Abstract
Crystallization remains a critical step in X-ray structure determination. Because it is not generally possible to rationally predict crystallization conditions, commercial screens have been developed which sample a wide range of crystallization space. While this approach has proved successful in many cases, a significant number of proteins fail to crystallize despite being soluble and monodispersed. It is established that chemical modification can facilitate the crystallization of otherwise intractable proteins. Here we describe a method for the reductive methylation of lysine residues which is simple, inexpensive, and efficient, and report on its application to ten proteins. We describe the effect of methylation on the physico-chemical properties of these proteins, and show that it led to diffraction-quality crystals from four proteins and structures for three that had hitherto proved refractory to crystallization. The method is suited to both low- and high-throughput laboratories.
Main Text
Introduction
Crystallography remains the leading method for solving in atomic detail the structures of macromolecules and their complexes. However, success rests entirely on obtaining diffraction-quality crystals and the process of crystallization remains a hit-and-miss affair, typically involving screening hundreds of conditions. It appears from a number of large-scale analyses (see, for instance, Fogg et al., 2006 and Canaves et al., 2004) that at present the success rate remains no more than 50%.
It is self-evident that molecular flexibility will tend to be detrimental to the formation of a highly ordered crystal lattice. For proteins, such flexibility can arise from large-scale motions of domains or portions of the polypeptide chain such as surface loops and chain termini. Such motions may be reduced or eliminated by redesign of the protein construct (for instance, to remove disordered terminal extensions) or by binding to a “crystallization chaperone” which might be as large as an antibody fragment (Iwata et al., 1995) or as small as a chemical inhibitor (Ren et al., 1995). In addition, if poorly ordered crystals are obtained, diffraction can sometimes be improved by controlled dehydration (see, for instance, Esnouf et al., 1998 and Heras et al., 2003). Bioinformatics and experimental approaches can be used to detect and perhaps control large-scale disorder (Esnouf et al., 2006, Yang et al., 2005, Geerlof et al., 2006, Stroh et al., 2005) and the use of thermostable versions of proteins may also help (Adams et al., 2003, Lesley et al., 2002), as these might be expected to be more rigid at temperatures lower than experienced in vivo.
At the other extreme, small motions such as those due to flexible, solvent-exposed amino acid side chains can be equally disruptive to a well-ordered crystal lattice. One approach to this problem has been to selectively mutate these residues to smaller hydrophobic residues. Initially described by Lawson et al. (1991), where the diffraction quality of human ferritin crystals was greatly improved by the mutation of an exposed lysine to a glutamine, microengineering of flexible surface residues has produced some remarkable crystallization successes. The Derewenda laboratory has championed such approaches and the state of the art is described in a recent review (Derewenda and Vekilov, 2006). Particular attention has been given to the mutation of large charged residues such as lysine (Longenecker et al., 2001, McElroy et al., 1992) and glutamate (Mateja et al., 2002). While such “surface-entropy-reduction mutagenesis” is now an established technique, it remains a time- and labor-intensive procedure which requires the production of modified expression constructs. In addition, in the absence of structural knowledge, it is quite possible that key stabilizing salt bridges might be inadvertently targeted for destruction.
Surface engineering of proteins by chemical modification offers a radically different approach to reducing surface entropy and has led to some dramatic successes, none more so than the first application by the Rayment laboratory to a fragment of myosin (Rayment et al., 1993). This approach offers several advantages: first, recloning, expression, and purification steps are eliminated; second, modification of the mature protein avoids the possibility of inducing misfolding of the nascent polypeptide chain (Rypniewski et al., 1993); and third, only those residues on the exposed surface of the protein will be modified, not those protected in the protein core or buried in strong protein-protein interfaces. Conversely, such a “shotgun” method will indiscriminately label residues which may be critical for biological function. The most common approach to chemical modification has been the reductive methylation of free amino groups in which primary amines (i.e., lysine residues and the N terminus) are modified to tertiary amines. Originally used as a method of isotope labeling (Means and Feeney, 1968, Rice and Means, 1971), this has proven value for protein crystallization, allowing, in certain instances, the formation of diffraction-quality crystals for proteins that had previously proved refractory to crystallization (Kobayashi et al., 1999, Kurinov et al., 2000, Rayment et al., 1993, Saxena et al., 2006, Schubot and Waugh, 2004).
