MV-H (underlined in Figure 2), and thus represents an appealing place for interactions. Indeed, both MV receptors bind to overlapping sites [54] that are now mapped in this region. In contrast, MV-H carries a glycan linked to Asn215 facing toward the center of the β-propeller [27••]. Thus, the region used by both the HN and NiV-G proteins for interactions with their receptors is effectively closed off for binding in MV-H. Compared with the NiV-G protein, MV-H therefore uses a different strategy to engage its cellular receptors. It does not modulate the binding surface at the top of the β-propeller, but instead creates a new one at its side. Glycans are being used as determinants of receptor binding in MV-H by shielding one possible binding region and exposing another.
Figure 3.
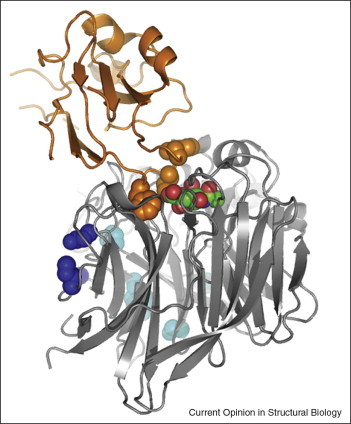
Receptor-binding regions in paramyxovirus cell attachment proteins. Ribbon drawing of the β-propeller domain of the NDV-HN structure (gray) [12], representative of the paramyxovirus attachment proteins, with a bound sialic acid molecule (carbons in green and oxygens in red). A ribbon drawing of the EFNB2 receptor bound to the NiV-G protein is shown in orange [29••]. The position of EFBN2 was generated by superposing the structures of the NiV-G and the NDV-HN β-propeller domains. NiV-G is not shown. The side chains of hydrophobic residues in the GH-loop of the EFBN2 receptor that penetrate into the recessed center of the β-propeller are represented with orange spheres. Side chains of solvent-exposed residues in the HN structure that align with CD46-binding and SLAM-binding residues in the MV-H protein are represented with light and dark blue spheres, respectively.
Ganglioside receptor recognition by the polyomaviruses
Polyomaviruses are a group of small, nonenveloped DNA viruses that can infect birds, rodents, and primates. Members of the group include simian virus 40 (SV40) and murine polyomavirus (Polyoma) as well as a number of human polyomaviruses such as the BK and JC viruses (BKV and JCV, respectively). Recently, a new human polyomavirus was found to be linked to Merkel cell carcinoma, an aggressive type of skin cancer [55•]. All polyomavirus capsids are constructed from 360 copies of the major coat protein, VP1, arranged in pentamers on a T = 7 icosahedral lattice [56]. The cell surface receptors for SV40, Polyoma, and BKV are gangliosides, complex, sialic acid containing sphingolipids that reside primarily in lipid rafts. SV40 uses the ganglioside GM1, whereas BKV binds GD1b and GT1b, and Polyoma attaches to GD1a and GT1b [57, 58]. Structures of complete Polyoma particles and of Polyoma VP1 pentamers in complex with ganglioside receptor fragments [9, 59] revealed that VP1 binds the oligosaccharide portions of the gangliosides in shallow surface pockets that are formed by extensive loops at the outer edge of the capsid. The recently determined structure of the structurally conserved SV40 VP1 pentamer in complex with GM1 [28••] shows that SV40 recognizes its ligand at a similar location on the outer surface of VP1. The Polyoma and SV40 receptors both feature a terminal sialic acid connected to a galactose via an α2,3 glycosidic linkage, and this structural motif is bound by essentially the same region of the capsid.
