Abstract
Over the past several years a wide variety of molecular assays for the detection of respiratory viruses has reached the market. The tests described herein range from kits containing primers and probes detecting specific groups of viruses, to self-contained systems requiring specialized instruments that extract nucleic acids and perform the polymerase chain reaction with little operator input. Some of the tests target just the viruses involved in large yearly epidemics such as influenza, or specific groups of viruses such as the adenoviruses or parainfluenza viruses; others can detect most of the known respiratory viruses and some bacterial agents.
Keywords: Respiratory viruses, Laboratory diagnosis, Molecular methods, Polymerase chain reaction
Key points
-
•
Molecular tests for the detection of respiratory viruses are more sensitive and can detect more viruses than the traditional methods of culture and antigen detection.
-
•
There are now several molecular assays available, cleared by the Food and Drug Administration, which differ with respect to the viruses detected, instrumentation, throughput, hands-on time, the need for separate nucleic acid extraction, and sensitivity for certain groups of viruses.
-
•
Issues associated with molecular tests for respiratory viruses include: possible false-negative results due to sequence variants; the inability of many assays to discriminate rhinoviruses from enteroviruses; the ability to detect viral nucleic acids in asymptomatic individuals; and the increased prevalence of coinfections.
Introduction
The purpose of this review is to provide an update on recent advances in molecular testing for respiratory viruses, focusing primarily on commercially available assays that have been cleared by the Food and Drug Administration (FDA) for use in the United States. Rather than a detailed look at technical aspects of the assays, attention is paid herein to practical aspects of the tests such as viruses detected, nucleic acid extraction requirements, throughput, and so forth. References are provided for more detailed descriptions of sensitivity and specificity, as well as technical aspects. Issues pertinent to both clinicians and laboratorians regarding molecular assays in general for respiratory virus detection are also discussed.
For the purposes of this article, the following viruses are considered agents of viral respiratory tract infections: influenza A and B; respiratory syncytial virus (RSV); parainfluenza virus types 1 to 4; rhinoviruses; human coronaviruses (NL63, HKU1, 229E, OC43); human metapneumovirus; and adenoviruses. Although other viruses such as human bocavirus and the WU and KI polyomaviruses may be detected in human respiratory tract specimens, their role in causing respiratory tract disease has not been firmly established, and therefore is not covered.
Respiratory tract disease caused by infection with these viruses imposes a significant burden on human society. Community-based studies have revealed the high prevalence of viral respiratory infections, with young children experiencing the greatest number of infections annually and the number of infections decreasing with age. Such studies have also noted that rhinoviruses, influenza viruses and coronaviruses tend to cause the highest number of infections.1, 2 Viral respiratory tract infections are also an important cause of morbidity and mortality in adults, with the elderly being particularly at risk.3, 4 In addition to the very young and elderly, other groups with underlying conditions such as solid-organ and hematopoietic stem-cell transplant recipients,5, 6 patients with chronic obstructive pulmonary disease,7 and those with asthma8, 9 are also known to be at higher risk for severe complications from viral respiratory tract infections. As an example of the importance of viral respiratory infections, in the relatively small country of the Netherlands, a country-wide study led to the estimate that 900,000 individuals visit their physicians annually with an acute respiratory tract infection.2
Because they are obligate intracellular pathogens, the ability to provide a useful laboratory diagnosis for viruses has historically lagged behind that for bacterial infections. Before the introduction of molecular methods into the diagnostic virology laboratory, the traditional methods of cell culture and antigen detection were the predominant methods for the detection of respiratory viruses in clinical specimens. Conventional cell-culture detection of respiratory viruses involves the selection of a range of cell types known to support the growth of respiratory viruses, then observation of the cells by microscopy to look for morphologic changes, referred to as cytopathic effects, which are characteristic for different viruses. A major drawback to conventional viral culture is that it can take as long as 2 weeks for results, seriously compromising the clinical usefulness of this method.10 Although variations of conventional cell culture have been developed that shorten the time to detection,11 there are still several respiratory viruses that grow poorly or not at all in culture, also limiting the usefulness of these methods.
The development of antigen-detection methods was considered to be a major advance in the laboratory diagnosis of viral respiratory infections. By detecting viral antigens directly in patient specimens, either by fluorescent antibody methods or enzyme immunoassay–based techniques, it was possible to obtain a result within a clinically useful time period. Antigen-detection tests remain an important tool in viral diagnostics; however, this method also has shortcomings, with some methods having poor sensitivity for some viral targets, particularly in adult populations. In addition, owing to the lack of conserved antigens there are several respiratory viruses, such as rhinoviruses and coronaviruses, for which antigen-detection assays do not exist.
Molecular detection of viral nucleic acids has revolutionized the laboratory diagnosis of viral infections. Before the development of nucleic acid amplification technologies there were attempts to detect viral nucleic acids in clinical specimens by methods such as dot-blot hybridization, but these were largely unsuccessful because of a lack of sensitivity. The publication of the first description of a nucleic acid amplification method, the polymerase chain reaction (PCR), in 1986,12 describing how nucleic acids could be specifically and exponentially amplified to a readily detected level, was soon followed by numerous publications describing the successful application of this method to the detection of viral nucleic acids in clinical specimens. Today, PCR and other nucleic acid amplification tests (NAATs) are beginning to supplant the traditional laboratory methods to the point that in the future, these methods will likely be the primary laboratory methods in viral diagnostics.
There are advantages and disadvantages associated with the use of NAATs for the diagnosis of viral respiratory infections (Box 1 ). Advantages include the extreme sensitivity of these techniques and the fact that viral viability does not have to be maintained prior to testing, allowing for the potential to ship specimens long distances to testing laboratories; this is in contrast to the problems associated with shipping of specimens for culture, especially for viruses such as RSV, which is known to rapidly lose viability. Another advantage is the ability to detect viruses, such as the human coronaviruses, for which no practical culture or antigen-detection methods exist. The rapid, sensitive, and specific results afforded by molecular testing also allow for the timely institution of antiviral therapy and the proper cohorting of patients admitted to the hospital. There are, however, disadvantages associated with NAAT testing as well. For example, there is the potential for the appearance of sequence variants, which can produce false-negative results. It has also become well established that molecular tests can detect viral nucleic acids in respiratory specimens obtained from asymptomatic individuals, complicating the interpretation of results. NAAT assays can also be more expensive than nonmolecular methods, although with the introduction of ever more assays to the market it is expected that prices will drop. These and other molecular testing issues are discussed in greater depth later.
Box 1. Advantages and disadvantages of NAATs for the detection of respiratory viruses.
Advantages of NAATs for the detection of respiratory viruses
-
•
The ability to identify viruses that are not detected by conventional culture and antigen detection methods
-
•
Extreme sensitivity
-
•
Rapidity and accuracy of results allow for timely institution of antiviral therapy and appropriate infection control
-
•
Because viral viability does not need to be maintained, specimen transport conditions can be relaxed, allowing specimens to be sent to distant testing sites
Disadvantages of NAATs for the detection of respiratory viruses
-
•
False-negative results due to the existence of sequence variants
-
•
NAATs more often result in detection of viruses in asymptomatic individuals than other methods
-
•
Higher cost
NAATs for the detection of respiratory viruses
NAATs for the detection of respiratory viruses have evolved from user-developed and laboratory-developed tests using conventional PCR technology with electrophoresis-gel detection of products, to user-developed real-time PCR-based tests whereby amplified products are detected by technologies primarily involving the production of luminescent signals that are proportional to the amount of target amplified. A problem with these technologies is that they are limited with respect to the number of targets that can practically be amplified and detected, or “multiplexed,” in a single reaction. For many of the traditional real-time platforms 3 or 4 targets are the maximum, a real drawback when one wants to comprehensively detect up to 20 or more different targets. Other technologies have now appeared in commercial formats, which are able to surmount the limitations of the other platforms. For a detailed description of these technologies, the reader is referred to an excellent review dealing with this topic.13
There is an extensive literature describing noncommercial user-developed NAATs for the detection of respiratory viruses. Because this review deals with commercially available assays for the detection of respiratory viruses, the reader is referred to reviews dealing with this topic.14, 15 However, some brief observations can be made concerning such assays. There are many examples in the literature of comparisons of molecular assays for a particular virus with conventional methods such as culture and/or antigen detection. For some viruses, such as RSV and influenza viruses, there appears to be a modest but real increase in sensitivity, whereas for others such as human metapneumovirus (hMPV) and human rhinoviruses, there is a more significant increase in detection by molecular methods. In addition, user-developed assays tend to be developed to detect one virus, or one group of related viruses, and are constrained by the previously mentioned limitations of conventional real-time assays to a maximum of 3 or 4 targets. Because the signs and symptoms of respiratory virus infections can overlap between the different viruses, to confidently rule out all potential viruses it would be necessary to run a battery of assays. One instance in which single directed assays may be useful is in geographic areas where there are annual epidemics of influenza and RSV. During such epidemics it may make sense to test just for these viruses before testing for other agents.