Here we describe a simple generic protocol for the methylation of lysines residues, developed in the Oxford Protein Production Facility (OPPF) and the Division of Structural Biology at Oxford University, which has been adopted as a routine crystallization rescue strategy. The procedure is cheap and robust, and mass spectrometry provides a straightforward readout of the extent of methylation. We describe its application to a set of ten proteins. The process yielded a homogeneous modification for nine of these; four have formed diffraction-quality crystals leading to three high-resolution structures to date.
The Protocol
The method used is derived from that published previously (Rayment, 1997, Rayment et al., 1993). The methylation reaction is performed overnight in 50 mM HEPES (pH 7.5), 250 mM NaCl at protein concentrations of 1 mg/ml or less. Twenty microliters freshly prepared 1 M dimethylamine-borane complex (ABC; Fluka product 15584) and 40 μl 1 M formaldehyde (made from 37% stock; Fluka product 33220) are added per ml protein solution, and the reactions are gently mixed and incubated at 277K for 2 hr. A further 20 μl ABC and 40 μl formaldehyde are added and the incubation continued for 2 hr. Following a final addition of 10 μl ABC, the reaction is incubated overnight at 277K. In some cases, the reaction leads to a significant amount of precipitated protein (occasionally over 50%), which is removed by centrifugation before purification of the soluble methylated protein by size-exclusion chromatography columns (S75 or S200 Superdex 16/60 on Äkta express FPLC systems; GE Healthcare) pre-equilibrated in 20 mM Tris-HCl (pH 7.5), 200 mM NaCl. Appropriate peak fractions are pooled and concentrated in centrifugal concentrators (Vivascience) to appropriate concentrations (as determined by a precrystallization test; PCT; Hampton Research). The concentrated protein is then often buffer exchanged into 20 mM Tris-HCl using a micro Bio-Spin column (Bio-Rad), and crystallization experiments are set up immediately.
Compared to previous protocols, the concentration of the protein in the reaction is markedly reduced, to 1 mg/ml or less, as higher concentrations often led to protein precipitation and significant loss of material. As a consequence, the formaldehyde and reductant are in greater excess over the number of free amines on the protein, producing no adverse effects and perhaps helping to ensure the homogeneous modification observed (see below). Similarly, the reaction is not quenched with a high concentration of ammonium sulfate, as this also frequently led to precipitation; instead, quenching is performed by the Tris-HCl buffer used for the postreaction size-exclusion chromatography by reaction with the amines of the buffer (in line with the observation of Rayment, 1997 that any primary amine can be used to quench). This purification step also separates the protein from excess reagents and monitors protein quality and oligomeric state.
Application to a Test Set of Proteins
To test whether our protocol for the reductive methylation of lysines could be used as a generic rescue strategy, a cohort of ten proteins which had proved refractory to structural analysis was selected. These proteins, from a number of source organisms, were selected on the basis that they were soluble, stable, and, in seven out of the ten cases, did not crystallize. Eight of the test set (both native and selenomethionine [SeMet] derivatives) were produced by the OPPF pipeline (described in both Ren et al., 2005 and Siebold et al., 2005) and two came from the Division of Structural Biology. The proteins are listed in Table 1. All were >95% pure (as judged by SDS-PAGE analysis) before the methylation reaction.
Table 1.
Data for the Cohort of Ten Proteins Subject to Methylation
| Name of Proteina | MW (kDa) N/Mb | pI Cb/N/M | No. of Sites R (P)c | Protein Conc. N/Md | Oligomeric State N/Me | Crystal N/M | Diffraction N/M | Comments |
|---|---|---|---|---|---|---|---|---|
| BA BA0252 (alanine racemase) | 43.8/44.4 | 6.2/5.5/5.2 | 20 (21) | 22/15 | M/M | no/yes | NA/2.8 Å | Structure solved. Methylation essential for crystallization. |
| MHV NSP9 | 13.2/13.5 | 8.5/9.5/8.6 | 11 (11) | 14/8 | D/D | no/yes | NA/2.3 Å | Structure solved. Methylation essential for crystallization. |
| MVEV NS5A (methyl transferase) | 31.2/31.7 | 9.5/>10/9.2 | 19 (22) | 9/6 | M/M | yes/yes | no/2.1 Å | Structure solved. Protein precipitates partially in reaction. Methylation dramatically improves crystal quality. |
| Bacteriophage Ф8 P1 | 86.8/87.8 | 6.1/5.6/5.8 | 36 (38) | 10/4 | −f | yes/yes | 4.0 Å/4.0 Å | Crystal structure not solved due to large unit cell. Cell dimensions change on methylation. |
| SARS NSP1 domain | 14.6/14.8 | 5.9/−/− | 7 (7) | 8/9 | M/M | no/no | NA | |
| SARS NSP10 | 16.9/17.1 | 6.9/−/− | 7/8 (8) | −/4 | M/M | no/no | NA | Methylated as two species. |
| SARS NSP15 | 38.1/38.8 | 6.0/−/− | 26 (26) | 12/8 | M/M | no/no | NA | |
| FoxP2 337–716 | 43.0/43.5 | 5.8/−/− | 16 (16) | 8/6 | D/M | no/no | NA | |
| FoxP2 337–433 | 12.8/13.0 | 7.6/−/− | 7 (7) | 24/9 | XX/D | no/no | NA | |
| RSVN | 44.4/45.2 | 6.5/−/− | 28 (31) | 12/8 | X/X | yes/yes | 10 Å/10 Å | Changed crystal morphology and condition. |
NA, not applicable.