Although the VP1 surface loops exhibit relatively high sequence variability among polyomaviruses, key residues between SV40 and Polyoma are conserved. For example, a central contact between the sialic acid and Polyoma VP1 is mediated by Arg77, which is replaced with Lys67 in SV40. One would therefore expect to also see at least partially similar contacts in both cases. Strikingly, however, the sialic acid–galactose moieties are bound in completely different orientations to the two proteins (Figure 4 ). In Polyoma, the sialic acid carboxylate faces away from the fivefold axis of the pentamer, forming a key salt bridge with Arg77, and the glycerol side chain points away from the virion into solution. In SV40, the sialic acid glycerol chain in GM1 faces toward VP1, whereas its carboxylate group faces toward the fivefold axis and does not engage in a salt bridge that would neutralize its charge. The Lys67 side chain in SV40 does not contact the sialic acid but instead forms the ridge that separates the binding pockets for the two branches of GM1. The residues that stabilize the conformation of Arg77 in Polyoma, Gln59 and Tyr72, correspond to Asn57 and Gln62 in SV40, respectively, which are unable to make similar interactions with Lys67. As a result, Lys67 is held in place by Asp81, which is equivalent to Gly91 in Polyoma. Residues at equivalent positions in the sequence and in space therefore perform drastically different tasks in SV40 and Polyoma. Thus, two highly homologous viruses that probably have a similar origin evolved to use distinct receptor-binding motifs for the recognition of highly similar receptor molecules.
Figure 4.
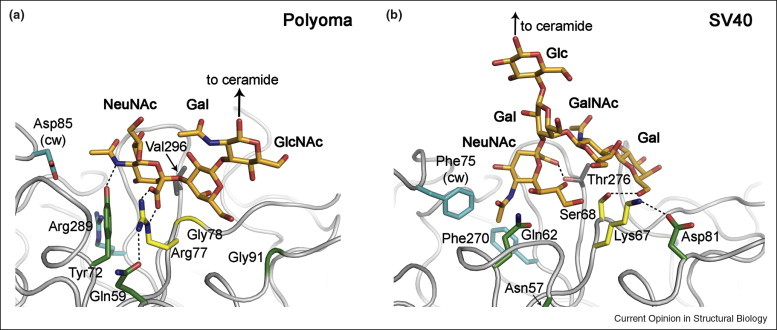
SV40 and Polyoma bind sialic acid in different orientations. (a) Interactions between SV40 VP1 and the oligosaccharide portion of ganglioside GM1 [28••]. (b) Interactions between Polyoma VP1 and an oligosaccharide fragment that corresponds to the terminal portion of ganglioside receptors GD1a and GT1b [9]. The views into the VP1-binding sites are similar in both panels, but the sialic acid moieties are bound in completely different orientations by the two viruses. VP1 residues at equivalent positions are shown in the same color. The Arg–Gly motif of Polyoma and its SV40 counterpart are colored yellow, and the residues holding them in place are shown in green. Hydrophobic residues lining the deep cavity in SV40 and their charged Polyoma equivalents are turquoise.
Conclusions
Within a family of viruses, one frequently finds a number of virus subtypes that vary in cellular tropism and pathogenicity. In many cases, these altered properties can be directly linked to small changes in the coat protein structure that switch specificity from one receptor to another. Structural data on one virus–receptor complex rarely allow for a prediction of how a closely related virus would engage a different receptor. However, the comparison of similar viruses in complex with different receptors has become possible through the structure determination of several virus–receptor complexes. As we have shown here, such a comparison does provide some clues about the strategies by which viruses switch their specificity. We find that, in several cases, exceedingly small changes in surface structure are used to modulate receptor-binding interactions. Binding sites can be deconstructed by simply inserting one residue into a loop, as seen in the Ads that no longer bind CAR, or by subtle modifications that change the binding specificity from sialic acid to a protein receptor, as seen in the paramyxovirus glycoproteins HN and G. Similarly, binding to receptors can be modulated, in part, through the introduction of glycans at strategic positions, as seen in the MV-H glycoprotein. Finally, there exists surprising variability in the mode of viral coat protein binding to sialylated oligosaccharides. These compounds can be recognized by structurally very similar proteins, such as the variants of the polyomavirus VP1 molecules, through highly distinct binding motifs. The available structural database is becoming large enough to perhaps also advance an understanding of specificity switching in related cases where detailed structural information is still lacking.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgments
The authors wish to thank David Persson and Ursula Neu for help with figure preparation, and members of the Stehle laboratory for critical reading of the manuscript. This work has been supported by grants from the Deutsche Forschungsgemeinschaft (SFB685 and STE-1463) to TS and from the Ministerio de Ciencia e Innovacion of Spain (BFU2005-05972 and BFU2008-00971) to JMC.
