Abstract
Recent cases of emergent diseases have renewed interest in the evolutionary and ecological mechanisms that promote parasite adaptation to novel hosts 1, 2, 3, 4, 5, 6. Crucial to adaptation is the degree of mixing of original, susceptible hosts, and novel hosts. An increase in the frequency of the original host has two opposing effects on adaptation: an increase in the supply of mutant pathogens with improved performance on the novel host 7, 8, 9; and reduced selection to infect novel hosts, caused by fitness costs commonly observed to be associated with host switching 10, 11, 12, 13, 14, 15, 16, 17. The probability of disease emergence will therefore peak at intermediate frequencies of the original host. We tested these predictions by following the evolution of a virus grown under a range of different frequencies of susceptible (original) and resistant (novel) host bacteria. Viruses that evolved to infect resistant hosts were only detected when susceptible hosts were at frequencies between 0.1% and 1%. Subsequent experiments supported the predictions that there was reduced selection and mutation supply at higher and lower frequencies, respectively. These results suggest that adaptation to novel hosts can occur only under very specific ecological conditions, and that small changes in contact rates between host species might help to mitigate disease emergence.
Keywords: HUMDISEASE, MICROBIO
Results and Discussion
We investigated how the degree of mixing between susceptible and resistant host bacteria affected adaptation of viruses to the resistant host. We established replicate bacterial populations with a range of susceptible host frequencies (between 0% and 90%), but with the same total population density, in glass tubes containing nutrient-rich media, and then inoculated 105 isogenic lytic phage particles into the bacteria populations. Phages were evolved by transferring a fraction (1%) of the phage population to fresh bacteria and media every 48 hr, during which time they reached maximal densities of approximately 109 ml−1 in the most permissive treatment (90% susceptible hosts; Figure S1, available online). We re-established initial frequencies of susceptible and resistant bacteria (from frozen stocks) at the beginning of each transfer, to ensure the different mixing treatments imposed consistent selection pressures on the phages. Note that although the percentage of susceptible hosts varied during 48 hr growth, there was a near-perfect positive correlation between the percentage of susceptible hosts between 0 and 48 hr when measured across treatments (see Supplemental Data). The presence of phage clones that could infect the susceptible and “resistant” bacteria were assessed on a daily basis, and densities of phage genotypes that could infect the resistant or susceptible host were determined at the end of the 10 transfer experiment.
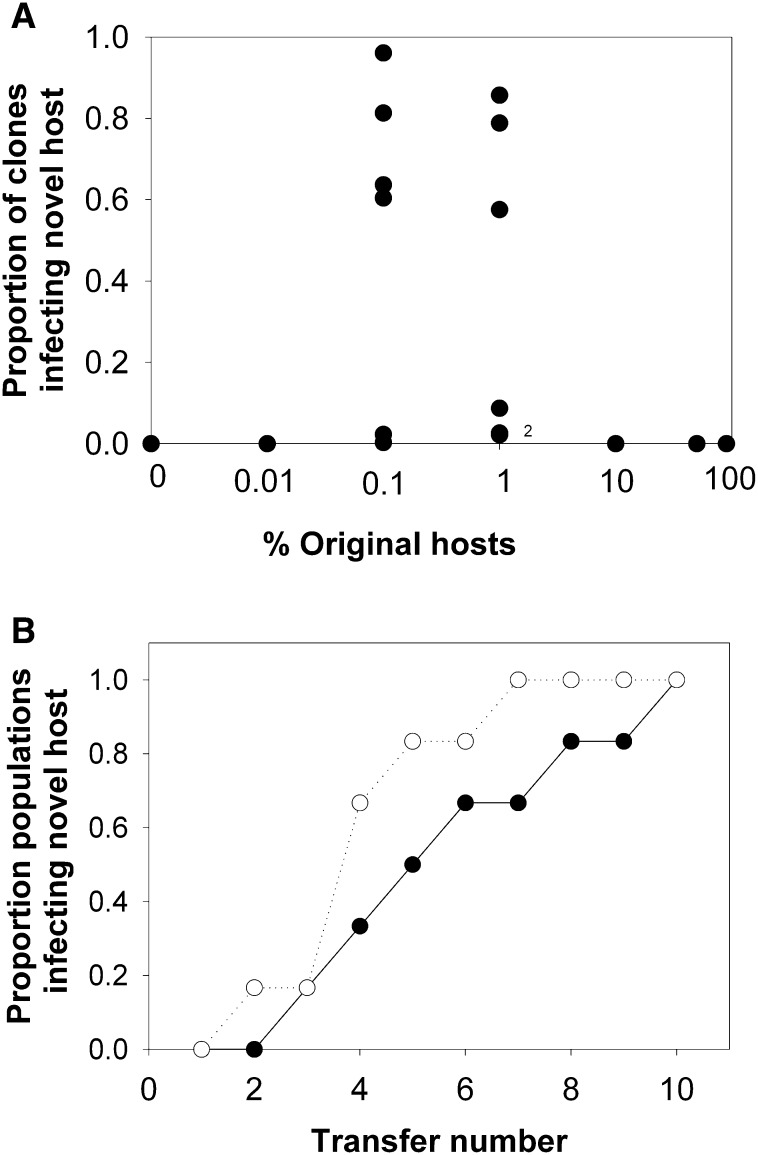
Phages persisted in all populations throughout the experiment, except under the 0% susceptible clone treatment, in which the phages were driven extinct in all replicates by the fourth transfer. As hypothesized, adaptation to the novel host, as measured by the proportion of phages that could infect the novel host at the end of the experiment, peaked at an intermediate susceptible host frequency ( Figure 1). Populations evolved under the 1% and 0.1% treatments contained approximately 50% of individual phages that could infect the novel host, with mutant phages present in all replicates. No mutant phages were detected in any other treatment (Figure 1).