Commercially available NAATs for respiratory viruses
Hologic Assays
The company starting out as Prodesse, later acquired by Gen-Probe and now Hologic (Hologic Gen-Probe Inc, San Diego, CA), offers 5 assays for molecular detection of respiratory viruses (Box 2 ). The assays are all based on TaqMan real-time PCR technology; require that nucleic acids be extracted by either the Roche MagNA Pure (Roche Diagnostics, Indianapolis, IN) or bioMerieux easyMAG (bioMerieux Inc, Durham, NC) automated extractors; and require that the reactions be run on a Cepheid SmartCycler (Cepheid, Sunnyvale, CA). The kits contain all of the reagents necessary to run the reaction, including an internal control, and have the benefit of being cleared for in vitro diagnostic use by the FDA. Publications have described the use of the assays as comparators for other tests, with favorable results.16, 17 In a comparison of 3 influenza PCR assays the ProFlu+ failed to detect 2 of 29 seasonal influenza A H1 viruses but was 100% sensitive for similar numbers of influenza H3, 2009 H1N1, and influenza B specimens.18 In addition, the company Web site provides performance data and links to data presented in poster presentations. These assays might be particularly useful for those wishing to screen during yearly epidemics of influenza and RSV, or to determine the cause of RSV-like illness in young children testing negative for RSV for which hMPV or parainfluenza virus might be a consideration. The ProFAST+ assay that subtypes influenza A could be useful for guiding antiviral therapy. Drawbacks to these assays include the necessity to run 4 separate assays to rule out all of the targets, and the fact that they do not detect parainfluenza type 4 or any of the coronaviruses. In addition, the requirement to use the Cepheid SmartCycler would limit throughput to 16 samples per SmartCycler module.
Box 2. Respiratory virus NAAT kits offered by Hologic (Gen-Probe/Prodesse).
-
•Prodesse Pro hMPV+ Assay
-
○Detects human metapneumovirus
-
○
-
•Prodesse ProAdeno+ Assay
-
○Detects human adenoviruses associated with respiratory infections
-
○
-
•Prodesse ProFAST+
-
○Discriminates subtypes of Influenza A including:
-
▪Seasonal influenza A/H1
-
▪Seasonal influenza A/H3
-
▪2009 H1N1 influenza A
-
▪
-
○
-
•Prodesse ProFlu+ Assay
-
○Detects influenza A, influenza B, and RSV
-
○
-
•Prodesse ProParaflu+ Assay
-
○Detects and differentiates human parainfluenza virus 1, 2, and 3
-
○
Quidel Assays
Quidel Corp (San Diego, CA) markets 2 FDA-cleared assays for the detection of respiratory viruses. The Influenza A+B assay detects both influenza A and B viruses, and the hMPV Assay detects human metapneumovirus. Both assays are sold in a kit format containing primers and fluorescently labeled probes, master mix, and an internal processing control. Quidel recommends the inclusion of a positive control for each assay, either in the form of a commercial product, which they sell separately, or through the use of known previously positive specimens. Both assays are approved for nasal or nasopharyngeal swab specimens extracted on the bioMerieux NucliSENS easyMAG automated extractor, and require amplification on either a Cepheid SmartCycler or ABI 7500 Fast Dx for the Influenza A+B assay or the ABI 7500 Fast Dx for the hMPV test.
To date there are no peer-reviewed evaluations of the Quidel assays available, but performance data are available for download from the company Web site.19 Advantages of the assays include: molecular detection of 3 important and common respiratory viruses; 2° to 8°C kit storage temperature (no need to store frozen); room temperature setup; and 2-year shelf-life of reagents. Disadvantages include inability to detect other respiratory viruses and the lack of peer-reviewed evaluations of the assays.
Cepheid Assay
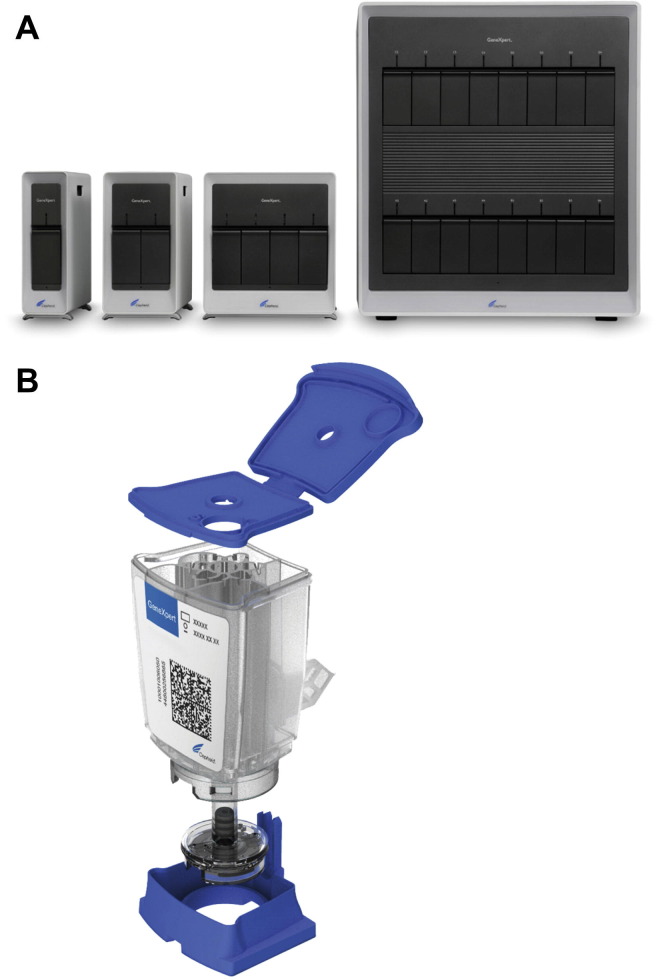
The Cepheid GeneXpert Flu Assay (Cepheid, Sunnyvale, CA) consists of a single-use disposable cartridge and associated instrument, including a computer with analysis software (Fig. 1 ). Following addition of specimen and placement in the instrument, extraction of nucleic acids and PCR take place within the cartridge. An independent evaluation of the Xpert Flu test reported that the assay has 2 minutes of hands-on time, with the run being completed in 76 minutes.20 The assay detects and differentiates influenza A and B, identifies influenza A 2009 H1N1 if present, and includes an internal control. The assay is FDA-cleared for use with nasal aspirates and washes, as well as nasopharyngeal swabs. The instrument itself comes in different iterations capable of accommodating 1, 2, 4, or 16 cartridges at a time. In addition to the influenza assay, Cepheid offers several cartridges for the detection of other targets.
Fig. 1.
Cepheid GenXpert system. (A) Instruments with 1- to 16-cartridge capacity. (B) Exploded view of GenXpert cartridge.
(Courtesy of Cepheid, Sunnyvale, CA; with permission.)
The Cepheid GeneXpert system was the first technology to allow for democratization of molecular testing. When molecular testing was first introduced into the diagnostic virology laboratory it was viewed as a technically demanding method that required highly trained technologists. The “specimen in, answer out” format of the GeneXpert system allows for molecular testing by laboratories with little or no previous experience in the area, with Clinical Laboratory Improvement Amendments classifying the Xpert Flu assay as “moderately complex.”
