Abstract
In both within-host and epidemiological models of pathogen dynamics, the basic reproductive ratio, , is a powerful tool for gauging the risk associated with an emerging pathogen, or for estimating the magnitude of required control measures. Techniques for estimating , either from incidence data or in-host clinical measures, often rely on estimates of mean transition times, that is, the mean time before recovery, death or quarantine occurs. In many cases, however, either data or intuition may provide additional information about the dispersal of these transition times about the mean, even if the precise form of the underlying probability distribution remains unknown. For example, we may know that recovery typically occurs within a few days of the mean recovery time. In this paper we elucidate common situations in which is sensitive to the dispersal of transition times about their respective means. We then provide simple correction factors that may be applied to improve estimates of when not only the mean but also the standard deviation of transition times out of the infectious state can be estimated.
Keywords: Basic reproductive ratio, Virus dynamics, Survival probability, Weibull distribution, Gamma distribution, Epidemiology
1. Introduction
The basic reproductive ratio, , is a fundamental concept in both within-host and epidemiological models of pathogen dynamics. Defined as the number of secondary infections caused by a single infectious individual in a susceptible population, is used to gauge the magnitude of the risk associated with a novel pathogen, and to estimate the degree of control that will be necessary to contain an outbreak. For example, in the last several years has been a key factor in our understanding of emerging diseases such as Severe Acute Respiratory Syndrome (SARS) (Choi and Pak, 2003, Lipsitch et al., 2003, Lloyd-Smith et al., 2003, Riley et al., 2003) and avian influenza (Stegeman et al., 2004, Ferguson et al., 2005, Longini et al., 2005), livestock diseases such as Bovine Spongiform Encephalitus (Woolhouse and Anderson, 1997, Ferguson et al., 1999, de Koeijer et al., 2004) and Foot and Mouth Disease (Ferguson et al., 2001, Matthews et al., 2003), as well as vector-borne disease such as Dengue (Luz et al., 2003), Malaria (Hagmann et al., 2003, Smith and Ellis McKenzie, 2004) and West Nile Virus (Wonham et al., 2004, Cruz-Pacheco et al., 2005). Recent analyses of HIV therapy (Smith, 2006), river blindness (Filipe et al., 2005) and in cattle (Matthews et al., 2006) also rely on this important concept (see Heffernan et al., 2005 for review).
Given precise knowledge of the probability distributions for times in the exposed or infectious states, the mathematical derivation of is straight-forward (Diekmann and Heesterbeek, 2000). In practical terms, however, the estimation of from incidence or clinical data is far more difficult. Systems of ordinary differential equations (ODEs) or other simplifying arguments are used to derive estimates of in terms of measurable parameters (Dietz, 1975, Anderson and May, 1991, Dietz, 1993, Mollison, 1995, Diekmann and Heesterbeek, 2000, Hethcote, 2000, Brauer, 2002).
These estimates typically assume, however, that the lifetimes of individuals in each state are exponentially distributed. In an ODE formulation, this is a direct result of the Markov property that each state is memoryless, that is, the probability of making a transition to a new state does not depend on the length of time an individual has spent in the current state. For example, in all ODE models the probability of death is independent of the age of the individual or the time since infection; likewise the probability of recovery is independent of the time since infection. (For simplicity we use the word lifetime to mean the time spent in a single state or class such as “infected”; we do not restrict this word to mean the entire lifetime of the individual.)
As noted by many others, this assumption of constant transition probabilities is mathematically convenient, but is often difficult to defend on biological grounds (Lloyd, 2001a, Lloyd, 2001b, Lloyd, 2001c; Heesterbeek and Dietz, 1996, Perelson and Nelson, 1999, Nelson et al., 2004). Previous studies have considered the effects of non-exponential lifetime distributions on the dynamics and persistence of childhood diseases (Grossman, 1980, Keeling and Grenfell, 1997, Keeling and Grenfell, 1998; Lloyd, 2001b, Lloyd, 2001c). Non-exponential distributions have also been used to describe the mean incubation period (Anderson, 1988, Lui et al., 1988) and infectious period (Blythe and Anderson, 1988, Castillo-Chavez et al., 1989, Mittler et al., 1998, Nelson et al., 2004) of Human Immunodeficiency Virus (HIV). For example, Lloyd (2001a) elucidated the sensitivity of to the probability distributions describing the viral life cycle.
Unfortunately, for any practical study, the underlying probability distributions for transition times are almost certainly unknown. Nonetheless, in many cases the data at hand (or practical intuition) may provide more information than simply the mean lifetime. For example, while death from natural causes may be assumed to be exponential during an epidemic, recovery from the disease is almost certainly less dispersed. If the standard deviation of the recovery times is also known, we may be able to use this information to derive a more accurate estimate of . We might also improve our estimate of if the standard deviation is not known precisely, but is known to lie within a certain range.
The goal of this paper is to derive improved estimates of for situations in which not only the mean, but some understanding of the dispersal of transition times is available to the clinical or epidemiological practitioner. To focus this investigation, we will restrict our attention to transition times out of the infectious state. We thus consider a large number of processes such as natural death, death due to disease, recovery and removal to quarantine, or for in-host models, loss of infected cells due to natural death, lysis, immune-mediated cell death, etc. The probability distributions of other parameters, such as infectivity, are also critical to disease dynamics (Nelson et al., 2004, Lloyd, 2001c) and we hope to address such issues in future work.
We use the concept of “competing processes” to identify situations in which is sensitive to the dispersal of transition times. We then use the two most common lifetime distributions—the Weibull and the Gamma (see the Appendix)—to derive correction factors for , based on both the mean and the standard deviation of the appropriate lifetimes. The correction factor is modest when one of the competing processes is exponentially distributed, but approaches 200% when both processes are narrowly dispersed and have similar means. We motivate this study by first discussing several recent examples from the literature.