Figure 1.
Virus Adaptation to Host Frequencies
(A) Proportion of susceptible clones infecting the novel host at the end of the experiment as a function original host frequency; 2 indicates two data points.
(B) Proportion of populations through time containing clones that can infect the susceptible host in the 0.1% (closed symbol) and 1% (open symbol) susceptible host frequency treatments.
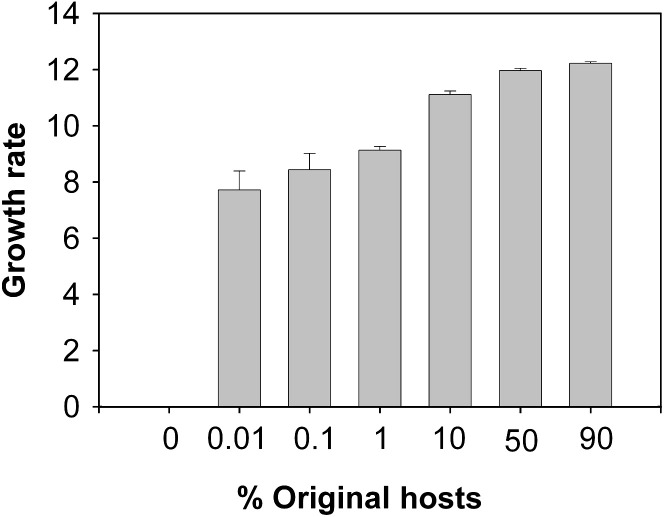
The inability of phages to adapt to the novel hosts at low susceptible host frequencies was likely to be due to reduced population growth rate of the phages limiting the supply of beneficial mutations 7, 8, 9. Consistent with this view, we found that the growth rate of the ancestral phage did indeed increase with increasing susceptible host densities ( Figure 2, quadratic term F 1,45 = 11.0, p = 0.002). Furthermore, the relationship between phage growth rate and starting susceptible host frequencies demonstrates that starting host frequency is a predictable determinant of phage epidemiology.
Figure 2.
Ancestral Virus Growth Rates
Mean and standard error of the mean (±SEM; n = 6) growth rate (estimated Malthusian parameter over 48 hr) of the ancestral phage on the different frequencies of the original host.
