Abstract
An investigation into the causes of canine infectious respiratory disease was carried out in a large rehoming kennel. Tissue samples taken from the respiratory tract of diseased dogs were tested for the presence of coronaviruses using RT–PCR with conserved primers for the polymerase gene. Sequence analysis of four positive samples showed the presence of a coronavirus with high similarity to both bovine and human coronavirus (strain OC43) in their polymerase and spike genes, whereas there was a low similarity to comparable genes in the enteric canine coronavirus. This canine respiratory coronavirus (CRCV) was detected by RT–PCR in 32/119 tracheal and 20/119 lung samples, with the highest prevalence being detected in dogs with mild clinical symptoms. Serological analysis showed that the presence of antibodies against CRCV on the day of entry into the kennel decreased the risk of developing respiratory disease.
Keywords: Coronavirus, Canine respiratory disease, Kennel cough
Introduction
Canine infectious respiratory disease (CIRD) is a highly contagious disease, especially in dogs housed in groups in rehoming centers and boarding or training kennels. Many dogs suffer from a mild cough and recover after a short time; however, in some cases a severe bronchopneumonia can develop (Appel and Binn, 1987). In addition to causing distress to the dogs, CIRD also delays rehoming at rescue centers and causes disruption of schedules in training kennels as well as incurring considerable treatment costs.
CIRD is considered to be a multifactorial disease because it usually occurs when dogs from different origins are brought together, a situation that exposes the dogs to a variety of different microorganisms as well as an unfamiliar environment. The infectious agents that have been considered the major causative pathogens involved are canine parainfluenzavirus (CPIV) (Binn et al., 1967), canine adenovirus type 2 (CAV-2) (Ditchfield et al., 1962), and the bacterium Bordetella bronchiseptica Bemis et al 1997a, Keil and Fenwick 1998. Also, canine herpesvirus, human reovirus, and mycoplasma species have been isolated from dogs with symptoms of CIRD Karpas et al 1968, Lou and Wenner 1963, Randolph et al 1993. Experimental infection of dogs with single infectious agents has been shown to cause only mild respiratory symptoms but failed to reproduce the severe disease that can be seen in natural outbreaks, supporting the theory that the pathogenesis of CIRD is multifactorial Appel and Percy 1970, Karpas et al 1968, Bemis et al 1997b.
Vaccines are available against some of the infectious agents that have been found to be associated with this disease, namely, Bordetella bronchiseptica as well as CPIV and CAV-2. Despite the use of these vaccines, CIRD is still prevalent in kennels worldwide, suggesting that additional viruses or bacteria may be involved in the disease.
Members of the family Coronaviridae are enveloped viruses, 80–160 nm in diameter, containing a linear positive-stranded RNA genome. The structural proteins of coronaviruses are the spike glycoprotein, the membrane glycoprotein, and the nucleocapsid protein. The hemagglutinin/ esterase glycoprotein is found only on the surface of group 2 coronaviruses (e.g., bovine coronavirus and murine hepatitis virus) (Spaan et al., 1988).
The polymerase gene of coronaviruses is known to be highly conserved. It has therefore previously been used for phylogenetic analysis of this virus family (Stephensen et al., 1999). A possible role of Coronaviridae in the pathogenesis of CIRD was investigated in this study because members of this family are known to cause respiratory disease in humans as well as cattle, swine, and poultry Mäkelä et al 1998, Pensaert et al 1986, Ignjatovic and Sapats 2000. In cattle, bovine respiratory coronavirus is associated with shipping fever, a multifactorial respiratory disease similar to CIRD (Storz et al., 2000). Canine coronaviruses are reported to cause acute diarrhea mainly in young dogs (Tennant et al., 1993). However, one study reports the detection of canine coronavirus in dogs with respiratory disease and describes the isolation of the virus from one lung sample and three intestinal samples (Binn et al., 1979).
This investigation sought to detect coronaviruses associated with CIRD in a large kenneled dog population with a history of endemic respiratory disease, using virus culture and PCR techniques as well as serology on paired serum samples.
Results
PCR using consensus primers for the coronavirus RNA polymerase gene
Using the primers Conscoro5 and Conscoro6, we analyzed the cDNA obtained from 40 tracheal samples by RT–PCR. Of these, 7 were found to be positive by PCR and subsequent hybridization (17.5%). The PCR products were cloned and sequenced and the sequence data were compared to available viral sequences using the FASTA similarity search program (Pearson, 1990).