2. Recent estimates of are sensitive to lifetime distributions
SARS: The estimation of for SARS received considerable attention (Donnelly et al., 2003, Lipsitch et al., 2003, Lloyd-Smith et al., 2003, Riley et al., 2003, Nishiura et al., 2004). Lipsitch et al. (2003) used an SEIR approach to derive , where is the mean daily rate at which infectious cases who are not in quarantine are detected and isolated, q is the fraction of persons contacted but not infected by an infectious person, where k is the baseline daily number of contacts per capita and b is the probability of transmission per contact between a susceptible and an infectious, m is the per capita death rate due to disease and is the per capita recovery rate.
Fig. 1 shows the sensitivity of to the CoV of the recovery and death time distributions; we chose the value of b such that , the mean value estimated by Lipsitch et al. (2003), when lifetimes are distributed exponentially (dot). These estimates of are consistent with the range reported in the literature. We find that is quite sensitive to the dispersal of the relevant distributions.
Fig. 1.
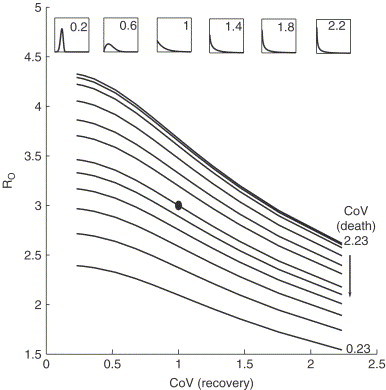
for SARS is plotted with respect to the coefficient of variation of the recovery time distribution for various values of the CoV of the death time distribution (lines). For reference, the Weibull distribution is also plotted for different values of the CoV (see insets). Parameters values: , , and . The value of b is chosen so that (dot). See Donnelly et al. (2003) and Lipsitch et al. (2003) for details.
HIV epidemiology: for the epidemiological spread of HIV has also been derived from ODE models. For example, Hyman and Li (2000) derive , where is the transmission probability per contact and r is the contact rate. The parameter gives the rate of progressing to AIDS, while gives the rate at which individuals are removed from the infectious compartment by other causes, for example, ceasing to be sexually active. The contact rate for individuals who have progressed to AIDS is assumed to be negligible; HIV is no longer transmitted at this stage.
As an ODE formulation, this model assumes that the rate of progressing to AIDS is independent of the time since infection. In Fig. 2 (a) we demonstrate the sensitivity of to this assumption. Again we plot versus the CoV of a Weibull distribution of transition times to AIDS; the dot gives the case when these times are exponentially distributed.
Fig. 2.
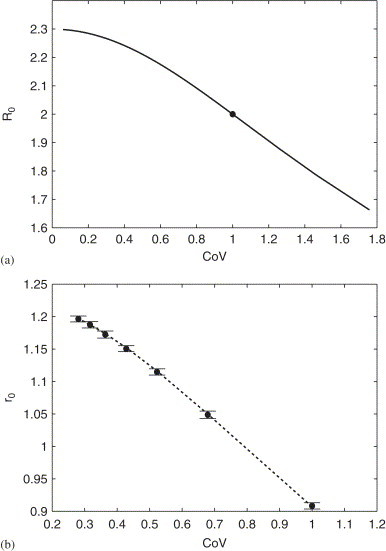
(a) for the epidemiological spread of HIV is plotted with respect to the CoV of the transition time to AIDS. The dot denotes the case when these transition times are exponentially distributed; the distribution is described by a Weibull distribution with shape parameter and mean (see text for details). All other lifetime distributions are assumed to be exponential. (Blower et al., 2001), (Blower et al., 2001) and such that for the ODE model is 2 (Blower and McLean, 1994, Valesco-Hernandez et al., 2002). (b) The initial growth rate for the in-host HIV model is plotted as a function of the CoV of the Weibull distribution of cell death times (dotted line); both uninfected and infected cells were assumed to have the same shape parameter although different means. Here is the solution to Eq. (23) when and , when . The dots and error bars give one standard error of the mean as computed by Monte Carlo simulation (see Heffernan and Wahl, 2005). Parameters values: cells , , , , , and (Perelson and Nelson, 1999, Wahl and Nowak, 2000, Smith and Wahl, 2004).
HIV immunology: is also central to the quantitative study of within-host HIV dynamics. The basic model of in-host infection consists of a set of ODEs that describe the interactions between susceptible cells, x, infected cells, y, and infectious virions (Ho et al., 1995, Wei et al., 1995, Perelson et al., 1996, Nowak and May, 2000):
| (1) |
Susceptible cells are produced at a constant rate with mean from a pool of precursor cells and die with rate d. Virions are budded with rate k and have probability Q of being infectious. Infectious virions can infect healthy cells via mass action dynamics where describes the efficacy of this process. Infected cells die with rate a and virions are cleared at rate u.
The basic reproductive ratio for this system depends on , the probability that an infectious virion budded at time zero produces an infected cell that is still alive at time t. In terms of the underlying distributions we see that:
| (2) |
where denotes the probability that the virion is still in circulation at time r, denotes the rate at which a virion, which has existed for time r, infects and is the probability that an infected cell lives to be age . Convolution is denoted by . Substituting into Eq. (22) (see Appendix A.2) with and assuming that , and are distributed exponentially, we find that , where is the mean number of uninfected cells at the uninfected equilibrium.
Eqs. (2) and (22) may also be used, however, when any of the underlying distributions are non-exponential. As an example, we consider the case when cell death time distributions are less dispersed than exponential, while all other processes are exponentially distributed. It is straight-forward to demonstrate through Eq. (22) that the basic reproductive ratio is insensitive to this change. However, the initial growth rate of the virus strongly depends on these cell lifetime distributions, as demonstrated in Fig. 2(b). This point will be taken up again in Section 4.
Note that we have not considered any change in the infection time of an infectious virion since this case is not included in our restricted focus on distributions of transition times out of the infected state only. We likewise do not explore the delay between the infection of a cell and virion production (Herz et al., 1996, Culshaw and Ruan, 2000, Nelson and Perelson, 2002, Dixit and Perelson, 2004).