To facilitate testing during the outbreak of 2009 H1N1 influenza A, an initial version of the Xpert Flu assay detecting only influenza A with differentiation of 2009 H1N1 influenza A was granted Emergency Use Authorization in December 2009. Published comparisons of this version of the Xpert Flu assay with conventional methods, laboratory-developed techniques, and commercial PCR assays for the detection of influenza A viruses revealed acceptable sensitivity and specificity, with somewhat reduced sensitivity for the detection of 2009 H1N1 viruses.21, 22, 23 An evaluation using a collection of retrospectively tested respiratory specimens and a subsequent FDA-cleared version of the Xpert Flu assay with the added ability to detect influenza B reported 100% sensitivities for the detection of seasonal H1N1 and H3N2 influenza A viruses and influenza B viruses, but only 77% sensitivity for 2009 H1N1, compared with a laboratory-developed PCR test with up-front automated nucleic acid extraction.20 Despite the reduced sensitivity for 2009 H1N1 viruses, the investigators indicated that the rapid turnaround time of the Xpert Flu assay make it a reasonable option for laboratory testing. Two other peer-reviewed studies comparing the FDA-cleared version of the Xpert Flu assay with culture, direct fluorescent antibody staining, antigen immunoassays, and reference molecular tests report generally enhanced sensitivity relative to conventional methods and acceptable performance relative to reference molecular methods, with both studies supporting the use of the Xpert Flu assay for the rapid diagnosis of influenza.24, 25
The Cepheid Xpert Flu assay offers laboratories the advantage of a rapid, sensitive molecular test for influenza that is simple to perform. The disposable, self-contained cartridge system, in addition to abrogating the need for up-front nucleic acid extraction, also greatly reduces the opportunity for false positives attributable to amplicon contamination that can affect other NAATs. Disadvantages of the assay include, depending on the instrument configuration, limited throughput and the fact that the assay only detects influenza viruses.
IQuum Assay
IQuum (Marlborough, MA) offers a molecular assay called the Liat Influenza A/B assay. The assay is FDA-cleared for nasopharyngeal swabs, and detects and differentiates influenza A and B. The assay is in a sample-in-answer-out format, and has a turnaround time of only 20 minutes with about 1 minute of hands-on time, according to information provided by the company.26 The assay consists of a single-use disposable Liat Influenza A/B Assay Tube containing all the test reagents and the associated instrument, called the Liat Analyzer. The assay uses TaqMan probe real-time PCR chemistry and includes all appropriate controls. Because of the simplicity of operation and rapidity of results, the assay is being considered for possible use as a point-of-care test. Performance data provided by the manufacturer indicate a sensitivity and specificity relative to culture of 100% and 96.8% for influenza A and 100% and 94.1% for influenza B. At the time of writing there were no peer-reviewed evaluations of the assay available.
Focus Diagnostics Assays
Focus Diagnostics Inc (Cypress, CA) markets 3 FDA-cleared molecular diagnostic assays for the detection of influenza viruses and RSV. The Simplexa Influenza A H1N1 (2009) assay detects influenza A viruses and differentiates 2009 influenza A H1N1. The Simplexa Flu A/B & RSV assay detects and differentiates influenza A and B viruses and RSV. Both assays require the use of the 3M Integrated Cycler and its associated computer and analysis software (Fig. 2 ). The assays are real-time PCR assays that use a bifunctional fluorescent probe-primer combined with a reverse primer, to specifically amplify and detect viral sequences and include an internal control. Both assays have requirements regarding acceptable specimen types and nucleic acid extraction methods (Box 3 ). The 3M Integrated Cycler uses a novel single-use Universal Disk that contains 96 reaction positions. Of note, Focus markets other molecular assays, all of which use the same reaction conditions so that different assays can be simultaneously run on the same disk. The single independent evaluation of the Simplexa Flu A/B & RSV available at this time reported that the test requires 45 minutes of hands-on time, with results being available in approximately 2.5 hours.27 The same investigators reported that in a comparison with another FDA-cleared NAAT (Nanosphere Verigene RV+) using both retrospective and prospective specimens, the Simplexa assay had lower a sensitivity for all 3 targets, which was statistically significant for influenza A and B.27 This result was noted to be at odds with the manufacturer’s sensitivity data, with the investigators indicating that strain differences could be the explanation for their observation. The manufacturer’s performance data are available for download from the company Web site.28
Fig. 2.
3M Integrated Cycler and Focus Simplexa kit.
(Courtesy of Focus Diagnostics, Cypress, CA; with permission.)
Box 3. Respiratory NAAT kits offered by Focus Diagnostics.
-
•Simplexa Influenza A H1N1 (2009)
-
○Detects influenza A with differentiation of 2009 H1N1
-
○FDA-cleared for nasopharyngeal swabs and aspirates
-
○Requires nucleic acid extraction with either:
-
▪Automated Roche MagNA Pure LC System
-
▪Manual Qiagen QIAamp Viral RNA Mini Kit (Qiagen Inc, Valencia, CA)
-
▪
-
○Uses 96-well Universal Disk and 3M Integrated Cycler
-
○
-
•Simplexa Flu A/B & RSV
-
○Detects and differentiates influenza A and B and RSV
-
○FDA-cleared for nasopharyngeal swabs
-
○Requires nucleic acid extraction with either:
-
▪Automated Roche MagNA Pure LC System
-
▪Automated bioMerieux NucliSENS easyMag
-
▪
-
○Uses 96-well Universal Disk and 3M Integrated Cycler
-
○
-
•Simplexa Flu A/B & RSV Direct
-
○Detects and differentiates influenza A and B and RSV
-
○FDA-cleared for nasopharyngeal swabs
-
○Does not require up-front nucleic acid extraction
-
○Uses 8-well Direct Amplification Disk and 3M Integrated Cycler
-
○
Focus Diagnostics also offers the Simplexa Flu A/B & RSV Direct assay, which uses their Direct Amplification Disk, a disk with 8 sample positions that both extracts nucleic acids and amplifies viral sequences for influenza A and B and RSV. Through the use of adhesive foil covers for the reaction positions, it is not necessary to choose between having to run 8 specimens or waste unused positions; rather, the disk can be reused until 8 specimens have been run. The assay uses the same method as the other Simplexa assays, and requires the 3M Integrated Cycler. At the time of writing there were no independent evaluations of the assay, but manufacturer’s data are available for download from the Web site.28
The Simplexa assays offer the advantage of a multiplex assay for 2 of the most prevalent respiratory viruses, a high throughput using the 96-well disk, and a cycling program that allows for the simultaneous running of other Focus Diagnostic assays. The Simplexa Direct assay represents another choice for laboratories considering a sample-in-answer-out platform. Disadvantages include that the assays only test for influenza and RSV, and that there are only minimal peer-reviewed data available.
Nanosphere Assay
In addition to other infectious disease and genetics targets, Nanosphere Inc (Northbrook, IL) offers a commercial FDA-cleared assay, called the Verigene RV+, which detects influenza and RSV in nasopharyngeal swab specimens. The assay detects and differentiates influenza A and B and RSV, and also provides influenza A subtyping information (seasonal H1 and H3 and 2009 H1N1). In addition, the assay subtypes RSV, and a version of the product available outside the United States includes detection of the H275Y mutation conferring oseltamivir resistance in influenza A viruses. The assay is in a sample-in-answer-out format, therefore it does not require nucleic acid extraction and includes all the necessary reagents, including processing and inhibition controls.
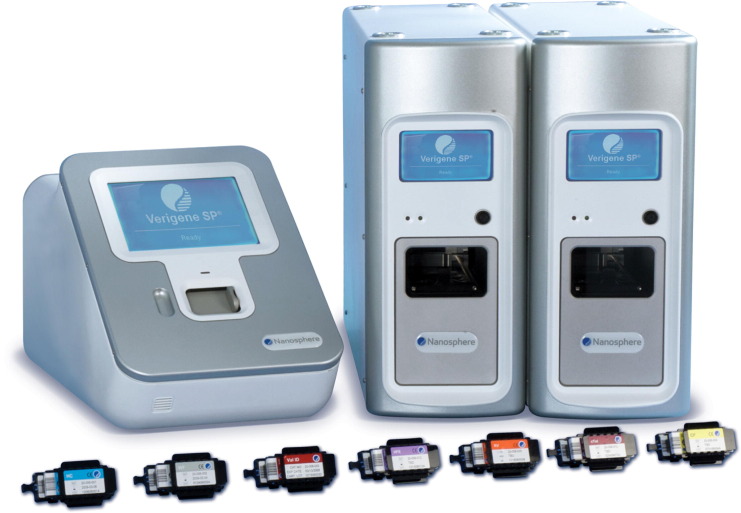
The assay, using a novel detection technology, takes place in a single-use cartridge that is placed in a Verigene Processor SP instrument where nucleic acids are extracted, purified, and target-amplified if present (Fig. 3 ). A hybridization step involving gold nanoparticles to which are bound probes specific for the amplified product is used as part of the detection process. The cartridge is then placed in the Verigene Reader, which reads the result by the detection of light scatter rather than other methods such as fluorescence. For those interested, Thaxton and colleagues29 have published a detailed description of the technology. An independent evaluation reported that the assay requires 5 minutes of hands-on time, and the results are ready in 2.5 hours.27
Fig. 3.