Comparison of the coronavirus cDNA polymerase sequence obtained from four of the canine tracheal samples to other coronavirus sequences revealed that they were most similar to sequence data from bovine coronavirus (GenBank Accession No. AF 220295) and human coronavirus strain OC 43 (GenBank Accession No. AF 124989). The identity in the analyzed 251-bp sequence was 98.8% for the bovine and 98.4% for the human coronavirus polymerase gene, whereas it was only 68.53% for canine coronavirus (strain 1–71).
An alignment of the novel sequence with the corresponding sequences of 11 coronaviruses and phylogenetic analysis using the maximum parsimony method resulted in the consensus tree shown in Fig. 1. The cDNA sequence obtained from a tracheal sample (T101) was found on a common branch with bovine coronavirus, human coronavirus-OC43, and hemagglutinating encephalomyelitis virus.
Fig. 1.
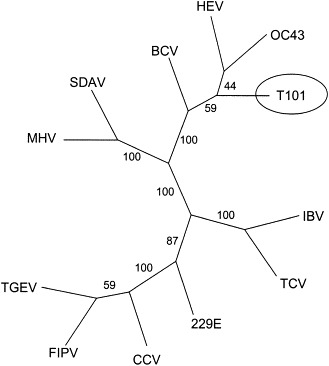
Consensus tree for cDNA sequences from a 251-nucleotide region of the polymerase gene of 12 coronaviruses. The sequence obtained from the canine respiratory coronavirus is designated T101. The numbers indicate bootstrap values obtained by analysis of 100 data sets. BCV, bovine coronavirus; CCV, canine coronavirus; FIPV, feline infectious peritonitis virus; HEV, hemagglutinating encephalomyelitis virus; IBV, infectious bronchitis virus; MHV, mouse hepatitis virus; OC43, human coronavirus strain OC43; SDAV, sialodacryoadenitis virus; TCV, turkey coronavirus; TGEV, transmissible gastroenteritis virus; 229E, human coronavirus strain 229E; T101, canine respiratory coronavirus (PCR product from tracheal sample T101).
The virus was provisionally called canine respiratory coronavirus (CRCV).
Sequencing of the spike gene
For further analysis of the RNA sequence of CRCV, an alignment of the RNA for the spike gene of the bovine coronavirus LY 138 strain and the human coronavirus OC43 strain was performed. Consensus regions were chosen for the selection of four primer pairs amplifying the complete spike gene in four overlapping fragments; the primer sequences are shown in Table 1. The cDNA obtained from tracheal sample T101 was used to perform RT–PCR and subsequent sequencing of the obtained spike fragments. The analysis of the sequencing data showed that the spike gene of CRCV is 4092 nucleotides long, corresponding to 1093 amino acids. It was determined that the cDNA sequence obtained from tracheal sample T101 had a 97.312% nucleotide identity with the spike gene of bovine coronavirus strain LY138 (GenBank Accession No. AF058942) and 96.921% identity with that of human coronavirus strain OC43 (GenBank Accession No. Z32768) in an overlap of 4092 nucleotides. When comparing the amino acid sequence obtained by translation of the cDNA sequence from T101 to the amino acid sequence of the BCV, HCV-OC43, and canine enteric coronavirus spike proteins the identities were 96, 95.2, and 21.2%.
Table 1.
Primers designed from an alignment of the spike genes of bovine coronavirus (GenBank Accession No. AF058942) and human coronavirus, OC43 (GenBank Accession No. L14643)
| Name | Sequence | Location in BCV spike gene |
|---|---|---|
| Sp 1 | 5′-CTT-ATA-AGT-GCC-CCC-AAA-CTA-AAT | 1637–1660 |
| Sp 2 | 5′-CCT-ACT-GTG-AGA-TCA-CAT-GTT-TG | 2258–2236 |
| Sp 3 | 5′-GTT-GGC-ATA-GGT-GAG-CAC-TG | 1666–1686 |
| Sp 4 | 5′-GCA-ATG-CTG-GTT-CGG-AAG-AG | 2107–2088 |
| Sp 5 | 5′-AAC-GGT-TAC-ACT-GTT-CAG-CC | 931–950 |
| Sp 6 | 5′-CAA-GTA-AAT-GAG-TCT-GCC-TG | 1121–1102 |
| Sp 7 | 5′-GGC-TGC-CAC-CTC-TGC-TAG-TC | 2919–2938 |
| Sp 8 | 5′-ATT-GTT-AAA-TGC-ATT-AGC-AAT-AAG-C | 3069–3045 |
| SpF | 5′-TTT-TTG-ATA-CTT-TTA-ATT-TCC-TTA-CC | 4–29 |
| SpR | 5′-GTC-GTC-ATG-TGA-WGT-TTT-RAT-TAC | 4089–4066 |
PCR using primers for the hemagglutinin/esterase gene
Bovine coronavirus and other group 2 coronaviruses contain an additional structural protein, the hemagglutinin/esterase (HE). Because of the high similarity of CRCV with BCV, we analyzed the presence of a HE gene in CRCV.