3. Correction factors for competing processes
3.1. Competing processes
It is clear that changes to the mean times associated with transitions out of the infectious state(s) directly affect . The fact that is also sensitive to the shapes of these distributions, even if the mean is unchanged, is also relatively well-known (see Diekmann and Heesterbeek, 2000).
In the previous examples we found that sensitively depends on the shape of the distribution for the progression to AIDS. Likewise, for the SARS example, changes in the death time distribution affected . However, the shape of the death time distribution for infected cells had no effect on for in-host HIV infection. This can be explained using the concept of competing processes. For SARS, individuals leave the infected state either through recovery or death; recovery and death are competing, in this sense, to end the infectious period. In the epidemiological model of HIV, the infectious period ends if individuals progress to AIDS, or if they cease to be sexually active. In the in-host HIV model, however, the death of an infected cell is not in competition with another process.
We define the “effective infectious period”, L, as the time for which an individual remains infectious, irrespective of the process by which the individual leaves this compartment. When two (or more) processes are in competition to end the effective infectious period, not only the mean but the entire shape of the probability distribution for each process will affect the mean of L, and thus affect .
Mathematically, consider two renewal processes which have transition times given by random variables A and B with means and , respectively. The probability distribution of A gives the distribution of times at which an infectious individual would leave the infectious compartment through process A; likewise for B. We assume that the two processes are independent, which is already implied in the system of differential equations. We also assume that the two processes are “competing”, by which we mean that an infectious individual will leave the infectious compartment through one and only one of the two processes. In this case the survival probability, the probability that an individual is still in the infectious compartment at time t, can be written as
| (3) |
since A and B are independent. Thus, is simply the product of the tail distributions for A and B. The effective infectious period is then found by simply integrating from zero to infinity.
Fig. 3 plots the effective infectious period, L versus the mean of B, when both A and B are exponentially distributed and the mean of A is fixed (top panel). The insets show examples of the two distributions when (far left), (middle) and (far right). When (closed circle), L is given by . When the means of the two distributions are very different (at the extremes to the left and right), L is dominated by one process and approaches either or . When a and b are of the same magnitude, however, the effective infectious period will be sensitive to both distributions.
Fig. 3.
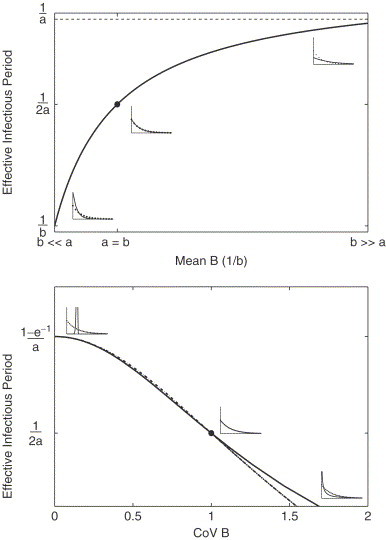
(Top panel) The effective infectious period is plotted versus the mean of distribution B, when B is exponentially distributed. It is assumed that A is exponentially distributed with mean . Three different scenarios are highlighted: when (left inset), (middle inset, dot) and (right inset). (Bottom panel) The effective infectious period is plotted versus the CoV of distribution B with mean when B is described by a Weibull distribution (solid line) and a Gamma distribution (dash dot line). It is assumed that A is exponentially distributed with mean . Three different scenarios are highlighted for B: less dispersed than exponential (left inset), exponential (middle inset) and more dispersed (right inset).
In the bottom panel we therefore examine the case when more closely. We now let the CoV of B vary, while A is exponentially distributed. B is described either by a Weibull distribution (solid line) or a Gamma distribution (dash dot line). When the CoV of B is 1, which corresponds to an exponential distribution, (dot, middle inset). When the CoV is greater than 1, L decreases. When the CoV is less than 1 such that B is less dispersed than exponential (see left inset), L increases toward a plateau. For the Gamma distribution this asymptotic value of L is given by
| (4) |
where k is the shape parameter, denoting the number of stages in the Gamma distribution. We also note that if (not shown), a similar limit is obtained, with as the CoV of B narrows. For the Weibull distribution with shape parameter , it is straight-forward to demonstrate that the same limits are approached as . In fact as demonstrated in the figure, the Weibull and Gamma distributions give only slightly different effective infectious periods for the same CoV.
3.2. Correction factors
We have found that the effective infectious period, and thus , is sensitive not only to distribution means but also to distribution shapes when two or more processes “compete” to end the infectious period, and when the mean times for the competing processes are of the same order of magnitude. As discussed previously, methods for estimating from incidence or clinical data typically assume that all the underlying processes are exponentially distributed. In the following section, we derive more accurate estimates for , assuming that one or both of the competing processes are non-exponentially distributed.
Rather than re-deriving for a number of models (SIR, SEIR, vector etc.), we report a “correction factor”, : the ratio of when the lifetimes are non-exponentially distributed to the value that would be calculated assuming exponential lifetimes. Where possible we report as a function of the CoV of the relevant distribution; in many cases we are able to derive limiting values of as this distribution narrows. We also find that can approach this asymptotic limit very quickly, and simple formulae can be used to correct estimated values of , or to gauge the sensitivity of to dispersion in the underlying distributions.
Throughout the following section we again assume that two renewal processes compete to end the infectious period. The transition times for these processes are given by random variables A and B, with means and , respectively. As stated previously, we assume throughout this derivation that other processes involved in the infection time course (for example, entering the infectious state) occur at constant rates. This implies, in particular, that infectiousness is constant throughout the infectious period, such that may be written as , where L is the effective infectious period and the transmission rate is constant. We are then able to estimate as the ratio of the true value of effective infectious period, L, to the value of L predicted by an ODE model, .
3.3. One process is exponentially distributed
In this section we assume that only random variable A is exponentially distributed. For example, it may be natural to assume that the recovery distribution is less dispersed than exponential, while the natural death rate is roughly constant over the time course of the infection. (In the latter case the correction factor would be modest, however, because the means of the two distributions are usually quite different.) Removing infectious individuals to quarantine is another process which competes with recovery and is more likely to have a mean of the same order of magnitude.