BA, Bacillus anthracis; MHV, murine hepatitis virus; MVEV, Murray Valley encephalitis virus; SARS, severe acute respiratory syndrome; FoxP2, Forkhead box P2; RSVN, respiratory syncytial virus nucleoprotein.
N, native, unmodified protein; M, methylated; C, calculated.
R, number of sites methylated; P, number of potential sites.
Protein concentration (mg/ml).
Oligomeric state was as estimated from size-exclusion chromatography. M, monomer; D, dimer; X, decamer; XX, possible 16-mer (MW ∼150 kDa).
The precise oligomeric state is ill determined, but reduced upon methylation.
Changes in the oligomeric state, following methylation, were observed during the postreaction size-exclusion chromatography purification step for three of the ten proteins (Table 1). Samples were removed pre- and postreaction for electrospray ionization mass spectrometry analysis (an increase in mass of 28 Da corresponding to the modification of one lysine residue or the N-terminal amine). For four of the ten proteins not all the potential sites (taken to be the number of lysine residues plus one) were methylated, implying that those residues remaining unmodified were inaccessible to the reagents, either buried in the protein core or masked by bound ligand at the active site in the case of the viral methyltransferase and bacterial alanine racemase (see Table 1). Certain of the proteins were also characterized by isoelectric focusing (IEF) (performed according to the manufacturer's instructions on a vertical system using pH 3–10 IEF precast gels with the manufacturer's buffers; Novex; Invitrogen) and broad-range isoelectric point (pI) markers (GE Healthcare). For those proteins with a high pI, the pI was reduced by approximately one unit upon methylation, while for those with a lower pI, there was little change (Table 1).
Crystallization experiments were set up and monitored using the crystallization facility in the OPPF (Brown et al., 2003, Walter et al., 2003, Walter et al., 2005). Of the ten proteins methylated, five gave crystals. For four of the proteins, the crystals were of diffraction quality. For two of these, no crystals had been previously obtained; and in one further case the crystal quality was markedly improved (see Table 1), resulting in three successful structure determinations. For one protein the unit cell dimensions have been reduced, presumably due to the reduction in oligomeric state, while in the last case there is a change in crystal morphology, and analysis is still in progress. We will briefly describe the three successful structural analyses.
Bacilus anthracis Alanine Racemase
The enzyme alanine racemase from Bacillus anthracis was targeted as part of the ongoing collaborative structural proteomics initiative between the OPPF and York Structural Biology laboratory (Au et al., 2006). An N-terminal His6-tagged form of the protein was expressed, purified and the tag removed, using established protocols (Ren et al., 2005, Siebold et al., 2005). The protein showed two distinct oligomeric states in gel filtration, assumed to be tetrameric and dimeric. Extensive crystallization trials (approximately 800 conditions) on both oligomeric forms (in the presence of the cofactor pyridoxal 5′-phosphate) at both 15 and 22 mg/ml yielded no crystal hits and so reductive methylation was attempted. Mass spectrometry analysis revealed that 20 out of 21 potential sites were fully methylated, reducing the pI from 5.5 to 5.2. Methylated dimeric protein at ∼15 mg/ml crystallized at 298K within a few hours in high salt conditions. Crystals grown from optimized conditions were used for data collection and the structure was determined by molecular replacement to 2.9 Å. In the crystal, four lysine residues are located at the crystallographic interfaces between the dimers. It is presumably the modification of these residues which allows crystal formation. A paper describing the structure in greater detail is currently in preparation (K.-F.A., J.R., T.S.W., C.M., K. Harlos, D.I.S., and R.M. Esnouf, unpublished data).