Nanosphere Verigene system. Verigene processor on the right, reader and cartridges on the left. Instrument footprints in inches: Processor, 7.6 width × 18.7 height × 22.9 depth; reader, 11.7 width × 12.4 height × 20.5 depth. Processors are stackable. No computer is required for operation.
(Courtesy of Nanosphere, Northbrook, IL; with permission.)
In the single peer-reviewed evaluation of the Verigene RV+, the assay had 97% sensitivity for influenza A, 100% sensitivity for influenza B, and 100% sensitivity for RSV, with good specificities for all targets as determined using a set of several hundred retrospective and prospective specimens that were tested with other commercial assays, with discrepant results resolved using a laboratory-developed NAAT.27
Advantages of the Verigene RV+ include offering another choice in the sample-in-answer-out format, with limited labor time as well as the reduced risk of contamination previously mentioned for other self-contained disposable systems. The ability to detect the most common epidemic respiratory viruses and the ability to subtype influenza A viruses are other assets. Disadvantages include the low throughput, although this can be increased by having more processor modules, and the fact that only influenza and RSV are detected by the assay.
GenMark Assay
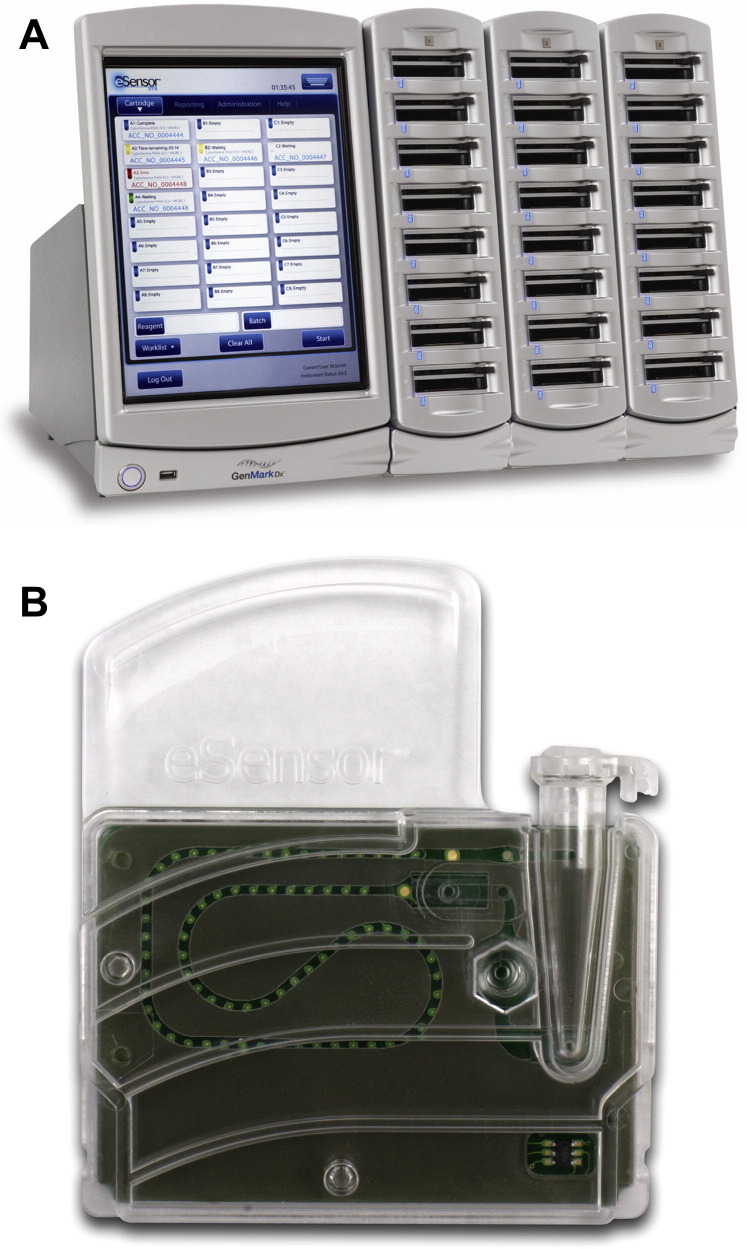
The GenMark eSensor Respiratory Viral Panel (GenMark Diagnostics, Carlsbad, CA) is a multiplex PCR assay, cleared by the FDA for nasopharyngeal swabs for the detection of 14 respiratory viruses (influenza A [seasonal H1, H3, 2009 H1], influenza B, RSV A and B, parainfluenza 1, 2, and 3, human metapneumovirus, rhinovirus, and adenovirus B/E and C). The kit provides all the reagents for performing the assay, and requires nucleic acid to be extracted on a bioMerieux NucliSENS easyMAG followed by amplification with a thermocycler using 0.2-mL tubes. Following amplification, potential amplicons are loaded into an eSensor cartridge, which is then placed in the XT-8 instrument where detection takes place by a novel technology involving production of electric current through specific binding of amplicons to a gold-plated electrode (Fig. 4 ). The instrument is of random access with spaces for up to 24 cartridges, and has an integrated computer with analysis software. An independent evaluation found that the assay required about 6.5 hours to test 6 specimens, with hands-on time of about 1 hour.30
Fig. 4.
GenMark eSensor system. (A) XT-8 instrument. (B) eSensor cartridge. Instrument footprint in inches: 15.75 width × 18.11 height × 16.14 depth. A computer is not required for operation.
(Courtesy of GenMark Diagnostics, Carlsbad, CA; with permission.)
A single peer-reviewed study evaluated the eSensor by testing 250 frozen specimens from pediatric patients and comparing the results with a panel of laboratory-developed PCR assays, with overall agreement between the methods being reported as 99.2%.30 Of note, the investigators reported that the GenMark assay was more sensitive and specific than their laboratory assay for the detection of rhinoviruses and, although they found that the assay may have higher sensitivity for some adenovirus types, the assay showed some cross-reactivity for adenoviruses other than those found in species B, C, and E.
Advantages of the eSensor Respiratory Viral Panel include: the ability to detect a wide range of respiratory viruses in a single assay; the ability to subtype influenza A and RSV viruses; potentially higher sensitivity for some adenovirus types; and higher specificity for rhinoviruses. Potential disadvantages to the assay include: the inability to detect coronaviruses and parainfluenza type 4; the requirement for up-front nucleic acid extraction; and potential contamination issues raised by the need to manipulate PCR products following amplification.
Luminex Assays
Luminex (Austin, TX) offers 2 multiplex molecular assays for the detection of respiratory viruses, the xTAG Respiratory Virus Panel (RVP) and the xTAG RVP FAST assays. The assays detect different numbers of respiratory viruses and are cleared for specific nucleic acid extraction methods (Box 4 ). Other versions of these assays that detect different targets are available in other global markets outside the United States.
Box 4. Luminex assays.