Assume that A is exponentially distributed and B is either described by a Gamma distribution or a Weibull distribution. If B is described by a Gamma distribution with k stages, the effective infectious period can be found by integrating Eq. (3):
| (5) |
and thus the correction factor is
| (6) |
If , which can be used to approximate the case when a and b are similar, this factor reduces to
| (7) |
Of course in practice we are unlikely to have in hand an estimate of k. Recall, however, that the CoV of the Gamma distribution is given by . Thus if C can be estimated, we can write the correction factor as
| (8) |
It is also quite useful to note that, in analogy with Eq. (4),
| (9) |
which reduces to if . In practice this limit is approached very quickly as the distribution of B becomes less dispersed, as shown in Fig. 4 . As a practical example, assume as in an ODE model that the rate at which individuals are moved to quarantine is independent of the time since infection, with a mean time of about 1 week. If we know in addition that the recovery time is 1 week days, the CoV is about 0.3, and is within about 2% of the limit given in Eq. (9). The approximation is even better if the CoV is smaller than 30%.
Fig. 4.
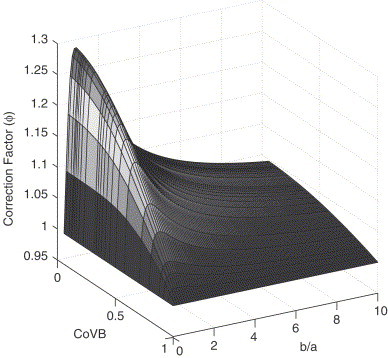
The correction factor is plotted versus the CoV of distribution B and the ratio of means when B is described by a Gamma distribution. It is assumed that A is exponentially distributed with mean .
Fig. 4 plots the correction factor for different ratios of the distribution means, assuming B is described by a Gamma distribution. We see that decreases with the CoV of B for all values of . Note that is maximized at when . This can be found numerically by solving , which is obtained by differentiating Eq. (9).
If B is assumed to be a Weibull distribution with shape parameter we obtain:
| (10) |
where is the Gamma function. This can only be solved numerically, however results (not shown) are extremely close to those illustrated in Fig. 4 for the Gamma distribution.
3.4. Neither process is exponentially distributed
In many cases, it may be more realistic to assume that neither of the processes in competition are distributed exponentially. For example, both recovery and death due to disease typically depend on the time that a host has been infected. The correction factor for such cases is more complex. If A and B are described by Weibull distributions with shape parameters and , respectively, we find
| (11) |
Similarly, when A and B are Gamma distributions with shape parameters and we find
| (12) |
Both of these expressions must be determined numerically.
These expressions can be simplified considerably, however, if the shapes of the two distributions are known to be similar. Recall, for example, that a shape parameter in the Weibull distribution corresponds to a distribution that is roughly Gaussian, with a CoV of around 30–40%. If processes A and B can be approximated by Weibull distributions with the same shape parameter , we find
| (13) |
This allows for three further simplifications: when, in addition to the shapes, the means of the two processes are roughly equal we find . If the means differ but the CoV is small (say less than about 30%), we can take the limit as to find . Finally, if the means are similar and the CoV is small, we have . These results are summarized in Table 1 . Although it may seem unlikely that the two relevant distributions would have exactly the same shape, the assumption that both shapes have a CoV of less than 30% may be met in many cases.
Table 1.
Approximations to the correction factor, , for competing processes A and B with means and , respectively
| B Weibull | B narrow | |
|---|---|---|
| when | or Gamma | |
| A exponential | ||
| A, B non-exponential | ||
| same shape | , or | |
| 2 |
C gives the CoV of the non-exponential distribution; for the Gamma distribution .
Entries in bold are simplifications for the case .
Similarly, if A and B can be approximated by Gamma distributions with different means but the same shape parameter k, the correction factor is
| (14) |
where is the generalized hypergeometric function. Although this expression is still unwieldy, when this reduces nicely to
| (15) |
since . Once again although k may not be known, we can substitute , where C is the CoV of the distribution, to find as a function of the CoV.
Fig. 5 plots the correction factor, determined numerically, versus the CoV of A and B when A and B are both described by Weibull distributions but do not have the same shape. In the top panel the distribution means are the same, ; in the middle panel . In each case the correction factor decreases as the CoVs of A and B increase. Note once again that the limit for small CoV gives a good approximation for whenever the CoV is less than about 30%.
Fig. 5.
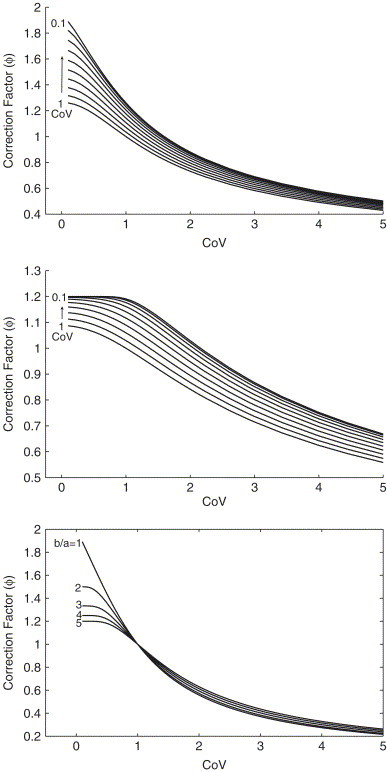
The correction factor is plotted versus the CoV of distributions A and B when A and B are both described by Weibull distributions with means and , respectively. The panels illustrate cases when the ratio of means changes: (top panel), (middle panel). Note that the correction factor determined when is symmetric to the case when . In the bottom panel we plot the correction factor when A and B have similar shapes for different ratios of .
In the bottom panel we plot the correction factor for the analytically simpler case when the distributions have similar shapes. We take the ratio of the means, to be between 1 and 5. Again we see that is very close to its limiting value whenever the CoV is small. The larger is, the more quickly this very simple limit is approached. Results for the Gamma distribution (not shown) were once again almost indistinguishable.