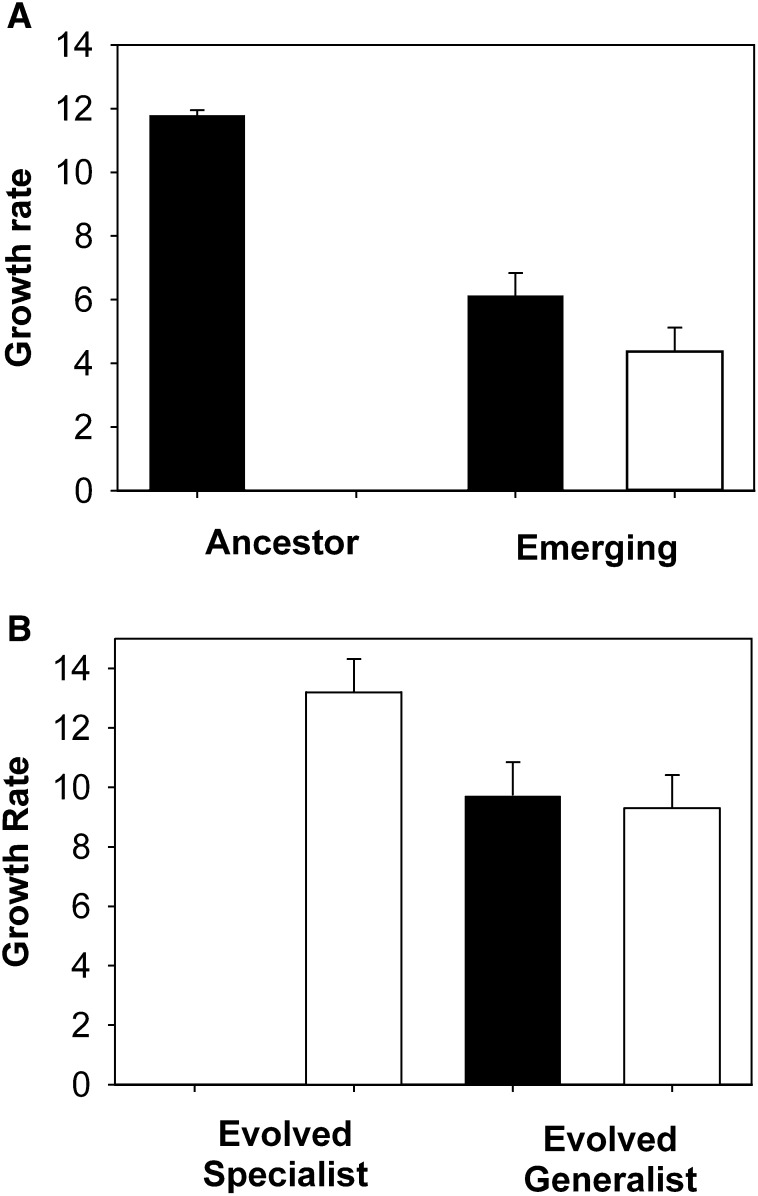
The lack of adaptation of phages at high susceptible host frequencies probably resulted from selection against variants that infect the resistant hosts. Costs associated with generalism can arise for two biologically plausible reasons. First, if viral yields produced by mutant phages in novel hosts are lower than those in the original host (an ecological cost of generalism) 10, 11. This scenario is analogous to that assumed in optimal foraging theory, where patches vary in quality [18]. Second, if the ability to infect novel hosts is associated with reduced performance on the original host 12, 13, 14, 15, 16, 17 (an evolutionary cost of generalism [19]). We tested these possibilities by isolating mutant phage and growing them on both the original and ancestral hosts. We found that growth of the newly arisen mutant phages did not differ significantly between the novel and original host ( Figure 3A; paired t = 1.35, p = 0.24), but that growth was considerably lower than the ancestral phages on the original host (Figure 3a; t = 7.95, p = 0.001).
Figure 3.
Costs of Infecting Novel Hosts
(A) Mean (±SEM; n = 6) growth rate (estimated Malthusian parameter over 48 hr) of the ancestral phage and newly arisen phage mutants that can infect the novel host on the original host (filled bar) and the novel host (open bar).
(B) Mean (±SEM; n = 6) growth rate of evolved generalist and specialist phage clones.
Selection against infecting the novel hosts therefore resulted from an evolutionary cost to generalism. These costs were ameliorated to some extent by the final time point of the experiment, but generalist phages still showed a growth rate cost relative to the ancestral phages on the susceptible host (Figure 3; t = 14.73, p < 0.001). Generalist phages showed marginally worse performance on the resistant host than ancestral phages on the susceptible host (Figure 3; t = 2.35, p = 0.065), but, strikingly, 78% of clones from the final time point had lost the ability to infect the susceptible host; no such specialists were observed when mutant phages were first detected. Consistent with a cost of generalism, these specialist phages performed substantially better on the novel host than did the evolved generalists (Figure 3b; t = 2.7, p = 0.024). We are not aware of other studies that have reported such rapid loss of a virus' ability to infect its original host, and subsequent experiments will explore the ecological and genetic determinants of virus host switching in contrast to the more frequently observed evolution of generalist viruses.
Despite the simplicity of the experimental system, the mechanisms ultimately responsible for this pattern—mutation supply and selection—will inevitably play important roles in many cases of pathogen evolution [20]. Increasing mutation supply rate (by increasing the density of susceptible hosts) will, by definition, increase the probability of a mutation appearing in the population that allows infection of the novel host. Crucial to the subsequent decline in viral adaptation with increasing susceptible host frequency are costs associated with the ability to infect and transmit from multiple hosts, and data from laboratory populations of viruses suggest that both ecological and genetic costs of generalism are common 10, 11, 12, 13, 14, 15, 16, 17. Crucially, natural populations of parasites can often infect and transmit from multiple host species [1], and initial costs associated with infecting novel hosts are also likely to be common 1, 6.