Contributor Information
Thilo Stehle, Email: thilo.stehle@uni-tuebingen.de.
José M Casasnovas, Email: jcasasnovas@cnb.csic.es.
References
- 1.Baranowski E., Ruiz-Jarabo C.M., Domingo E. Evolution of cell recognition by viruses. Science. 2001;292:1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- 2.Martinez M.A., Verdaguer N., Mateu M.G., Domingo E. Evolution subverting essentiality: dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc Natl Acad Sci U S A. 1997;94:6798–6802. doi: 10.1073/pnas.94.13.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reischl A., Reithmayer M., Winsauer G., Moser R., Gosler I., Blaas D. Viral evolution toward change in receptor usage: adaptation of a major group human rhinovirus to grow in ICAM-1-negative cells. J Virol. 2001;75:9312–9319. doi: 10.1128/JVI.75.19.9312-9319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson J.M., Mohanty J.G., Crowell R.L., St John N.F., Lublin D.M., Finberg R.W. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55) J Virol. 1995;69:1903–1906. doi: 10.1128/jvi.69.3.1903-1906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hueffer K., Parrish C.R. Parvovirus host range, cell tropism and evolution. Curr Opin Microbiol. 2003;6:392–398. doi: 10.1016/s1369-5274(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 6.LeCouturier V., Fayolle J., Caballero M., Carabana J., Celma M.L., Fernandez-Munoz R., Wild T.F., Buckland R. Identification of two amino acids in the hemagglutinin glycoprotein of measles virus (MV) that govern hemadsorption, HeLa cell fusion, and CD46 downregulation: phenotypic markers that differentiate vaccine and wild-type MV strains. J Virol. 1996;70:4200–4204. doi: 10.1128/jvi.70.7.4200-4204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 8.Weis W.I., Brown J.H., Cusack S., Paulson J.C., Skehel J.J., Wiley D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 9.Stehle T., Harrison S.C. High-resolution structure of a polyomavirus VP1–oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 1997;16:5139–5148. doi: 10.1093/emboj/16.16.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fry E.E., Lea S.M., Jackson T., Newman J.W., Ellard F.M., Blakemore W.E., Abu-Ghazaleh R., Samuel A., King A.M., Stuart D.I. The structure and function of a foot-and-mouth disease virus–oligosaccharide receptor complex. EMBO J. 1999;18:543–554. doi: 10.1093/emboj/18.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L., Luo Y., Wu Y., Tsao J., Luo M. Sialylation of the host receptor may modulate entry of demyelinating persistent Theiler's virus. J Virol. 2000;74:1477–1485. doi: 10.1128/jvi.74.3.1477-1485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crennell S., Takimoto T., Portner A., Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin–neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence M.C., Borg N.A., Streltsov V.A., Pilling P.A., Epa V.C., Varghese J.N., McKimm-Breschkin J.L., Colman P.M. Structure of the haemagglutinin–neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Burmeister W.P., Guilligay D., Cusack S., Wadell G., Arnberg N. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J Virol. 2004;78:7727–7736. doi: 10.1128/JVI.78.14.7727-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong P.D., Wyatt R., Robinson J., Sweet R.W., Sodroski J., Hendrickson W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bewley M.C., Springer K., Zhang Y.B., Freimuth P., Flanagan J.M. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 17.Carfi A., Willis S.H., Whitbeck J.C., Krummenacher C., Cohen G.H., Eisenberg R.J., Wiley D.C. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 18.Dormitzer P.R., Sun Z.Y., Wagner G., Harrison S.C. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002;21:885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullen M.M., Haan K.M., Longnecker R., Jardetzky T.S. Structure of the Epstein–Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol Cell. 2002;9:375–385. doi: 10.1016/s1097-2765(02)00465-3. [DOI] [PubMed] [Google Scholar]