4. Discussion
Our main results are summarized in Table 1. We find that simple closed-form expressions for the correction factor are available when one of two competing processes is described by an exponential distribution, while the other has a measurable CoV or can be assumed to have a CoV less than about 30% (first row of Table 1). If neither competing process is distributed exponentially, simple approximations are only possible when the two processes have roughly the same shape, although their means may of course differ (second row). The bold entries in the table give even simpler results for the case when the two competing processes have similar means. In the limiting case when the processes have similar means and similar shapes, we see that the correction factor approaches 2 as the shapes of the distributions narrow.
These derivations assume that any non-exponential distributions can be approximated by either a Gamma or a Weibull distribution, and that some knowledge of the dispersal of the distribution about its mean is available. We found very little difference in the effective infectious period when either a Gamma or Weibull was assumed (see Fig. 3). Interestingly, each of these distributions had analytical advantages, and reduced to simple closed-form expressions in different cases. We thus include results from both distributions in Table 1 where appropriate.
The formulae we derive are applicable to situations in which two processes compete to end the infectious period. Examples in epidemiology include death due to disease, recovery, natural death and quarantine; for in-host models, examples include the loss of infected cells due to natural death, lysis, or immune-mediated cell death. These formulae apply both to models with direct transmission from one host to another e.g. SARS, or to vector models where the pathogen is carried between hosts e.g. malaria. It is also straightforward to apply the correction factors provided here to situations in which more than two processes are in competition (as in the SARS example, Fig. 1).
The dispersal of lifetimes involved in entering the infectious state may also affect . For an SEIR model, a slightly different correction factor can be easily derived for transitions into the infected state from the exposed state. Likewise for a virus or other types of in-host pathogens, the transition from one infected state to another may involve transitions both out of and into an infected state. For example, in the in-host model of HIV, the infection of a cell is a transition out of the infectious state for a virion, but it also represents a transition into the infected state for an infected cell. These situations suggest clear avenues for future work. In future we also hope to relax the assumption that the transmissibility of the disease is constant throughout the infectious period.
We would also like to discuss, briefly, the effects of lifetime distributions for “non-competing” processes. For example, in Section 2 we found that lifetime distribution for the infected cells did not affect . This is because the death of infected cells is not in competition with any other process. Nonetheless, solving system (1) for the infected equilibrium we find
| (16) |
where is the initial number of uninfected T-cells (Nowak and May, 2000, Lloyd, 2001a). If, instead, we assume that the death time distributions for the uninfected and infected cells are distributed as arbitrary distributions and with means and , respectively, the infected equilibrium is given by
| (17) |
and
| (18) |
Thus, we find that the infected equilibrium will change even though is not affected when the death of infected cells is non-exponentially distributed. We also find that if and are known, can be directly calculated from disease data irrespective of this distribution. If the distributions of competing processes are non-exponential, however, the relationship between and the infected equilibrium will change.
For non-competing processes, the initial growth rate may be sensitive to lifetime distributions even though is unaffected, as shown in Fig. 2. Fig. 6 plots the time distribution of infectious virions budded from a single infected cell ( from Eq. (22)) when the distribution of cell lifetimes is exponential (dashed line) or approximately Gaussian (solid line). In both cases the area under the curve is the same, and thus does not change. When lifetimes are Gaussian-distributed, however, the infected cells bud the same number of virions in a shorter period of time, thus increasing the initial growth rate (Fig. 2(b)). In this case, the conventional set of differential equations (1) allows us to obtain the correct value for the basic reproductive ratio, but does not model the initial growth rate or subsequent dynamics accurately.
Fig. 6.
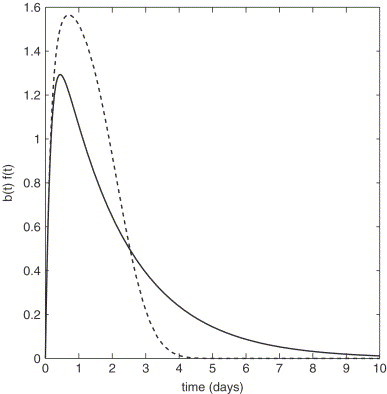
Infectious HIV virions budded per unit time, assuming two contrasting distributions for the death times of infected cells. The death time distribution of infected cells is given by a Weibull distribution with shape parameters 1 (exponential, dashed line) and 3 ( normal, solid line). The area under each curve is the same i.e. is the same, but changes. Parameters values are the same as in Fig. 2.
Like the infected equilibrium, can be determined from disease data for many infectious pathogens, and this value can be used to determine (see Hethcote and Tudor, 1980, Nowak and May, 2000, Lipsitch et al., 2003; Lloyd, 2001a, Lloyd, 2001b, Lloyd, 2001c for examples). We note that this method for estimating is not robust in the presence of non-exponential lifetime distributions for either competing or non-competing processes.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada. We are indebted to Baolai Ge and Gary Molenkamp for their invaluable technical expertise.
Appendix A.
A.1. Probability distributions
The Weibull distribution, first introduced by Wallodi Weibull to model failure times in reliability applications (Weibull, 1951), is one of the most widely used distributions in survival analysis. The Weibull distribution is also a natural model for lifetime (survival) probabilities in many instances, as it models waiting times until a critical event occurs.
We employ the two parameter Weibull distribution:
| (19) |
where is the shape parameter of the distribution and is the scale parameter. Here is the gamma function and is the mean of the distribution. Intuitively, this gives the distribution of times until a critical event occurs, such as cell death, the clearance of an infection or the transition from exposed to infectious. If the failure rate is increasing, that is, the probability that an individual will leave the state increases over time; if the failure rate is decreasing and if the failure rate is constant. A shape parameter of one corresponds to an exponential distribution and a shape parameter of three corresponds to a distribution that is approximately Gaussian. The CoV for the Weibull distribution with shape parameter is .