An alignment of the HE gene sequences of BCV and HCV OC43 was used to design the primers HE1 and HE2 (Table 2). Four tracheal samples that had previously been identified as positive for coronavirus RNA by RT–PCR with primers for the spike gene were tested by RT–PCR with the primer set for the HE gene. All four samples showed a PCR band of the expected size (Fig. 2).
Table 2.
Primers designed from an alignment of the hemagglutinin/esterase genes of BCV (GenBank Accession No. M84486) and HCV OC43 (GenBank Accession No. M76373)
| Name | Sequence | Location in BCV HE gene |
|---|---|---|
| HE 1 | 5′-TAT-CGC-AGC-CTT-ACT-TTT-GT | 418–437 |
| HE 2 | 5′-ACC-GCC-GTC-ATG-TTA-TCA-G | 914–896 |
Fig. 2.
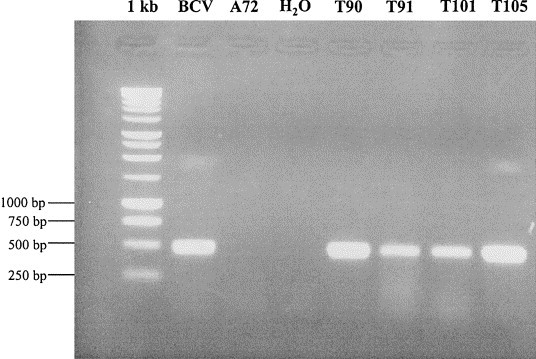
RT–PCR using consensus primers directed to the hemagglutinin/esterase gene of bovine and human coronavirus (strain OC43). The agarose gel electrophoresis shows PCR products of the expected size of 497 bp for the positive control (BCV) and four tracheal samples (T90–T105). BCV, bovine coronavirus positive control sample; A72, coronavirus-negative A72 cells; H2O, PCR mix without DNA; T90–T105, tracheal samples of study dogs; 1 kb, molecular size standard (Promega).
Detection of CRCV in tissue samples by RT–PCR and association of PCR-positive samples with respiratory signs
Using a nested set of primers for the spike gene (Sp1–2 and Sp3–4), tracheal and lung samples from 119 dogs were analyzed for CRCV. Of these, 42 were from dogs with no respiratory signs (grade 1), 18 dogs had shown mild respiratory signs (grade 2), 46 had shown moderate (grade 3), and 13 severe respiratory signs (grades 4 and 5). Grades 4 and 5 were merged due to the low case numbers in these groups.
In total 32 tracheal samples (26.9%) and 20 lung samples (16.8%) were found positive by nested RT–PCR. For eight dogs a positive PCR result was obtained for both trachea and lung. Table 3 shows the PCR results for coronavirus in dogs with different grades of respiratory disease.
Table 3.
RT–PCR results for tracheal and lung samples
| Respiratory signs | Trachea positive samples | Lung positive samples | Trachea and lung positive samples |
|---|---|---|---|
| None | 11/42 (26.2%) | 8/42 (19.1%) | 2/42 |
| Mild | 10/18 (55.6%) | 4/18 (22.2%) | 4/18 |
| Moderate | 9/46 (19.6%) | 8/46 (17.4%) | 2/46 |
| Severe | 2/13 (15.4%) | 0/13 | 0/13 |
RT–PCR results from tracheal and lung samples of 119 dogs with different respiratory signs (none to severe) using a nested PCR directed against the coronavirus spike gene. The table shows the number of positive samples out of total sample number and the percentage of positive samples in parentheses.
Sequence analysis of the nested PCR products obtained from tracheal tissues of six different dogs showed identical DNA sequences for all six cDNA samples.
Establishment of a serological assay for CRCV
Because of the homology of the spike gene region of CRCV to the spike region of bovine coronavirus, an ELISA antigen for BCV was used for serological analysis.
Sera from 5 dogs with no history of infectious respiratory disease that had not been housed in the investigated kennel were tested. The OD values ranged from −0.013 to 0.39, with an average OD value of 0.154. Furthermore, sera from 30 dogs admitted to a veterinary clinic for various reasons were tested for antibodies to coronavirus. Of these, 20 samples showed an OD of <0.4 (−0.46 to 0.396) and 10 samples showed an OD of >1.0 (1.012 to 1.949). Samples with an OD of 0.6 or above were subsequently considered positive.