A feature of our experimental system that probably differs markedly from most natural host-parasite systems is that here the two host species are thoroughly mixed within the environment. Interactions in natural populations are likely to be assortative, such that, for a given ratio of host species within an environment, hosts are more likely to interact with their own species, and hence transmit parasites, more than with other species. This can have a major impact in a purely epidemiological (nonevolutionary) context, with heterogeneity decreasing the probability of an epidemic [21] but increasing the severity if it does occur 22, 23. Surprisingly, the extent of assortative interactions appears to have relatively little impact on the probability of parasite adaptation to novel host [9], possibly because heterogeneity can both promote and inhibit parasite transmission and hence density. However, existing theory does not, as far as we are aware, consider the role of assortative interactions on selection against infecting novel hosts because of ecological or evolutionary costs of generalism.
Our results suggest that if initial costs of infecting novel hosts are high, parasite adaptation is likely to occur under relatively restricted host ecological conditions, and might actually be more likely if contact between original and novel hosts is decreased. It is conceivable that adaptation to humans by a number of disease-causing organisms is not fundamentally constrained, but is selected against. For example, avian influenza virus infects, but does not successfully transmit from, humans. Conversely, adaptation of zoonotic parasites to humans (such as SARS coronavirus) might have resulted from increased selection to infect humans because of reduced contact between humans and natural hosts. The precise levels of host mixing that promote, or prevent, adaptation of parasites to novel hosts in natural systems will of course be system specific. However, if low frequencies of original hosts cause the greatest risk of disease emergence, it could be that imperfect isolation or quarantine between host types is more risky, as a management tool to avoid disease emergence, than intermediate levels of mixing. Estimates of parasite transmission from original and novel hosts might allow accurate predictions of the impact of changes in natural or managed host population mixing on disease emergence.
Experimental Procedures
Study Organisms
We used the 40 kb lytic dsDNA bacteriophage SBW25Φ2 which can infect Pseudomonas fluorescens SBW25 [24]. The “original host” was an isogenic mutant of SBW25 that had been genetically marked by an entire deletion of the PanB gene (SBW25ΔpanB) [25]. The resistant host was a SBW25Φ2 clone that had previously evolved resistance to ancestral SBW25Φ2 after approximately 20 generations of coevolution.
Selection Environments
Six populations were initiated using 107 cells (as estimated by optical density at 600 nm) of P. fluorescens SBW25 at each of six different frequencies of the original host (0%, 0.01%, 0.1%, 1%, 10%, 50%, and 90%) in the total population of original and novel hosts, and 105 particles of phage SBW25Φ2. Bacteria densities increased up to 1000-fold in a 48 hr period. Cultures, with no spatial structure (static: shaken for 29:1 min), were grown in 30 ml glass universals with loose plastic caps with 6 ml King's Media B, supplemented with excess pantothenic acid (0.005%), at 28°C. Every 48 hr phages were isolated from the experimental populations by the addition of 100 ml chloroform to 1 ml of culture followed by centrifugation for 3 min at 13,000 rpm, then sampling from the supernatant. A sample of each phage population was stored at 4°C, and 60 μl (1% of each population) was transferred to media containing 107 of the original and novel host bacteria (grown overnight from freezer stocks) at the same ratios as the initial conditions for each phage population. The populations were propagated for a total of ten transfers.
Host Range Mutant Assays
To assay for adaptation to the novel host and persistence on the original hosts, we determined if phage populations formed plaques (i.e., replicated) on the different host types. We spotted 5 μl phages (0.083% of total phages in a tube) onto soft agar containing a lawn of exponentially growing bacteria (original or novel hosts), and determined the presence of plaques after incubation at 28°C for 16 hr. When phage clones that could infect the novel host were first detected in a population, a sample of these clones was assayed on the original host to determine if they retained their infectivity. At the end of the experiment, densities were estimated on both host types from the number of plaque-forming units. Ten clones from each population that formed plaques on the novel host were assayed on the original hosts to determine the frequencies of evolved generalists and specialists.
Growth Rate Assays under Selective Conditions
Growth rates of the ancestral phage were estimated using the same starting densities of bacteria and phages, and same frequencies of susceptible hosts, as for the selection experiment. The number of plaques was determined after 48 hr to ascertain phage population sizes and growth rates calculated as Malthusian parameters (m), m = ln (N f / N 0), where N 0 is initial density and N f final density [26]. Assays were replicated six times and means calculated.