The Gamma distribution is also used to model failure times in reliability applications. We use the two parameter Gamma distribution given by the following:
| (20) |
where is a scale parameter, is the mean and k is the shape parameter. This distribution, like the Weibull, includes the exponential distribution when .
The Gamma distribution is not as widely used as the Weibull distribution in reliability analysis, but it is commonly used to describe stages of infection in epidemic models since it is simply a sum of exponential distributions; the sums of independent and identically distributed exponential random variables have a Gamma distribution. The Gamma distribution, unlike the Weibull, cannot be used to approximate a normal distribution. The CoV for the Gamma distribution with shape parameter k is equal to .
A.2. The basic reproduction ratio,
Consider a large population and let be the probability that a newly infected individual remains infectious for at least time a. This is called the survival probability. Also, let denote the average number of newly infected individuals that an infectious individual will produce per unit time when infected for total time a. Then, is given by
| (21) |
See Heesterbeek and Dietz (1996) for a detailed description of this equation and a historical overview.
The definition of must be extended for cases in which a series of states are involved in the “reproduction” of an infected individual. For example, in within-host viral dynamics, an infectious virion produces an infected cell which produces more infectious virions; this complete cycle must be taken into account in our derivation of . One approach is to define in this case as
| (22) |
and is simply the average number of infectious virions produced by a cell which has been infected for time a. Note that this extension to the concept is typically assumed in epidemiology and HIV dynamics (see Heffernan et al., 2005, for review), but differs from the standard Next Generation Approach (Diekmann et al., 1990, Diekmann and Heesterbeek, 2000, van den Driessche and Watmough, 2002).
A.3. The initial growth rate,
While the basic reproductive ratio gives the overall number of new infections per infected individual, it gives no measure of when these infections occur. In contrast gives the number of new infections per infected individual per unit time, in a susceptible population (Heesterbeek and Dietz, 1996).
Using the survival probability , as derived above, we propose that can be determined by solving the following renewal equation:
| (23) |
Here denotes the incoming rate of new infected individuals, or the incoming rate of individuals to state 1 of an infection cycle, at time t. Eq. (23) thus implies that new infections at time t are produced by new infections at time which are still infectious s time units later. The term is a Dirac delta function, and is necessary because we wish to determine after the instantaneous input of one infectious individual at time 0.
Conveniently, Eq. (23) is in the form of a convolution integral, which lends itself to solution by a Laplace transform. Taking the Laplace transform of both sides we obtain , where and denote the Laplace transforms of and , respectively. Isolating and taking the inverse Laplace transform we find that
| (24) |
Thus, the initial growth rate can be found by determining the largest exponent in the inverse Laplace transform for .
References
- Anderson R.M. The epidemiology of HIV infection: variable incubation plus infectious periods and heterogeneity in sexual activity. J. R. Stat. Soc. Ser A. 1988;155:6698. [Google Scholar]
- Anderson R.M., May R.M. Oxford University Press; Oxford: 1991. Infectious Diseases of Humans: Dynamics and Control. [Google Scholar]
- Blower S.M., McLean S.R. Prophylactic vaccines, risk behaviour change and the probability of eradicating HIV in San Francisco. Science. 1994;265:1451–1454. doi: 10.1126/science.8073289. [DOI] [PubMed] [Google Scholar]
- Blower S.M., Koelle K., Kirschner D.E., Mills J. Live attenuated HIV vaccines: predicting the tradeoff between efficacy and safety. Proc. Natl. Acad. Sci. 2001;98(6):3618–3623. doi: 10.1073/pnas.061029998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe S.P., Anderson R.M. Distributed incubation and infectious periods in a model of the transmission dynamics of the human immunodeficiency virus (HIV) IMA J. Math. Appl. Med. Biol. 1988;5:1–19. doi: 10.1093/imammb/5.1.1. [DOI] [PubMed] [Google Scholar]
- Brauer, F., 2002. Basic ideas of mathematical epidemiology. In: Castillo-Chavez, C., van den Driessche, P., Kirschner, D., Yakubu, A-A. (Eds.), Mathematical Approaches for Emerging and Reemerging Infection Diseases: An Introduction. The IMA Volumes in Mathematics and its Applications, vol. 125. Springer, New York, pp. 31–65.