Comparison of the immune response to CRCV of dogs with and without respiratory disease
The BCV antigen ELISA was performed using paired sera of 111 dogs from the study kennel. Of these, 81 dogs had shown symptoms of respiratory disease during a period of 21 days and 30 had remained healthy.
Of the group of dogs which developed respiratory disease, 17 were positive for antibodies to CRCV on the day of entry into the kennel and 64 were negative.
Of the 64 dogs in this group which had no detectable antibodies to CRCV on day 1, 63 tested positive on day 21. All 46 dogs of these 63 for which a sample on day 7 was available tested negative on day 7. Therefore 63 dogs showed a seroconversion during the study period, whereas only 1 dog remained negative.
Of the 30 dogs that had remained healthy, 17 had antibodies to CRCV on the day of entry. All of the 13 dogs that were negative on day 1 tested negative on day 7 but showed a seroconversion until day 21.
Thus, of 34 dogs that were positive for antibodies to CRCV on arrival in the kennel, 17 developed respiratory disease (50%), whereas of 77 dogs that were negative on arrival, 64 developed respiratory signs during the study period (83.1%) (Fig. 3).
Fig. 3.
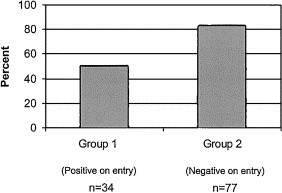
Comparison of the prevalence of respiratory disease in two groups of dogs: Dogs in group 1 were positive for serum antibodies to respiratory coronavirus on the day of entry into the kennel; dogs in group 2 were negative. The graph shows the percentage of dogs developing respiratory disease in group 1 compared to group 2 (P < 0.001). N is the total number of dogs in each group.
Therefore dogs that had no antibodies to CRCV on entry into the kennel had an increased probability of developing respiratory disease (P < 0.001).
Only 1 of the 77 dogs that were negative on arrival remained negative during the study period of 21 days, whereas 76 dogs showed a seroconversion.
Serology using canine enteric coronavirus (CECV) antigen
An ELISA assay using a canine coronavirus antigen was performed to investigate whether CRCV showed a serological cross-reaction to canine enteric coronavirus. Sera from 27 dogs, previously tested for antibodies to CRCV using the BCV antigen, were selected.
It was found that 8 dogs had antibodies to CECV on the day of entry into the kennel; of these 4 also had antibodies to CRCV. Nineteen dogs were found to be negative for CECV on day 1; 17 of these were also negative for CRCV. Of the 19 negative dogs, 5 showed a seroconversion to CECV during the 21-day period of the investigation and 17 showed a seroconversion to CRCV.
Analysis of the prevalence of respiratory disease in this group showed that 6 of the 8 dogs (75%) that were positive for antibodies to CECV on day 1 developed respiratory disease. Of the group of 19 dogs that had no detectable antibodies to CECV on day 1, 15 showed signs of respiratory disease (78.9%) (P = 0.594).
Virus isolation
Tracheal tissue samples from five dogs that had been identified as positive for coronavirus RNA by RT–PCR were inoculated on cell cultures of canine adult lung fibroblasts and MDCK cells. For three samples virus isolation was also attempted on A72 cells. The cultures showed no signs of a cytopathic effect during three passages. After the third passage, RNA was extracted from the cultures and found to be negative for coronavirus by RT–PCR.
Discussion
This study reports the detection of a canine coronavirus, provisionally called CRCV, in kenneled dogs with respiratory disease.
Coronaviruses have been reported to cause respiratory disease of man, cattle, swine, and poultry, but their presence in the respiratory tract of dogs and a possible association with CIRD has not been determined. Only one study has mentioned the isolation of canine coronavirus from four dogs with symptoms of respiratory disease (Binn et al., 1979). Therefore it was investigated in this study whether coronaviruses could be detected in dogs from a kennel with a high prevalence of CIRD. The disease was endemic in this kennel and could not be controlled by the use of vaccines recommended against “kennel cough,” strongly suggesting that additional agents were contributing to the disease syndrome observed. Samples taken from the respiratory tract of these dogs were examined using RT–PCR primers directed to the conserved polymerase gene of coronaviruses (Stephensen et al., 1999). Analysis of the cDNA sequences obtained from the canine samples revealed that CRCV had the highest similarity with the polymerase gene of bovine coronavirus (98.8%) and human coronavirus OC43 (98.4%) but only a very low similarity to the polymerase gene of the enteric canine coronavirus (strain 1–71, 68.53% similarity).