- 20.Verdaguer N., Fita I., Reithmayer M., Moser R., Blaas D. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat Struct Mol Biol. 2004;11:429–434. doi: 10.1038/nsmb753. [DOI] [PubMed] [Google Scholar]
- 21.Yuan P., Thompson T.B., Wurzburg B.A., Paterson R.G., Lamb R.A., Jardetzky T.S. Structural studies of the parainfluenza virus 5 hemagglutinin–neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Seiradake E., Lortat-Jacob H., Billet O., Kremer E.J., Cusack S. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fiber heads in complex with the D1 domain of coxsackie and adenovirus receptor. J Biol Chem. 2006;281:33704–33716. doi: 10.1074/jbc.M605316200. [DOI] [PubMed] [Google Scholar]
- 23•.Cao S., Lou Z., Tan M., Chen Y., Liu Y., Zhang Z., Zhang X.C., Jiang X., Li X., Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol. 2007;81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; The structure of the Norovirus (Norwalk-like virus) capsid protein in complex with carbohydrates provides information about receptor binding. Noroviruses are also known as ‘cruise-ship viruses’ and can cause gastroenteritis epidemics. The structure may help with the design of small-molecule reagents that exclusively interact with norovirus.
- 24.Blanchard H., Yu X., Coulson B.S., von Itzstein M. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*) J Mol Biol. 2007;367:1215–1226. doi: 10.1016/j.jmb.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 25••.Persson B.D., Reiter D.M., Marttila M., Mei Y.F., Casasnovas J.M., Arnberg N., Stehle T. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat Struct Mol Biol. 2007;14:164–166. doi: 10.1038/nsmb1190. [DOI] [PubMed] [Google Scholar]; The crystal structure of the Adenovirus type 11 knob in complex with two domains of its receptor, CD46, suggests that the receptor is in a profoundly altered conformation when interacting with its ligand. Such a conformational change may prove to be conserved among the other pathogens that use CD46 as a receptor.
- 26••.Colf L.A., Juo Z.S., Garcia K.C. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]; See annotation to Ref. [27••].
- 27••.Hashiguchi T., Kajikawa M., Maita N., Takeda M., Kuroki K., Sasaki K., Kohda D., Yanagi Y., Maenaka K. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Along with Ref. [26••], this work reports the first crystal structures of a noncarbohydrate binding paramyxovirus attachment protein, the measles virus hemagglutinin. A key finding in these studies is that the hemagglutinin resembles a ‘dead’ neuraminidase fold. The structures also provide a platform for mapping interactions with the measles virus receptors CD46 and SLAM, and show that glycans attached to the hemagglutinin are important for the modulation of virus–receptor interactions.
- 28••.Neu U., Woellner K., Gauglitz G., Stehle T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc Natl Acad Sci U S A. 2008;105:5219–5224. doi: 10.1073/pnas.0710301105. [DOI] [PMC free article] [PubMed] [Google Scholar]; The crystal structure of the capsid protein of SV40, VP1, in complex with the carbohydrate portion of its ganglioside receptor GM1 reveals that the receptor is bound in a shallow groove at the outer surface of the capsid via a complex network of interactions. Comparison with murine polyomavirus receptor complexes demonstrates that SV40 uses a different mechanism of sialic acid binding despite highly conserved architecture of the overall protein.
- 29••.Bowden T.A., Aricescu A.R., Gilbert R.J., Grimes J.M., Jones E.Y., Stuart D.I. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]; See annotation to Ref. [30••].