Growth Rates of Evolved Phages
Six clones were isolated from the first six populations in which mutants that could infect the novel host were detected (emerging clones), and six generalist and specialist clones that could infect the resistant host were isolated from independent populations at the final time point. A total of 105 phage particles of each clone and the ancestral phage were then grown on either 107 original or novel hosts. Densities and growth rates after 48 hr were determined as above. Assays on each population were replicated two or three times, and means calculated.
Acknowledgments
We thank the Leverhulme Trust, European Research Council, and the Royal Society for funding this work. We are grateful to Sam Brown, Jim Bull, and Sylvain Gandon for useful discussions.
Published online: April 16, 2009
Footnotes
Supplemental Data include Supplemental Experimental Procedures and one figure and can be found with this article online at http://www.cell.com/current-biology/supplemental/S0960-9822(09)00827-6.
Supplemental Data
References
- 1.Woolhouse M.E.J., Haydon D.T., Antia R. Emerging pathogens: The epidemiology and evolution of species jumps. Trends Ecol. Evol. 2005;20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morens D.M., Folkers G.K., Fauci A.S. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolhouse M.E.J., Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleaveland S., Laurenson M.K., Taylor L.H. Diseases of humans and their domestic mammals: Pathogen characteristics, host range and the risk of emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parrish C.R., Holmes E.C., Morens D.M., Park E.C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antia R., Regoes R.R., Koella J.C., Bergstrom C.T. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennehy J.J., Friedenberg N.A., Holt R.D., Turner P.E. Viral ecology and the maintenance of novel host use. Am. Nat. 2006;167:429–439. doi: 10.1086/499381. [DOI] [PubMed] [Google Scholar]
- 9.Yates A., Antia R., Regoes R.R. How do pathogen evolution and host heterogeneity interact in disease emergence? Proc. Biol. Sci. 2006;273:3075–3083. doi: 10.1098/rspb.2006.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heineman R.H., Springman R., Bull J.J. Optimal foraging by bacteriophages through host avoidance. Am. Nat. 2008;171:E149–E157. doi: 10.1086/528962. [DOI] [PubMed] [Google Scholar]
- 11.Guyader S., Burch C.L. Optimal foraging predicts the ecology but not the evolution of host specialisation in bacteriophages. PLoS ONE. 2008;3:e1946. doi: 10.1371/journal.pone.0001946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao L., Levin B.R., Stewart F.M. A complex community in a simple habitat: An experimental study with bacteria and phage. Ecology. 1977;58:369–378. [Google Scholar]
- 13.Novella I.S., Elena S.F., Moya A., Domingo E., Holland J.J. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J. Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crill W.D., Wichman H.A., Bull J.J. Evolutionary reversals during viral adaptation to alternating hosts. Genetics. 2000;154:27–37. doi: 10.1093/genetics/154.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner P.E., Elena S.F. Cost of host radiation in an RNA virus. Genetics. 2000;156:1465–1470. doi: 10.1093/genetics/156.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohannan B.J.M., Lenski R.E. Linking genetic change to community evolution: Insights from studies of bacteria and bacteriophage. Ecol. Lett. 2000;3:362–377. [Google Scholar]
- 17.Poullain V., Gandon S., Brockhurst M.A., Buckling A., Hochberg M.E. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution. 2008;62:1–11. doi: 10.1111/j.1558-5646.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- 18.Pyke G.H., Pulliam R., Charnov E.L. Optimal foraging: A selective review of theory and tests. Q. Rev. Biol. 1977;52:137–154. [Google Scholar]
- 19.Futuyma D.J., Moreno G. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 1988;19:207–233. [Google Scholar]
- 20.Grenfell B.T., Pybus O.G., Gog J.R., Wood J.L.N., Daly J.M., Mumford J.A., Holmes E.C. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd-Smith J.O., Schreiber S.J., Kopp P.E., Getz W.M. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May R.M., Anderson R.M. The transmission dynmaics of human immunodeficiency virus (HIV) Philos. Trans. R. Soc. Lond. B Biol. Sci. 1988;321:565–607. doi: 10.1098/rstb.1988.0108. [DOI] [PubMed] [Google Scholar]
- 23.Gupta S., Anderson R.M., May R.M. Networks of sexual contacts: Implicatiosn for the pattern of spread of HIV. AIDS. 1989;3:807–817. [PubMed] [Google Scholar]
- 24.Buckling A., Rainey P.B. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rainey P.B., Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 26.Lenski R.E., Rose M.R., Simpson S.C., Tadler S.C. Long term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. Am. Nat. 1991;138:1315–1341. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.