Phylogenetic analysis of the polymerase sequences of 11 coronaviruses showed CRCV to be located on a common branch with 3 group 2 viruses: bovine coronavirus (BCV), human coronavirus strain OC43 (HCV-OC43), and hemagglutinating encephalomyelitis virus. However, canine enteric coronavirus, a group 1 coronavirus, was shown to be only distantly related.
Canine respiratory coronavirus therefore is a novel coronavirus of dogs that is most closely related to BCV and HCV-OC43, both of which are known to cause respiratory disease.
Sequence analysis of the spike gene confirmed the high similarity of CRCV with BCV and HCV-OC43, both of which are members of the group 2 of the Coronaviridae family. Group 2 coronaviruses contain an additional structural protein, HE. We were able to demonstrate the presence of a HE gene in the cDNA obtained from canine tracheal samples by PCR. CRCV therefore has a HE gene and belongs to group 2 of the coronavirus family.
Attempts to isolate CRCV from tissue of the respiratory tract, using either canine lung fibroblasts or a kidney epithelial cell line, have been unsuccessful so far. However, the isolation of CRCV may require the use of fetal canine cells or tracheal organ culture. Also it is possible that the virus was inactivated during storage or due to freezing and thawing.
By PCR, CRCV was detected in tracheal and lung tissue and therefore appears to infect the upper and lower respiratory tracts of dogs. The possible presence of viral RNA in other tissues needs to be analyzed in further studies and the target cells of CRCV have to be identified using in situ hybridization or immunohistochemistry.
The presence of antibodies to CRCV was analyzed using an ELISA based on a BCV antigen because of the high sequence similarity of the two viruses in the spike gene and because CRCV antigen could not be obtained by virus culture. The ELISA results confirmed the presence of a virus similar to BCV in the study population.
The prevalence of antibodies was 30% at the time of entry into the kennel and 99% after 21 days. Almost all dogs negative on the day of entry into the kennel showed a seroconversion to CRCV within 3 weeks, indicating that the virus is highly contagious.
Serology using an antigen for CECV showed a much lower prevalence of antibodies to CECV on day 21. Therefore the BCV ELISA results did not reflect an infection with canine enteric coronavirus and the cross-reactivity between the two antigens seems to be low. Serum antibodies to CRCV were present in about 30% of dogs of various origins, including dogs entering a rehoming kennel as well as pet dogs. The presence of CRCV is therefore not limited to the investigated kennel and the virus seems to be established in the dog population.
Within the kenneled population, CRCV RNA was detected in 27.3% of dogs with all grades of respiratory disease as well as in 26.2% of dogs that were apparently healthy at the time of euthanasia. However, CRCV RNA was most frequently found in the trachea of dogs with mild cough (55%). Studies using the human coronavirus strain 229E have shown that coronaviruses can cause disruption of the respiratory epithelium and ciliary dyskinesia (Chilvers et al., 2001). If one supposes that an infection with CRCV may have a similar effect then the virus could play an important role in the early stages of the pathogenesis of CIRD. By damage of the respiratory epithelium and disruption of the ciliary clearance CRCV could facilitate the entry of other viral or bacterial pathogens causing the more severe respiratory symptoms. The less frequent detection of CRCV in tissue samples from dogs with more severe disease may be explained by the destruction of the respiratory epithelium by other microorganisms in the advanced stages of CIRD. At the same time it is likely that the immune response that is induced by CRCV in almost all dogs would help to clear the infection, causing the prevalence of CRCV RNA to be lower in later stages of the disease.
Furthermore, serological analysis revealed that dogs with antibodies to CRCV on the day of entry into the kennel developed respiratory disease less frequently than dogs without antibodies (P < 0.001). Therefore the presence of antibodies to CRCV had a protective effect against respiratory disease in this population, indicating a possible role of the virus in the pathogenesis of CIRD. However 50% of the dogs that were seropositive for CRCV on day 1 still developed respiratory disease. It was not determined if dogs which developed disease had lower antibody titers than those that stayed healthy as the ELISA was performed using a single serum dilution. Alternatively, the disease in seropositive dogs may well have been caused by other respiratory pathogens present in this population. Studies determining the prevalence of other pathogens are currently under way.
As CIRD is a complex disease involving a variety of microorganisms, further epidemiological studies are required to determine the exact role of CRCV in this syndrome. It is likely that infections with CRCV alone may cause only subclinical or mild respiratory symptoms but in conjunction with other pathogenic agents severe respiratory disease may occur. Alternatively, the presence of CRCV may exacerbate disease caused by other agents.