- 30••.Xu K., Rajashankar K.R., Chan Y.P., Himanen J.P., Broder C.C., Nikolov D.B. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci U S A. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Along with Ref. [29••], this work presents crystal structures of both Nipah and Hendra attachment glycoproteins in complex with their human receptors, EFNB2 and EFNB3. These are the first structures of any paramyxovirus attachment protein in complex with a protein receptor. They have relevance for paramyxovirus attachment and provide a structural template for the design of antivirals that can block virus binding to receptors. Such an approach has proven to be successful for the homologous influenza virus neuraminidase, which uses a structurally related site for receptor binding.
- 31•.Yang Z., Bjorkman P.J. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc Natl Acad Sci U S A. 2008;105:10095–10100. doi: 10.1073/pnas.0804551105. [DOI] [PMC free article] [PubMed] [Google Scholar]; UL18 is a human cytomegalovirus class I MHC homolog that can present peptides. Despite sharing only 25% sequence identity, the structure of UL18 and its mode of peptide binding are surprisingly similar to those of host MHC-I molecules. The crystal structure of a complex between UL18 and the human receptor LIR-1 reveals UL18's strategy of attachment to LIR-1 while avoiding interactions with bound peptides and association with most MHC-I-binding proteins.
- 32.Bu W., Mamedova A., Tan M., Xia M., Jiang X., Hegde R.S. Structural basis for the receptor binding specificity of Norwalk virus. J Virol. 2008;82:5340–5347. doi: 10.1128/JVI.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Choi J.M., Hutson A.M., Estes M.K., Prasad B.V. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc Natl Acad Sci U S A. 2008;105:9175–9180. doi: 10.1073/pnas.0803275105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structures of the Norwalk capsid protein in complex with A-type and H-type histo-blood group antigens reveal that both are bound in a similar manner. The structures explain the narrow specificity of Norwalk virus and suggest a mechanism by which other, less specific viruses in the norovirus family can infect broader human populations. See also Ref. [23•].
- 34••.Kirchner E., Guglielmi K.M., Strauss H.M., Dermody T.S., Stehle T. Structure of reovirus sigma1 in complex with its receptor junctional adhesion molecule-A. PLoS Pathog. 2008;4:e1000235. doi: 10.1371/journal.ppat.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reoviruses engage cells by binding to the immunoglobulin superfamily member junctional adhesion molecule-A (JAM-A). The crystal structure of reovirus attachment protein sigma 1 in complex with JAM-A shows that sigma 1 disrupts the native JAM-A dimer, engaging a single JAM-A molecule via virtually the same interface that is used for JAM-A homodimerization. Thus, reovirus takes advantage of the adhesive nature of an immunoglobulin-superfamily receptor by usurping the ligand-binding site of this molecule to attach to the cellular surface.
- 35.Horwitz M.S. Adenoviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. edn 4. Lippincott-Raven; 2001. pp. 2301–2326. [Google Scholar]
- 36.Roelvink P.W., Lizonova A., Lee J.G., Li Y., Bergelson J.M., Finberg R.W., Brough D.E., Kovesdi I., Wickham T.J. The coxsackievirus–adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnberg N., Edlund K., Kidd A.H., Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor. J Virol. 2000;74:42–48. [PMC free article] [PubMed] [Google Scholar]
- 38.Gaggar A., Shayakhmetov D.M., Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nat Med. 2003;9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 39.Segerman A., Atkinson J.P., Marttila M., Dennerquist V., Wadell G., Arnberg N. Adenovirus type 11 uses CD46 as a cellular receptor. J Virol. 2003;77:9183–9191. doi: 10.1128/JVI.77.17.9183-9191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marttila M., Persson D., Gustafsson D., Liszewski M.K., Atkinson J.P., Wadell G., Arnberg N. CD46 is a cellular receptor for all species B adenoviruses except types 3 and 7. J Virol. 2005;79:14429–14436. doi: 10.1128/JVI.79.22.14429-14436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Persson B.D., Muller S., Reiter D.M., Schmitt B.B., Marttila M., Sumowski C.V., Schweizer S., Scheu U., Ochsenfeld C., Arnberg N., Stehle T. An arginine switch in the species B adenovirus knob determines high-affinity engagement of the cellular receptor CD46. J Virol. 2009;83:673–786. doi: 10.1128/JVI.01967-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes a molecular switch in the interaction between adenoviruses and their receptor CD46. An arginine side chain in the adenovirus knob plays a critical role in mediating interactions with CD46 in knobs that use CD46 as a receptor. Although it is conserved in other, non-CD46 binding knobs, the arginine cannot mediate the same contact in these cases.