-
•Luminex xTAG RVP Assay
-
○12 targets including: RSV A and B; influenza A (H1, H3); influenza B; parainfluenza 1, 2, and 3; human metapneumovirus; adenoviruses; rhinovirus/enterovirus
-
○Cleared for nasopharyngeal swabs
-
○Requires:
-
▪Extraction on either:
-
•Qiagen QIAamp MiniElute (manual)
-
•bioMerieux EasyMAG (automated)
-
•bioMerieux MiniMAG (manual)
-
•
-
▪Thermocycler accommodating 0.2-mL tubes and 96-well plates
-
▪Specific lots of TAQ enzyme
-
▪
-
○Time to result ∼8.5 hours (including extraction)
-
○
-
•Luminex xTAG RVP FAST Assay
-
○8 targets including: RSV; influenza A (H1, H3); influenza B; human metapneumovirus; adenoviruses; rhinovirus/enterovirus
-
○Cleared for nasopharyngeal swabs
-
○Requires:
-
▪Extraction on:
-
•bioMerieux EasyMAG
-
•
-
▪Thermocycler accommodating 0.2-mL tubes and 96-well plates
-
▪
-
○Time to result ∼5 hours (including extraction)
-
○
Both assays use a multiplex real-time PCR performed in a thermocycler using 0.2-mL reaction tubes, and include extraction and assay controls. To overcome the technical issues associated with detecting products of large multiplex PCR assays, both assays use the proprietary Universal Tag sorting system whereby virus-specific oligonucleotide tags are added to viral amplicons, after which the tagged amplicons are hybridized to a liquid suspension of microsphere bead sets in a 96-well plate. Each bead set has a virus-specific antitag bound to its surface, with each bead set uniquely identified by the ratio of dyes impregnated into the beads. Each colored bead set thus represents a specific virus through the bead-antitag-tag-amplicon interaction. Following hybridization, the beads are then read by the Luminex 100/200 instrument (Fig. 5 ), a flow cell using dual lasers, one identifying the bead set and the other determining whether or not an amplicon is bound to the bead through the emission of a phycoerythrin reporter. The run time for the RVP assay is about 8.5 hours, including extraction. The RVP FAST assay decreases run time through the elimination of some of the assay steps; an independent assessment reports hands-on time for the FAST assay of 60 to 80 min, with the time to results, including extraction, requiring 5 hours.31 For those wishing more information, a detailed description of RVP technology has been published by the company.32
Fig. 5.
Luminex 200 instrument. Luminex 200 (right) and SD sheath fluid module (left). Instrument footprints in inches: Luminex 200, 17.00 width × 9.50 height × 20.00 depth; SD Module, 8.00 width × 9.75 height × 11.75 depth. A computer is required for operation.
(Courtesy of Luminex Corporation, Austin, Texas; with permission.)
The xTAG RVP assay was the first large multiplex respiratory virus assay to enter the market as an FDA-cleared assay, and as such there are several reports demonstrating increased or comparable sensitivity for the detection of respiratory viruses relative to antigen detection/culture methods and other molecular methods.33, 34, 35, 36 Other studies have demonstrated cost savings realized by implementing the RVP assay in place of conventional methods.37, 38
Although the RVP assay has increased sensitivity relative to conventional methods, when compared with other multiplex molecular assays the RVP assay has demonstrated reduced sensitivity for some targets such as parainfluenza viruses39 and RSV.40 Likewise, the RVP FAST assay, although demonstrating increased sensitivity relative to antigen detection/culture methods, has been reported to have reduced sensitivity for some targets relative to other molecular assays. Gharabaghi and colleagues39 reported reduced sensitivity for influenza B and adenoviruses, with another study also reporting lower sensitivity for adenoviruses.41 A recent study noted lower sensitivity for influenza B and RSV in comparison with the BioFire FilmArray.31
Advantages of the xTAG RVP assay include: the ability to detect a wide array of viruses in a single assay; high throughput; increased sensitivity relative to traditional methods; and the potential for cost savings. Disadvantages include: inability to detect some viruses such as the coronaviruses; a relatively large amount of hands-on time; reported reduced sensitivity for some targets relative to other assays; and the need for multiple user interventions, including opening tubes containing amplicons, which increase the potential for contamination to occur. The xTAG RVP FAST has the obvious advantage of requiring less hands-on time, but has the disadvantage that the current FDA-cleared version has fewer viral targets.
BioFire Assay
The BioFire (formerly Idaho Technology) FilmArray Respiratory Panel (RP) (BioFire Diagnostics, Salt Lake City, UT) is a multiplex real-time PCR assay capable of detecting 17 viral respiratory agents including: adenovirus; coronaviruses 229E, OC43, NL63 and HKU1; metapneumovirus; influenza A, H3, H1, and 2009 H1; parainfluenza viruses 1, 2, 3, and 4; RSV; and rhinovirus/enterovirus. Of note, the assay can also detect the bacterial agents Bordetella pertussis, Mycoplasma pneumoniae, and Chlamydophila pneumoniae. The assay also includes an RNA and DNA control, both of which must be positive for the assay to produce a result.
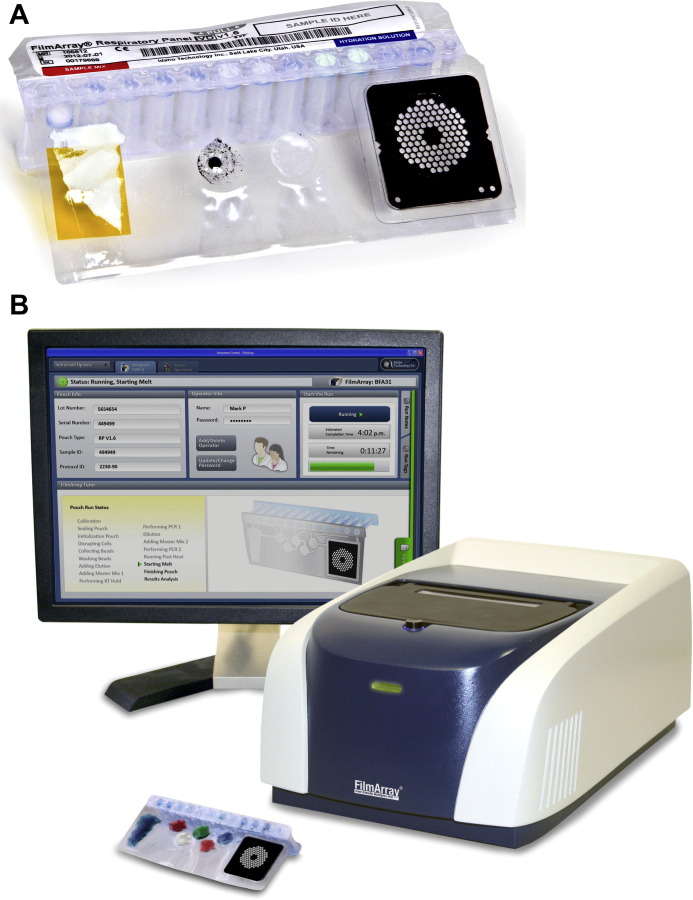
The FilmArray RP is a sample-in-answer-out test that uses a single-use disposable pouch containing lyophilized reagents with an associated instrument and computer, which has been cleared by the FDA for nasopharyngeal swabs (Fig. 6 ). After the specimen is introduced to the pouch, nucleic acids are extracted followed by reverse transcription and a 2-stage nested PCR reaction. The final PCR reaction takes place in a multiwell array, with products detected by melt-curve analysis. The associated instrument has room for a single pouch at a time. For those interested, a detailed description of the FilmArray technology has been published.42 The manufacturer notes that the RP assay cannot reliably discriminate rhinoviruses from enteroviruses, that the coronavirus OC43 component may cross-react with some strains of coronavirus HKU1, and that the assay has reduced sensitivity for adenovirus species C serotypes 2 and 6. An updated version of the assay with increased sensitivity for the detection of the adenovirus C serotypes is due out in 2013 (BioFire, personal communication, 2012).
Fig. 6.
BioFire FilmArray respiratory panel assay. (A) FilmArray RP Pouch. (B) FilmArray instrument and pouch. Instrument footprint in inches: 10.00 width × 6.5 height × 15.5 depth. A computer is required for operation.
(Courtesy of BioFire Diagnostics Inc, Salt Lake City, UT; with permission.)
Several studies have demonstrated that the FilmArray RP has greater sensitivity than antigen detection/culture methods and/or sensitivity generally comparable to that of other molecular methods.16, 31, 40, 43, 44, 45 Confirming the manufacturer’s information, some studies have noted decreased sensitivity of the FilmArray FP for adenoviruses.16, 44 Pierce and colleagues44 also reported that, using a collection of previously characterized adenovirus types, in addition to types 2 and 6 the FilmArray RP also had reduced sensitivity for detection of adenovirus types 20, 35, 37, and 41, although the investigators noted that these types have not been considered important causes of respiratory illnesses. Some of these same studies also noted increased sensitivity of the RP assay relative to other molecular tests for targets such as RSV, parainfluenza viruses, and influenza B.16, 31, 40
Advantages of the FilmArray RP include: sample-in-answer-out format for the largest number of viral respiratory agents currently on the market; rapid result with minimal hands-on time; performance generally comparable to that of other molecular methods. Disadvantages include: limited throughput; decreased sensitivity for some adenovirus types.