This study describes the detection of a canine respiratory coronavirus in dogs with CIRD, which is genetically and antigenically distinct from the previously described enteric coronavirus. The pathogenesis of “kennel cough” has not been thoroughly investigated since the 1970s, when B. bronchiseptica, canine adenovirus type 2, and canine parainfluenza were determined to be the main causes of the disease. However, the vaccination of all dogs against CPIV, CAV-2, and distemper virus did not help to control the disease in this kennel despite evidence that the majority of dogs responded to the vaccine within 21 days (data not shown). A B. bronchiseptica vaccine had been used in the past prior to this study but was discontinued because it failed to protect against respiratory disease. Most rehoming and training kennels frequently have to deal with outbreaks of CIRD despite regular vaccination of all dogs. The aetiology of CIRD therefore needs to be reevaluated and the role of novel microorganisms or microorganisms previously not associated with the disease should be established.
Materials and methods
Study population
Dogs from a well-established rehoming kennel with a history of endemic respiratory disease were monitored for this study. On entry into the kennel, all dogs were vaccinated with KAVAK DA2 PiP69 (Fort Dodge), a live attenuated vaccine for distemper virus, canine adenovirus type 2, canine parainfluenzavirus, and canine parvovirus. Also, a killed leptospirosis vaccine was used (Fort Dodge). The health status of each dog was assessed twice a day by a veterinary clinician and the respiratory signs were graded as follows: (1) no respiratory signs, (2) mild cough, (3) cough and nasal discharge, (4) cough, nasal discharge, and inappetence, (5) evidence of bronchopneumonia. The overall health status of the dogs was graded as follows: (1) good health, (2) poor health, (3) very poor health. The age, breed, and sex of the dogs were recorded.
One hundred and nineteen dogs from the kennel population were euthanased for welfare reasons, ranging from behavioral problems to signs of severe respiratory disease For these dogs, a full postmortem examination was performed. The tissue samples were stored at −70°C until further use.
Serum samples were collected from 111 dogs on the day of entry into the rehoming kennel. For 81 dogs a follow-up serum was available on day 7 and for 111 dogs a serum was available on day 21 after entry. Of the 111 dogs, 30 remained healthy during the 21 days between the first and the last serum samples, whereas 81 dogs developed respiratory disease.
Sera from 35 dogs housed elsewhere were obtained from the diagnostic service of the Royal Veterinary College. These sera had been submitted for biochemical analysis for various reasons. Five of these sera were from 18-month-old beagles with no history of respiratory disease. Sera were routinely stored at −20°C.
RNA extraction and RT–PCR
RNA was extracted from tracheal and lung tissue of 119 dogs using TriReagent (Sigma). Approximately 25–50 mg of homogenized tissue was used and RNA was extracted as recommended by the manufacturer.
Synthesis of cDNA was performed using Random Hexameres (Roche) and ImPromII reverse transcriptase (Promega).
For the detection of coronaviruses a modification of the primers 2Bp and 4Bm directed against the polymerase gene as described by Stephensen et al. (1999) were used (Conscoro5, 5′-ACT-CAR-ATG-AAT-TTG-AAA-TAT-GC; Conscoro6, 5′-TCA-CAC-TTA-GGA-TAR-TCC-CA).
PCR was performed using Taq polymerase (Promega) in the provided reaction buffer containing a final concentration of 2.5 mM MgCl2 and 0.5 μM primers. For PCR with the primers Conscoro5 and Conscoro6 the following temperature profile was used: After denaturation at 95°C for 5 min, 10 cycles were carried out at 95°C for 1 min, annealing at 37°C for 1 min, and extension at 72°C for 1 min. This was followed by 10 cycles using an annealing temperature of 45°C, 10 cycles at an annealing temperature of 50°C, and 10 cycles at an annealing temperature of 53°C, followed by a final extension at 72°C for 10 min.
A 20-μl fraction of the PCR product was analyzed on a 1.5% agarose gel and blotted onto a nylon membrane after electrophoresis. The nylon membrane was hybridized with an oligonucleotide probe specific for the PCR product at 37°C overnight (Probe Conscoro, AAG-TTT-TAT-GGY-GGY-TGG-GA). The probe was 3′A-tailed with digoxigenin–dUTP and was detected using anti-digoxigenin conjugate and CSPD chemoluminescent substrate (Roche).
Primers for the HE gene were chosen using an alignment of the HE genes of BCV strain LY-138 (GenBank Accession No. M84486) and HCV strain OC43 (GenBank Accession No. M76373). The sequence and location of the primers are shown in Table 2. The following temperature profile was used for the PCR: denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 1 min, 50°C for 40 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. The expected size of the PCR product was 497 bp.