- Castillo-Chavez C., Cooke K., Huang W., Levin S.A. On the role of long incubation periods in the dynamics of acquired immunodeficiency syndrome (AIDS) J. Math. Biol. 1989;27:373–398. doi: 10.1007/BF00290636. [DOI] [PubMed] [Google Scholar]
- Choi B.C.K., Pak A.W.P. A simple approximate mathematical model to predict the number of severe acute respiratory syndrome cases and deaths. J. Epidemiol. Commun. 2003;57(10):831–835. doi: 10.1136/jech.57.10.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Pacheco G., Esteva L., Montano-Hirose J.A., Vargas C. Modelling the dynamics of West Nile Virus. Bull. Math. Biol. 2005;67:1157–1172. doi: 10.1016/j.bulm.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Culshaw R.V., Ruan S. A delay-differential equation model of HIV infection of CD4 T-cells. Math. Biosci. 2000;165:27–39. doi: 10.1016/s0025-5564(00)00006-7. [DOI] [PubMed] [Google Scholar]
- de Koeijer A., Heesterbeek H., Schreuder B., Oberthür R., Wilesmith J., van Roermund H., de Jong M. Quantifying BSE control by calculating the basic reproduction ratio for the infection among cattle. J. Math. Biol. 2004;48:1–22. doi: 10.1007/s00285-003-0206-x. [DOI] [PubMed] [Google Scholar]
- Diekmann O., Heesterbeek J.A.P. Wiley; New York: 2000. Mathematical Epidemiology of Infectious Diseases: model Building, Analysis and Interpretation. [Google Scholar]
- Diekmann O., Heesterbeek J.A.P., Metz J.A.J. On the definition and the computation of the basic reproduction ratio in models for infectious diseases. J. Math. Biol. 1990;35:503–522. doi: 10.1007/BF00178324. [DOI] [PubMed] [Google Scholar]
- Dietz K. Transmission and control of arbovirus diseases. In: Ludwig D., Cooke K.L., editors. Epidemiology. Society for Industrial and Applied Mathematics; Philadelphia: 1975. pp. 104–121. [Google Scholar]
- Dietz K. The estimation of the basic reproduction number for infectious diseases. Stat. Methods Med. Res. 1993;2:23–41. doi: 10.1177/096228029300200103. [DOI] [PubMed] [Google Scholar]
- Dixit N.M., Perelson A.S. Complex patterns of viral load decay under antiretroviral therapy: influence of pharmacokinetics and intracellular delay. J. Theor. Biol. 2004;226:95–109. doi: 10.1016/j.jtbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Donnelly C.A., Ghani A.C., Leung G.M., Hedley A.J., Fraser C., Riley S., Abu-Raddad L.J., Ho L.-M., Thach T.-Q., Chau P., Chan K.-P., Lam T.-H., Tse L.-Y., Tsang T., Liu S.-H., Kong J.H.B., Lau E.M.C., Ferguson N.M., Anderson R.M. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N.M., Donnelly C.A., Woolhouse M.E.J., Anderson R.M. Estimation of the basic reproduction number of BSE: the intensity of transmission in British cattle. Proc. R. Soc. London B. 1999;266:23–32. doi: 10.1098/rspb.1999.0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N.M., Donnelly C.A., Anderson R.M. The foot and mouth epidemic in Great Britain: pattern of spread and impact of interventions. Science. 2001;292:1155–1160. doi: 10.1126/science.1061020. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M., Cummings D.A.T., Cauchemez S., Fraser C., Riley S., Meeyai A., Iamsirithaworn S., Burke D.S. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- Filipe J.A.N., Boussinesq M., Renz A., Collins R.C., Vivas-Martinez S., Grillet M.E., Little M.P., Basanez M.G. Human infection patterns and heterogeneous exposure in river blindness. Proc. Natl. Acad. Sci. USA. 2005;102:15265–15270. doi: 10.1073/pnas.0502659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman Z. Oscillatory phenomena in a model of infectious diseases. Theor. Popul. Biol. 1980;18:204–243. doi: 10.1016/0040-5809(80)90050-7. [DOI] [PubMed] [Google Scholar]
- Hagmann, R., Charlwood, J.D., Gil, V., Conceicao, F., do Rosario, V., Smith, T.A., 2003. Malaria and its possible control on the Island of Principe. Malar. J. 2(15), published online 2003 June 18. [DOI] [PMC free article] [PubMed]
- Heesterbeek J.A.P., Dietz K. The concept of in epidemic theory. Stat. Neerl. 1996;50:89–110. [Google Scholar]
- Heffernan J.M., Wahl L.M. Monte Carlo estimates of natural variation in HIV infection. J. Theor. Biol. 2005;236:137–153. doi: 10.1016/j.jtbi.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Heffernan J.M., Smith R., Wahl L.M. Perspectives on the basic reproductive ratio. J. R. Soc. Interface. 2005;2(4):281–293. doi: 10.1098/rsif.2005.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hethcote H.W. The mathematics of infectious diseases. SIAM Rev. 2000;42(4):599–653. [Google Scholar]
- Hethcote H.W., Tudor D.W. Integral equation models for endemic infectious diseases. J. Math. Biol. 1980;9:37–47. doi: 10.1007/BF00276034. [DOI] [PubMed] [Google Scholar]
- Herz A.V.M., Bonhoeffer S., Anderson R.M., May R.M., Nowak M.A. Viral dynamics in vivo: limitations on estimates of intracellular delay and virus decay. Proc. Natl. Acad. Sci. USA. 1996;93:7247–7251. doi: 10.1073/pnas.93.14.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D.D., Neumann A.U., Perelson A.S., Chen W., Leonard J.M., Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Hyman J.M., Li J. An intuitive formulation for the reproductive number for the spread of diseases in heterogenous populations. Math. Biosci. 2000;167:65–86. doi: 10.1016/s0025-5564(00)00025-0. [DOI] [PubMed] [Google Scholar]
- Keeling M.J., Grenfell T. Disease extinction and community size: modeling the persistence of measles. Science. 1997;275:65–67. doi: 10.1126/science.275.5296.65. [DOI] [PubMed] [Google Scholar]
- Keeling M.J., Grenfell T. Effect of variability in infection period on the persistence and spatial spread of infectious diseases. Math. Biosci. 1998;147:207–226. doi: 10.1016/s0025-5564(97)00101-6. [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Cohen T., Cooper B., Robins J.M., Ma S., James L., Gopalakrishna G., Chew S.K., Tan C.C., Samore M.H., Fisman D., Murray M. Transmission dynamics and control of sever acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A.L. The dependence of viral parameter estimates on the assumed viral life cycle: limitations of studies of viral load data. Proc. R. Soc. London B. 2001;268:847–854. doi: 10.1098/rspb.2000.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A.L. Destabilization of epidemic models with the inclusion of realistic distributions of infectious periods. Proc. R. Soc. London B. 2001;268:985–993. doi: 10.1098/rspb.2001.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A.L. Realistic distributions of infectious periods in epidemic models: changing patterns of persistence and dynamics. Theor. Popul. Biol. 2001;60:59–71. doi: 10.1006/tpbi.2001.1525. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith J.O., Galvani A.P., Getz W.M. Curtailing transmission of severe acute respiratory syndrome within a community and its hospital. Proc. R. Soc. London B. 2003;270:1979–1989. doi: 10.1098/rspb.2003.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longini I.M., Nizam A., Xu S.F., Ungchusak K., Hanshaoworakul W., Cummings D.A.T., Halloran M.E. Containing pandemic influenza at the source. Science. 2005;309:1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- Lui K.J., Darrow W.W., Rutherford G.W. A model-based estimate of the mean incubation period for AIDS in homosexual men. Science. 1988;240:1333–1335. doi: 10.1126/science.3163848. [DOI] [PubMed] [Google Scholar]
- Luz P.M., Codeco C.T., Massad E., Struchiner C.J. Uncertainties regarding dengue modeling in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. 2003;98(7):871–878. [PubMed] [Google Scholar]
- Matthews L., Haydon D.T., Shaw D.J., Chase-Topping M.E., Keeling M.J., Woolhouse M.E.J. Neighbourhood control policies and the spread of infectious diseases. Proc. R. Soc. London B. 2003;270:1659–1666. doi: 10.1098/rspb.2003.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L., Low J.C., Gally D.L., Pearce M.C., Mellor D.J., Heesterbeek J.A.P., Chase-Topping M., Naylor S.W., Shaw D.J., Reid S.W.J., Gunn G.J., Woolhouse M.E.J. Heterogeneous shedding of O157 in cattle and its implications for control. Proc. Natl. Acad. Sci. USA. 2006;103:547–552. doi: 10.1073/pnas.0503776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler J.E., Sulzer B., Neumann A.U., Perelson A.S. Influence of delayed viral production on viral dynamics in HIV-1 infected patients. Math. Biosci. 1998;152:143–163. doi: 10.1016/s0025-5564(98)10027-5. [DOI] [PubMed] [Google Scholar]
- Mollison, D. (Ed.), 1995. Epidemic Models: Their Structure and Relation to Data. Cambridge University Press, Cambridge.