Issues associated with molecular testing for respiratory viruses
False-Negative Results due to Sequence Variants
With the exception of the adenoviruses, the majority of the known viruses causing respiratory tract disease are RNA viruses, a group of viruses whose genomes are generally considered to exhibit higher rates of mutations. Mutation rates for some RNA viruses such as HIV1 and influenza A have been extensively studied, and there exist large sequence databases for these viruses. However, for some important respiratory viruses such as RSV and parainfluenza 1 the number of available sequences, and hence knowledge of sequence variation, is very limited, with some groups now undertaking projects sequencing more genomes of these important agents.46, 47 Because most molecular assays rely on hybridization between a primer and target sequence to detect the virus, lack of knowledge of the full extent of sequence variation could result in the inability to detect a particular viral strain or newly arising mutant. Such an event has already occurred in Sweden, where a strain of Chlamydia trachomatis with a deletion mutation rendering it nondetectable by 2 widely used commercial molecular assays spread around the country, likely aided by the production of false-negative results by what were presumed to be gold-standard diagnostic tests.48 With more laboratories switching to molecular assays for the detection of respiratory agents, it will be necessary for both laboratorians and clinicians to remain vigilant for cases of what appear to be viral respiratory tract infections with negative molecular results. With no formal surveillance system currently in place, sentinel laboratories with culture and high-throughput sequencing capabilities would be one solution for monitoring the emergence of sequence variants not detectable by standard molecular assays.
Reporting of Rhinovirus/Enterovirus Results
Before the availability of sequence data, rhinoviruses and enteroviruses were classified in separate genera of the family Picornaviridae. Sequencing of the majority of the genomes in both groups revealed that they were so closely related that both genera have now been combined into the single genus Enterovirus.49 The close genomic similarity between the enteroviruses and rhinoviruses has made it difficult to design PCR assays that can reliably discriminate between the 2 groups, with most multiplex assays reporting specimens positive for “rhinovirus/enterovirus,” a situation that can cause confusion for people receiving the results who may be thinking in terms of rhinoviruses classically causing common-cold–like infections and enteroviruses causing aseptic meningitis and other systemic illnesses. Unpublished data from the authors’ own laboratory, where more than 100 positive “rhinovirus/enterovirus” from multiplex PCR testing of upper and lower respiratory tract specimens were subjected to sequencing, revealed all the positives to be rhinovirus types, with one exception. This exception was an isolate of enterovirus 68, an enterovirus type known to be a cause of respiratory tract infections.50, 51, 52 Furthermore, other enterovirus types have also been implicated as causes of respiratory tract illness.49 Therefore, in most cases where a “rhinovirus/enterovirus” result is reported from a patient suffering from a viral respiratory-like illness, it is likely to be due to a rhinovirus or an enterovirus capable of causing a respiratory tract infection, although the presence of an asymptomatically shed enterovirus cannot be ruled out. Virus-testing laboratories will play an important role in educating other health care workers about the changes in picornavirus taxonomy and the interpretation of results reported as positive for “rhinovirus/enterovirus.”
Detection of Viral Nucleic Acids in Specimens from Asymptomatic Individuals
With the advent of molecular testing assays for respiratory virus, the paradigm of considering viral agents of respiratory disease to always be pathogenic when detected in specimens has changed. There are now numerous examples of the ability to detect respiratory virus nucleic acids in specimens from asymptomatic individuals. The primary viruses detected in asymptomatic patients are rhinoviruses/enteroviruses, which have been detected in normal children and adults, as well as immunocompromised patients.53, 54, 55, 56, 57, 58, 59, 60 Other viruses such as parainfluenza viruses, coronaviruses, adenoviruses, and human metapneumoviruses have been less commonly found in asymptomatic individuals.61, 62, 63, 64, 65, 66, 67, 68 Other studies have found viruses such as RSV, influenza, and metapneumovirus to be only rarely detected in asymptomatic individuals.55, 61, 65, 69 Jansen and colleagues55 speculated that detection of virus in asymptomatic people could be due to 3 reasons: (1) detection of the virus during the acute phase of infection before the development of symptoms; (2) detection of virus still being shed after resolution of symptoms; and (3) detection of virus in specimens from individuals experiencing a subclinical infection. The same investigators also quantified the level of viral nucleic acid in specimens, and found that that in general there were higher levels of virus in cases relative to asymptomatic controls.55 However, it should be noted that none of the currently available FDA-cleared assays have the ability to quantify virus in specimens. In addition, there are issues with standardizing collection methods for respiratory specimens to produce meaningful quantitative results.
The detection of viruses in asymptomatic persons is of concern, especially when rhinovirus/enteroviruses are detected in patients for whom other causes of their symptoms are being considered or in cases where coinfections are detected (see next section). It is suggested that laboratorians familiarize themselves with the issue of viral detection in asymptomatic people so as to be able to provide guidance to other health care workers.
Detection of Coinfections
It is well established that once multiplex molecular testing is used for the detection of respiratory viruses, specimens containing more than 1 virus (coinfections) are encountered at frequencies from approximately 10% to 30%.70, 71, 72, 73, 74, 75, 76 Ascribing significance to coinfections with respect to causing more severe illness is complicated, because researchers have studied different patient populations using assays capable of detecting different sets of viruses. Some studies have found certain combinations of viruses to be associated with more severe illness,73 whereas others have reported that infection with some viruses may interfere with infection with other viruses.77 It is beyond the scope of this review to go into this topic in depth, so the reader is referred to the article by Paranhos-Baccala and colleagues74 for a more detailed discussion.
There are likely several methods by which a patient could become infected by more than 1 respiratory virus, and these in turn could affect the outcome. If a patient is unfortunate enough to become acutely infected with 2 or more viruses at the same time, depending on the host and the virus(es), the outcome could be a more severe illness. If, on the other hand, an individual had a previous infection with a virus such as a rhinovirus and was still asymptomatically shedding virus and then became acutely superinfected with a second virus, such a scenario likely would not produce a more significant illness despite the fact that 2 viruses would be detected. In any event, laboratories instituting multiplex molecular testing for respiratory viruses can expect to see an increase in the incidence of multiple viral infections, and they should be prepared to answer questions from clinicians using the tests.
Summary
Over the past several years the market has gone from virtually no FDA-cleared molecular assays for the detection of respiratory viruses to the wide variety discussed in this review. It should be mentioned that in addition to the tests listed in this article there are a number of other tests available outside the United States or within the United States that are sold in research-use-only or other non-cleared formats. The tests described here range from kits containing primers and probes detecting specific groups of viruses to self-contained systems requiring specialized instruments that extract nucleic acids and perform PCR with little operator input. Some of the tests target just the viruses involved in large yearly epidemics such as RSV and influenza or specific groups of viruses such as the adenoviruses or parainfluenza viruses, while others can detect most of the known respiratory viruses as well as some bacterial agents. Some systems utilize 96-well plate formats with the corresponding high through-put as compared to others which have much more limited throughput. All of these things represent factors that have to be taken into account when deciding to use one of these assays. It is expected that there will be more cleared tests for more analytes reaching the market in the future thereby increasing competition and the options available. We are also likely to see the more complicated tests requiring trained technologists replaced by easy-to-perform sample-in-answer-out format tests. Some of these tests will be so simple that they may be granted waived status and will therefore be found in provider offices and other point-of-care situations where individuals such as microbiologists and infectious disease physicians may not be overseeing their use. While this will bring the ability to rapidly and sensitively detect respiratory viruses to places where it was not previously possible to do so, it is hoped that this simplification of molecular tests will not cause those using them to lose sight of some of the important issues associated with these tests such as false negative results due to sequence variants, the ability to detection of viruses in individuals not exhibiting symptoms and the significance of the detection of co-infections.