Primer sequences specific for the spike gene were derived from an alignment of the spike region of bovine coronavirus strain LY-138 (GenBank Accession No. AF 058942) and human coronavirus strain OC43 (GenBank Accession No. L14643).
For sequencing of the complete spike gene the primers Sp1–Sp8, SpF, and SpR were designed. Table 1 shows the primer sequences. A PCR was performed using Pfu polymerase (Promega) and the following temperature profile: denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 1 min, 50°C for 40 s, and 72°C for 3 min. The final extension was performed at 72°C for 10 min.
The PCR products were separated on an agarose gel and purified using the Qiaquick gel purification kit (Qiagen). PCR products were cloned into the pT7blue2 blunt vector (Novagen) and sequenced using the Thermo sequenase fluorescent labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia) using Cy5-labeled primers.
For the detection of coronavirus RNA in tissue samples, the cDNA was tested by PCR with the primers Sp1 and Sp2, followed by a nested PCR using the primers Sp3 and Sp4 and 2 μl of the product of the first amplification.
The temperature profile used was denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 1 min, 55°C for 40 s, and 72°C for 1 min. The final extension was performed at 72°C for 10 min. The nested PCR produced a 442-bp fragment.
Phylogenetic analysis
Nucleic acid similarity searches were performed using FASTA (Pearson, 1990), with a gap open penalty of 16 and a gap extension penalty of 4. Protein similarities were determined using FASTA or GCG (Genetics Computer Group, Wisconsin, USA) with a gap open penalty of 8 and a gap extension penalty of 2.
Sequence alignments were performed using ClustalX (Thompson et al., 1997).
The phylogenetic relationship to known coronaviruses was analyzed using the Phylip 3.6 package (Felsenstein, 1989). The alignments were followed by a bootstrap analysis using the Seqboot program. The data sets obtained were used for a maximum parsimony analysis using the DNApars program and a consensus tree was calculated using Consense. The resulting trees were drawn using the Treeview program (Page, 1996).
GenBank Accession numbers
The CRCV polymerase partial sequence has been assigned GenBank Accession No. AY150273; the CRCV spike gene sequence has been assigned GenBank Accession No. AY150272.
ELISA
ELISA antigen for bovine coronavirus or canine coronavirus (Churchill Applied Biosciences, Huntingdon, UK) was resuspended in PBS at the concentration recommended by the manufacturer and incubated on 96-well plates (Falcon) overnight at 37°C. The plates were washed with PBS and blocked with PBS containing 5% skimmed milk powder for 30 min. The sera were diluted 1:100 in blocking buffer and incubated on the plates for 1 h. After washing with PBS/0.05% Tween 20 (Sigma), a peroxidase-labeled rabbit anti-dog IgG conjugate (Sigma) was added (1:5000 in PBS/0.05% Tween 20) for 1 h. The plates were incubated with color substrate (OPD, Sigma) for 10 min and the reaction was stopped by adding 2 M H2SO4. The adsorption was determined in an ELISA photometer at 492 nm.
Virus culture
Virus isolation was attempted on canine adult lung fibroblasts (passages 3 to 7) and MDCK and A72 cells. The lung fibroblasts were maintained in MEM with 20% fetal calf serum (FCS); MDCK and A72 cells were maintained in MEM with 5% FCS. Tracheal tissue samples (approx 25 mg) were homogenized using a scalpel and mixed vigorously in 1 ml of MEM containing penicillin (100 U/ml), streptomycin (0.1 mg/ml), amphotericin B (2.5 μg/ml), and trypsin (1 μg/ml). The samples were centrifuged at 13,000 rpm for 10 min and the supernatant was used to inoculate cell cultures. After 30 min at 37°C the supernatant was removed and maintenance medium was added to the cultures. The cultures were passaged three times in the absence of a cytopathic effect. Then, RNA was extracted from the cells and RT–PCR for coronavirus was performed using the nested PCR for the spike gene.
Statistical analysis
The data were analysed using the χ2 test or Fisher’s exact test and P values below 0.05 were considered statistically significant.
Acknowledgements
The authors are most grateful to the Dogs Home Battersea for funding to Professor J. Brownlie and for support and guidance. We thank the veterinary nurses and clinicians at the Dogs Home Battersea for technical assistance. We also thank Dr. V. Chalker and Dr. M. Collins for critical reading of the manuscript and S. Oliver for providing a control sample for bovine coronavirus.