- 42.Freimuth P., Springer K., Berard C., Hainfeld J., Bewley M., Flanagan J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J Virol. 1999;73:1392–1398. doi: 10.1128/jvi.73.2.1392-1398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamb R.A., Parks G.D. Paramyxoviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Strauss S.E., editors. Fields Virology. edn 5. Lippincott, Williams and Wilkins; 2006. pp. 1449–1496. [Google Scholar]
- 44.Parks G.D., Lamb R.A. Folding and oligomerization properties of a soluble and secreted form of the paramyxovirus hemagglutinin–neuraminidase glycoprotein. Virology. 1990;178:498–508. doi: 10.1016/0042-6822(90)90347-t. [DOI] [PubMed] [Google Scholar]
- 45.Thompson S.D., Laver W.G., Murti K.G., Portner A. Isolation of a biologically active soluble form of the hemagglutinin–neuraminidase protein of Sendai virus. J Virol. 1988;62:4653–4660. doi: 10.1128/jvi.62.12.4653-4660.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaitsev V., von Itzstein M., Groves D., Kiefel M., Takimoto T., Portner A., Taylor G. Second sialic acid binding site in Newcastle disease virus hemagglutinin–neuraminidase: implications for fusion. J Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colman P.M., Lawrence M.C. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 48.Negrete O.A., Levroney E.L., Aguilar H.C., Bertolotti-Ciarlet A., Nazarian R., Tajyar S., Lee B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 49.Negrete O.A., Wolf M.C., Aguilar H.C., Enterlein S., Wang W., Muhlberger E., Su S.V., Bertolotti-Ciarlet A., Flick R., Lee B. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006;2:e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dörig R.E., Marcil A., Chopra A., Richardson C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 51.Naniche D., Varior-Krishnan G., Cervoni F., Wild T.F., Rossi B., Rabourdin-Combe C., Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tatsuo H., Ono N., Tanaka K., Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–896. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 53•.Bowden T.A., Crispin M., Harvey D.J., Aricescu A.R., Grimes J.M., Jones E.Y., Stuart D.I. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82:11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides evidence for a conformational change in the Nipah Virus G protein upon engagement of the ephrin receptor.
- 54.Santiago C., Björling E., Stehle T., Casasnovas J.M. Distinct kinetics for binding of the CD46 and SLAM receptors to overlapping sites in the measles virus hemagglutinin protein. J Biol Chem. 2002;277:32294–32301. doi: 10.1074/jbc.M202973200. [DOI] [PubMed] [Google Scholar]
- 55•.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides support for an association between a human polyomavirus and Merkel cell carcinoma, a rare but aggressive type of skin cancer. The identified virus, Merkel cell polyomavirus, may be a contributing factor in the pathogenesis of Merkel cell skin carcinoma.
- 56.Liddington R.C., Yan Y., Moulai J., Sahli R., Benjamin T.L., Harrison S.C. Structure of simian virus 40 at 3.8-Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 57.Tsai B., Gilbert J.M., Stehle T., Lencer W., Benjamin T.L., Rapoport T.A. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 2003;22:4346–4355. doi: 10.1093/emboj/cdg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Low J.A., Magnuson B., Tsai B., Imperiale M.J. Identification of gangliosides GD1b and GT1b as receptors for BK virus. J Virol. 2006;80:1361–1366. doi: 10.1128/JVI.80.3.1361-1366.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stehle T., Yan Y., Benjamin T.L., Harrison S.C. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]