References
- 1.Monto A.S. Epidemiology of viral respiratory infections. Am J Med. 2002;112(Suppl 6A):4S–12S. doi: 10.1016/s0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- 2.van Gageldonk-Lafeber A.B., Heijnen M.L., Bartelds A.I. A case-control study of acute respiratory tract infection in general practice patients in The Netherlands. Clin Infect Dis. 2005;41(4):490–497. doi: 10.1086/431982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenberg S.B. Respiratory viral infections in adults. Curr Opin Pulm Med. 2002;8(3):201–208. doi: 10.1097/00063198-200205000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg S.B. Viral respiratory infections in elderly patients and patients with chronic obstructive pulmonary disease. Am J Med. 2002;112(Suppl 6A):28S–32S. doi: 10.1016/S0002-9343(01)01061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renaud C., Campbell A.P. Changing epidemiology of respiratory viral infections in hematopoietic cell transplant recipients and solid organ transplant recipients. Curr Opin Infect Dis. 2011;24(4):333–343. doi: 10.1097/QCO.0b013e3283480440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weigt S.S., Gregson A.L., Deng J.C. Respiratory viral infections in hematopoietic stem cell and solid organ transplant recipients. Semin Respir Crit Care Med. 2011;32(4):471–493. doi: 10.1055/s-0031-1283286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sethi S. Molecular diagnosis of respiratory tract infection in acute exacerbations of chronic obstructive pulmonary disease. Clin Infect Dis. 2011;52(Suppl 4):S290–S295. doi: 10.1093/cid/cir044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gern J.E. Viral respiratory infection and the link to asthma. Pediatr Infect Dis J. 2004;23(Suppl 1):S78–S86. doi: 10.1097/01.inf.0000108196.46134.a6. [DOI] [PubMed] [Google Scholar]
- 9.Tan W.C. Viruses in asthma exacerbations. Curr Opin Pulm Med. 2005;11(1):21–26. doi: 10.1097/01.mcp.0000146781.11092.0d. [DOI] [PubMed] [Google Scholar]
- 10.Shetty A.K., Treynor E., Hill D.W. Comparison of conventional viral cultures with direct fluorescent antibody stains for diagnosis of community-acquired respiratory virus infections in hospitalized children. Pediatr Infect Dis J. 2003;22(9):789–794. doi: 10.1097/01.inf.0000083823.43526.97. [DOI] [PubMed] [Google Scholar]
- 11.LaSala P.R., Bufton K.K., Ismail N. Prospective comparison of R-mix shell vial system with direct antigen tests and conventional cell culture for respiratory virus detection. J Clin Virol. 2007;38(3):210–216. doi: 10.1016/j.jcv.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saiki R.K., Gelfand D.H., Stoffel S. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 13.Wittwer C.T., Kusukawa N. Real-time PCR and melting analysis. In: Persing D.H., editor. Molecular microbiology. ASM Press; Washington, DC: 2011. pp. 63–82. [Google Scholar]
- 14.Buller R.S., Arens M.Q. Molecular detection of respiratory viruses. In: Persing D.H., editor. Molecular microbiology. 2nd edition. ASM Press; Washington, DC: 2011. pp. 605–630. [Google Scholar]
- 15.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21(4):716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeffelholz M.J., Pong D.L., Pyles R.B. Comparison of the FilmArray respiratory panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J Clin Microbiol. 2011;49(12):4083–4088. doi: 10.1128/JCM.05010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Y.W., Lowery K.S., Valsamakis A. Validation of a PLEX-ID Flu device for simultaneous detection and identification of influenza viruses A and B. J Clin Microbiol. 2012;51:40–45. doi: 10.1128/JCM.01978-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selvaraju S.B., Selvarangan R. Evaluation of three influenza A and B real-time reverse transcription-PCR assays and a new 2009 H1N1 assay for detection of influenza viruses. J Clin Microbiol. 2010;48(11):3870–3875. doi: 10.1128/JCM.02464-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Available at: http://www.quidel.com/molecular/documents/us.php. Accessed January 28, 2013.
- 20.Popowitch E.B., Rogers E., Miller M.B. Retrospective and prospective verification of the Cepheid Xpert influenza virus assay. J Clin Microbiol. 2011;49(9):3368–3369. doi: 10.1128/JCM.01162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenny S.L., Hu Y., Overduin P. Evaluation of the Xpert Flu A Panel nucleic acid amplification-based point-of-care test for influenza A virus detection and pandemic H1 subtyping. J Clin Virol. 2010;49(2):85–89. doi: 10.1016/j.jcv.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller S., Moayeri M., Wright C. Comparison of GeneXpert FluA PCR to direct fluorescent antibody and respiratory viral panel PCR assays for detection of 2009 novel H1N1 influenza virus. J Clin Microbiol. 2010;48(12):4684–4685. doi: 10.1128/JCM.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambol A.R., Iwen P.C., Pieretti M. Validation of the Cepheid Xpert Flu A real time RT-PCR detection panel for emergency use authorization. J Clin Virol. 2010;48(4):234–238. doi: 10.1016/j.jcv.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Dimaio M.A., Sahoo M.K., Waggoner J. Comparison of Xpert Flu rapid nucleic acid testing with rapid antigen testing for the diagnosis of influenza A and B. J Virol Methods. 2012;186(1–2):137–140. doi: 10.1016/j.jviromet.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novak-Weekley S.M., Marlowe E.M., Poulter M. Evaluation of the Cepheid Xpert Flu Assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B viruses. J Clin Microbiol. 2012;50(5):1704–1710. doi: 10.1128/JCM.06520-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Available at: http://www.iquum.com/products/faba.shtml. Accessed January 24, 2013.
- 27.Alby K., Popowitch E.B., Miller M.B. Comparative evaluation of the Nanosphere Verigene RV+ Assay with the Simplexa Flu A/B & RSV Kit for the detection of influenza and respiratory syncytial viruses. J Clin Microbiol. 2013;51:352–353. doi: 10.1128/JCM.02504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Available at: http://www.focusdx.com/pdfs/pi/US/MOL2650.pdf. Accessed January 24, 2013.
- 29.Thaxton C.S., Georganopoulou D.G., Mirkin C.A. Gold nanoparticle probes for the detection of nucleic acid targets. Clin Chim Acta. 2006;363(1–2):120–126. doi: 10.1016/j.cccn.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 30.Pierce V.M., Hodinka R.L. Comparison of the GenMark diagnostics eSensor respiratory viral panel to real-time PCR for detection of respiratory viruses in children. J Clin Microbiol. 2012;50(11):3458–3465. doi: 10.1128/JCM.01384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babady N.E., Mead P., Stiles J. Comparison of the Luminex xTAG RVP Fast assay and the Idaho Technology FilmArray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol. 2012;50(7):2282–2288. doi: 10.1128/JCM.06186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krunic N., Merante F., Yaghoubian S. Advances in the diagnosis of respiratory tract infections: role of the Luminex xTAG respiratory viral panel. Ann N Y Acad Sci. 2011;1222:6–13. doi: 10.1111/j.1749-6632.2011.05964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balada-Llasat J.M., LaRue H., Kelly C. Evaluation of commercial ResPlex II v2.0, MultiCode-PLx, and xTAG respiratory viral panels for the diagnosis of respiratory viral infections in adults. J Clin Virol. 2011;50(1):42–45. doi: 10.1016/j.jcv.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahony J., Chong S., Merante F. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007;45(9):2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pabbaraju K., Tokaryk K.L., Wong S. Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J Clin Microbiol. 2008;46(9):3056–3062. doi: 10.1128/JCM.00878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong S., Pabbaraju K., Lee B.E. Enhanced viral etiological diagnosis of respiratory system infection outbreaks by use of a multitarget nucleic acid amplification assay. J Clin Microbiol. 2009;47(12):3839–3845. doi: 10.1128/JCM.01469-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dundas N.E., Ziadie M.S., Revell P.A. A lean laboratory: operational simplicity and cost effectiveness of the Luminex xTAG respiratory viral panel. J Mol Diagn. 2011;13(2):175–179. doi: 10.1016/j.jmoldx.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahony J.B., Blackhouse G., Babwah J. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol. 2009;47(9):2812–2817. doi: 10.1128/JCM.00556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gharabaghi F., Hawan A., Drews S.J. Evaluation of multiple commercial molecular and conventional diagnostic assays for the detection of respiratory viruses in children. Clin Microbiol Infect. 2011;17(12):1900–1906. doi: 10.1111/j.1469-0691.2011.03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rand K.H., Rampersaud H., Houck H.J. Comparison of two multiplex methods for detection of respiratory viruses: FilmArray RP and xTAG RVP. J Clin Microbiol. 2011;49(7):2449–2453. doi: 10.1128/JCM.