References
- Appel M, Binn L.N. Canine infectious tracheobronchitis short review: kennel cough. In: Appel M, editor. Virus Infections of Carnivores first ed. Elsevier Science Publishers; Amsterdam: 1987. pp. 201–211. [Google Scholar]
- Appel M, Percy D.H. SV-5-like parainfluenza virus in dogs. J. Am. Vet. Med. Assoc. 1970;156:1778–1781. [PubMed] [Google Scholar]
- Bemis D.A, Carmichael L.E, Appel M.J. Naturally occurring respiratory disease in a kennel caused by Bordetella bronchiseptica. Cornell Vet. 1977;67:282–293. [PubMed] [Google Scholar]
- Bemis D.A, Greisen H.A, Appel M.J. Pathogenesis of canine bordetellosis. J. Infect. Dis. 1977;135:753–762. doi: 10.1093/infdis/135.5.753. [DOI] [PubMed] [Google Scholar]
- Binn L.N, Alford J.P, Marchwicki R.H, Keefe T.J, Beattie R.J, Wall H.G. Studies of respiratory disease in random-source laboratory dogs: viral infections in unconditioned dogs. Lab. Anim. Sci. 1979;29:48–52. [PubMed] [Google Scholar]
- Binn L.N, Eddy G.A, Lazar E.C, Helms J, Murnane T. Viruses recovered from laboratory dogs with respiratory disease. Proc. Soc. Exp. Biol. Med. 1967;126:140–145. doi: 10.3181/00379727-126-32386. [DOI] [PubMed] [Google Scholar]
- Chilvers M.A, McKean M, Rutman A, Myint B.S, Silverman M, O’Callaghan C. The effects of coronavirus on human nasal ciliated respiratory epithelium. Eur. Respir. J. 2001;18:965–970. doi: 10.1183/09031936.01.00093001. [DOI] [PubMed] [Google Scholar]
- Ditchfield J, Macpherson L.W, Zbitnew A. Association of a canine adenovirus (Toronto A 26/61) with an outbreak of laryngotracheitis (“kennel cough”) Can. Vet. J. 1962;3:238–247. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP-Phylogeny Inference Package (Version 3.2c) Cladistics. 1989;5:164–166. [Google Scholar]
- Ignjatovic J, Sapats S. Avian infectious bronchitis virus. Rev. Sci. Tech. 2000;19:493–508. doi: 10.20506/rst.19.2.1228. [DOI] [PubMed] [Google Scholar]
- Karpas A, King N.W, Garcia F.G, Calvo F, Cross R.E. Canine tracheobronchitis: isolation and characterization of the agent with experimental reproduction of the disease. Proc. Soc. Exp. Biol. Med. 1968;127:45–52. doi: 10.3181/00379727-127-32618. [DOI] [PubMed] [Google Scholar]
- Keil D.J, Fenwick B. Role of Bordetella bronchiseptica in infectious tracheobronchitis in dogs. J. Am. Vet. Med. Assoc. 1998;15:200–207. [PubMed] [Google Scholar]
- Lou T.Y, Wenner H.A. Natural and experimental infection of dogs with reovirus, type1: pathogenicity of the strain for other animals. Am. J. Hyg. 1963;77:293–304. [Google Scholar]
- Mäkelä M.J, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimaki M, Blomqvist S, Hyypia T, Arstila P. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.D.M. Treeview: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pearson W.R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Pensaert M, Callebaut P, Vergote J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet. Q. 1986;8:257–261. doi: 10.1080/01652176.1986.9694050. [DOI] [PubMed] [Google Scholar]
- Randolph J.F, Moise N.S, Scarlett J.M, Shin S.J, Blue J.T, Bookbinder P.R. Prevalence of mycoplasmal and ureaplasmal recovery from tracheobronchial lavages and prevalence of mycoplasmal recovery from pharyngeal swab specimens in dogs with or without pulmonary disease. Am. J. Vet. Res. 1993;54:387–391. [PubMed] [Google Scholar]
- Spaan W, Cavanagh D, Horzinek M.C. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Stephensen C.B, Casebolt D.B, Gangopadhyay N.N. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 1999;60:181–189. doi: 10.1016/S0168-1702(99)00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J, Purdy C.W, Lin X, Burrell M, Truax R.E, Briggs R.E, Frank G.H, Loan R.W. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp. from cattle involved in two natural outbreaks of shipping fever. J. Am. Vet. Med. Assoc. 2000;216:1599–1604. doi: 10.2460/javma.2000.216.1599. [DOI] [PubMed] [Google Scholar]
- Tennant B.J, Gaskell R.M, Jones R.C, Gaskell C.J. Studies on the epizootiology of canine coronavirus. Vet. Rec. 1993;132:7–11. doi: 10.1136/vr.132.1.7. [DOI] [PubMed] [Google Scholar]
- Thompson J.D, Gibson T.J, Plewniak F, Jeanmougin F, Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]