- Nelson P.W., Perelson A.S. Mathematical analysis of delay differential equation models of HIV-1 infection. Math. Biosci. 2002;179:73–94. doi: 10.1016/s0025-5564(02)00099-8. [DOI] [PubMed] [Google Scholar]
- Nelson P.W., Gilchrist M.A., Coombs D., Hyman J.M., Perelson A.S. An age-structured model of HIV infection that allows for variations in the production rate of viral particles and the death rate of productively infected cells. Math. Biosci. Eng. 2004;1:267–288. doi: 10.3934/mbe.2004.1.267. [DOI] [PubMed] [Google Scholar]
- Nishiura H., Patanarapelert K., Sriprom M., Sarakorn W., Sriyab S., Ming Tang S. Modelling potential responses to severe acute respiratory syndrome in Japan: the role of initial attack size, precaution, and quarantine. J. Epidemiol. Commun. Health. 2004;58:186–191. doi: 10.1136/jech.2003.014894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M.A., May R.M. Oxford University Press; Oxford: 2000. Virus Dynamics. [Google Scholar]
- Perelson A.S., Nelson P. Mathematical analysis of HIV-1 dynamics in vivo. SIAM Rev. 1999;41:3–44. [Google Scholar]
- Perelson A.S., Neumann A.U., Markowitz M., Leonard J.M., Ho D.D. HIV-1 dynamics in vivo: virion clearance rate, infected cell lifespan, and viral generation time. Science. 1996;271:1582–1585. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- Riley S., Fraser C., Donnelly C.A., Ghani A.C., Abu-Raddad L.J., Hedley A.J., Leung G.M., Ho L.-M., Lam T.-H., Thach T.Q., Chau P., Chan K.-P., Lo S.-V., Leung P.-Y., Tsang T., Ho W., Lee K.-H., Lau E.M.C., Ferguson N.M., Anderson R.M. Transmission dynamics of the etiological agent SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- Smith, D.L., Ellis McKenzie, F., 2004. Statistics and dynamics of malaria infection in mosquitoes. Malar. J. 3, http://www.malariajournal.com/content/3/1/13. [DOI] [PMC free article] [PubMed]
- Smith R.J. Adherence to antiretroviral HIV drugs: how many doses can you miss before resistance emerges? Proc. R. Soc. London B. 2006;273:617–624. doi: 10.1098/rspb.2005.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.J., Wahl L.M. Distinct effect of protease and reverse transcriptase inhibition in an immunological model of HIV-1 infections with impulsive drug effects. Bull. Math. Biol. 2004;66:1259–1283. doi: 10.1016/j.bulm.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Stegeman A., Bouma A., Elbers A.R., de Jong M.C., Nodelijk G., de Klerk F., Koch G., van Boven M. Avian influenza A virus (H7N7) epidemic in The Netherlands in 2003: course of the epidemic and effectiveness of control measures. J. Infect. Dis. 2004;190:2088–2095. doi: 10.1086/425583. [DOI] [PubMed] [Google Scholar]
- Valesco-Hernandez J.X., Gershengorn H.B., Blower S.M. Could widespread use of combination antiretroviral therapy eradicate HIV epidemics? The Lancet Infect. Dis. 2002;2:487–493. doi: 10.1016/s1473-3099(02)00346-8. [DOI] [PubMed] [Google Scholar]
- van den Driessche P., Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 2002;180:29–48. doi: 10.1016/s0025-5564(02)00108-6. [DOI] [PubMed] [Google Scholar]
- Wahl, L.M., Nowak, M.A., 2000. Adherence and drug resistance: predictions for therapy outcome. Proc. R. Soc. London B 267, 835–843. [DOI] [PMC free article] [PubMed]
- Wei L.M., Ghosh S.K., Taylor M.E., Johnson V.A., Emini E.A., Deutsch P., Lifson J.D., Bonhoeffer S., Nowak M.A., Hahn B.H., Saag M.S., Shaw G.M. Viral dynamics in HIV-1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Weibull W. A statistical distribution function of wide applicability. J. Appl. Mech. 1951;18:293–297. [Google Scholar]
- Wonham M.J., de-Camino-Beck T., Lewis M.A. An epidemiological model for West Nile virus: invasion analysis and control applications. Proc. R. Soc. London B. 2004;271:501–507. doi: 10.1098/rspb.2003.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E.J., Anderson R.M. Understanding the epidemiology of BSE. Trends Microbiol. 1997;5:421–424. doi: 10.1016/S0966-842X(97)01146-3. [DOI] [PubMed] [Google Scholar]