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gadsby N.J., Hardie A., Claas E.C. Comparison of the Luminex Respiratory Virus Panel fast assay with in-house real-time PCR for respiratory viral infection diagnosis. J Clin Microbiol. 2010;48(6):2213–2216. doi: 10.1128/JCM.02446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poritz M.A., Blaschke A.J., Byington C.L. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PloS One. 2011;6(10):e26047. doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayden R.T., Gu Z., Rodriguez A. Comparison of two broadly multiplexed PCR systems for viral detection in clinical respiratory tract specimens from immunocompromised children. J Clin Virol. 2012;53(4):308–313. doi: 10.1016/j.jcv.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierce V.M., Elkan M., Leet M. Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J Clin Microbiol. 2012;50(2):364–371. doi: 10.1128/JCM.05996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renaud C., Crowley J., Jerome K.R. Comparison of FilmArray Respiratory Panel and laboratory-developed real-time reverse transcription-polymerase chain reaction assays for respiratory virus detection. Diagn Microbiol Infect Dis. 2012;74(4):379–383. doi: 10.1016/j.diagmicrobio.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck E.T., He J., Nelson M.I. Genome sequencing and phylogenetic analysis of 39 human parainfluenza virus type 1 strains isolated from 1997-2010. PloS One. 2012;7(9):e46048. doi: 10.1371/journal.pone.0046048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rebuffo-Scheer C., Bose M., He J. Whole genome sequencing and evolutionary analysis of human respiratory syncytial virus A and B from Milwaukee, WI 1998-2010. PloS One. 2011;6(10):e25468. doi: 10.1371/journal.pone.0025468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ripa T., Nilsson P.A. A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false-negative nucleic acid amplification tests. Sex Transm Dis. 2007;34(5):255–256. doi: 10.1097/OLQ.0b013e31805ce2b9. [DOI] [PubMed] [Google Scholar]
- 49.Tapparel C., Siegrist F., Petty T.J. Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol. 2013;14:282–293. doi: 10.1016/j.meegid.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 50.Jacobson L.M., Redd J.T., Schneider E. Outbreak of lower respiratory tract illness associated with human enterovirus 68 among American Indian children. Pediatr Infect Dis J. 2012;31(3):309–312. doi: 10.1097/INF.0b013e3182443eaf. [DOI] [PubMed] [Google Scholar]
- 51.Renois F., Bouin A., Andreoletti L. Enterovirus 68 in pediatric patients hospitalized for acute airway diseases. J Clin Microbiol. 2013;51:640–643. doi: 10.1128/JCM.02640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tokarz R., Firth C., Madhi S.A. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol. 2012;93(Pt 9):1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fry A.M., Lu X., Olsen S.J. Human rhinovirus infections in rural Thailand: epidemiological evidence for rhinovirus as both pathogen and bystander. PloS One. 2011;6(3):e17780. doi: 10.1371/journal.pone.0017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graat J.M., Schouten E.G., Heijnen M.L. A prospective, community-based study on virologic assessment among elderly people with and without symptoms of acute respiratory infection. J Clin Epidemiol. 2003;56(12):1218–1223. doi: 10.1016/S0895-4356(03)00171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jansen R.R., Wieringa J., Koekkoek S.M. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49(7):2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nokso-Koivisto J., Kinnari T.J., Lindahl P. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J Med Virol. 2002;66(3):417–420. doi: 10.1002/jmv.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peltola V., Waris M., Osterback R. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197(3):382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- 58.Srinivasan A., Flynn P., Gu Z. Detection of respiratory viruses in asymptomatic children undergoing allogeneic hematopoietic cell transplantation. Pediatr Blood Cancer. 2013;60(1):149–151. doi: 10.1002/pbc.24314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Benten I., Koopman L., Niesters B. Predominance of rhinovirus in the nose of symptomatic and asymptomatic infants. Pediatr Allergy Immunol. 2003;14(5):363–370. doi: 10.1034/j.1399-3038.2003.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright P.F., Deatly A.M., Karron R.A. Comparison of results of detection of rhinovirus by PCR and viral culture in human nasal wash specimens from subjects with and without clinical symptoms of respiratory illness. J Clin Microbiol. 2007;45(7):2126–2129. doi: 10.1128/JCM.02553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Advani S., Sengupta A., Forman M. Detecting respiratory viruses in asymptomatic children. Pediatr Infect Dis J. 2012;31(12):1221–1226. doi: 10.1097/INF.0b013e318265a804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Debiaggi M., Canducci F., Sampaolo M. Persistent symptomless human metapneumovirus infection in hematopoietic stem cell transplant recipients. J Infect Dis. 2006;194(4):474–478. doi: 10.1086/505881. [DOI] [PubMed] [Google Scholar]
- 63.Jartti T., Jartti L., Peltola V. Identification of respiratory viruses in asymptomatic subjects: asymptomatic respiratory viral infections. Pediatr Infect Dis J. 2008;27(12):1103–1107. doi: 10.1097/INF.0b013e31817e695d. [DOI] [PubMed] [Google Scholar]
- 64.Kalu S.U., Loeffelholz M., Beck E. Persistence of adenovirus nucleic acids in nasopharyngeal secretions: a diagnostic conundrum. Pediatr Infect Dis J. 2010;29(8):746–750. doi: 10.1097/INF.0b013e3181d743c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peck A.J., Englund J.A., Kuypers J. Respiratory virus infection among hematopoietic cell transplant recipients: evidence for asymptomatic parainfluenza virus infection. Blood. 2007;110(5):1681–1688. doi: 10.1182/blood-2006-12-060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prill M.M., Iwane M.K., Edwards K.M. Human coronavirus in young children hospitalized for acute respiratory illness and asymptomatic controls. Pediatr Infect Dis J. 2012;31(3):235–240. doi: 10.1097/INF.0b013e31823e07fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singleton R.J., Bulkow L.R., Miernyk K. Viral respiratory infections in hospitalized and community control children in Alaska. J Med Virol. 2010;82(7):1282–1290. doi: 10.1002/jmv.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walsh E.E., Peterson D.R., Falsey A.R. Human metapneumovirus infections in adults: another piece of the puzzle. Arch Intern Med. 2008;168(22):2489–2496. doi: 10.1001/archinte.168.22.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falsey A.R., Criddle M.C., Walsh E.E. Detection of respiratory syncytial virus and human metapneumovirus by reverse transcription polymerase chain reaction in adults with and without respiratory illness. J Clin Virol. 2006;35(1):46–50. doi: 10.1016/j.jcv.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Aberle J.H., Aberle S.W., Pracher E. Single versus dual respiratory virus infections in hospitalized infants: impact on clinical course of disease and interferon-gamma response. Pediatr Infect Dis J. 2005;24(7):605–610. doi: 10.1097/01.inf.0000168741.59747.2d. [DOI] [PubMed] [Google Scholar]
- 71.Canducci F., Debiaggi M., Sampaolo M. Two-year prospective study of single infections and co-infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. J Med Virol. 2008;80(4):716–723. doi: 10.1002/jmv.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franz A., Adams O., Willems R. Correlation of viral load of respiratory pathogens and co-infections with disease severity in children hospitalized for lower respiratory tract infection. J Clin Virol. 2010;48(4):239–245. doi: 10.1016/j.jcv.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kouni S., Karakitsos P., Chranioti A. Evaluation of viral co-infections in hospitalized and non-hospitalized children with respiratory infections using microarrays. Clin Microbiol Infect. 2012 doi: 10.1111/1469-0691.12015. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paranhos-Baccala G., Komurian-Pradel F., Richard N. Mixed respiratory virus infections. J Clin Virol. 2008;43(4):407–410. doi: 10.1016/j.jcv.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanner H., Boxall E., Osman H. Respiratory viral infections during the 2009-2010 winter season in Central England, UK: incidence and patterns of multiple virus co-infections. Eur J Clin Microbiol Infect Dis. 2012;31(11):3001–3006. doi: 10.1007/s10096-012-1653-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang G., Hu Y., Wang H. High incidence of multiple viral infections identified in upper respiratory tract infected children under three years of age in Shanghai, China. PloS One. 2012;7(9):e44568. doi: 10.1371/journal.pone.0044568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greer R.M., McErlean P., Arden K.E. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J Clin Virol. 2009;45(1):10–15. doi: 10.1016/j.jcv.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]