MVEV Methyltransferase
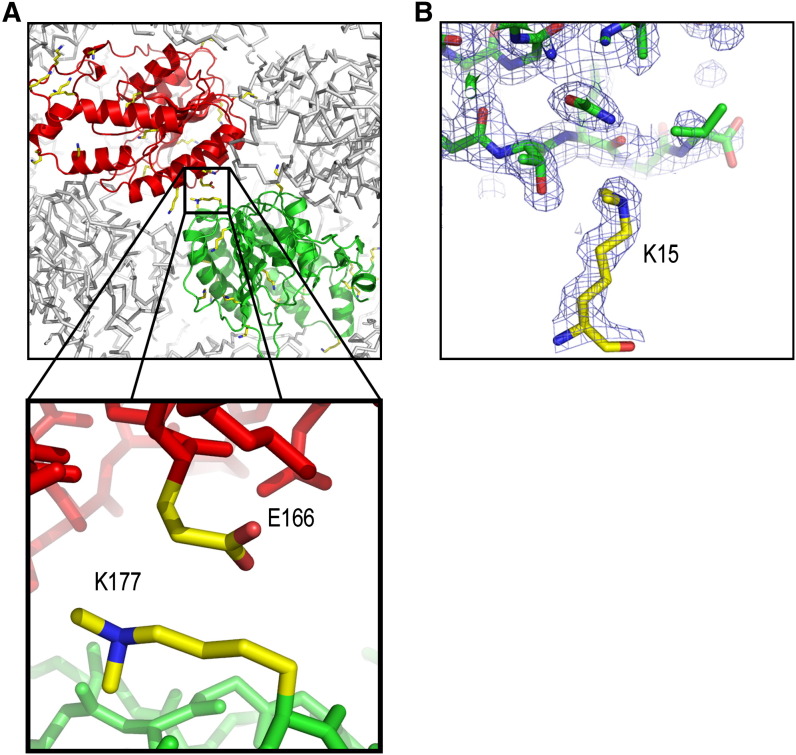
The flavivirus Murray Valley encephalitis virus (MVEV) is a target of the ongoing “Vizier” structural genomics initiative (http://www.vizier-europe.org/). The methyltransferase domain of nonstructural protein 5 (NS5) was chosen for structural analysis. Based on bioinformatic analysis (Albeck et al., 2006, Esnouf et al., 2006, Yang et al., 2005) and alignment with a related structure from Dengue virus (Egloff et al., 2002), a number of constructs were cloned and expressed. Although several constructs gave soluble protein, no crystals suitable for X-ray diffraction analysis were grown. Methylation of the protein modified 19 out of 22 sites and produced three different crystal forms, all of which diffracted to better than 2.1 Å. The majority of crystals grew from either 20% PEG3350 or high salt conditions. Upon methylation, the pI of the protein shifted from >10 to 9.2. In one crystal form, nine lysines are involved in crystal contacts ( Figure 1). A paper describing the structure of this protein is currently in preparation (R.A., J.R., A.V., T.S.W., D. Alderton, R. Hurrelbrink, S. Fuller, R.J.O., D.I.S., and J.M.G., unpublished data).
Figure 1.
Examples of Methylated Lysines from MVE Methyltransferase
(A) An example of the interaction between a methylated lysine and a glutamate residue at a crystal contact. An overview of the packing of molecules of MVE methyltransferase in one of the three crystal forms solved (space group P21, a = 46.4 Å, b = 68.3 Å, c = 82.0 Å, β = 98.4°) is drawn in the top panel, with the two molecules in the crystallographic asymmetric unit colored red and green, respectively. The close-up in the bottom panel shows the hydrophobic packing of the aliphatic components of the side chains lysine 177 and glutamate 166.
(B) An example of well-defined electron density for a methylated lysine (K15) involved in a crystal contact with the main chain of a neighboring molecule. The figure was produced using PyMOL (http://www.pymol.org).
MHV NSP9
As part of an ongoing structural proteomics project on coronaviruses (Meier et al., 2006), a cohort of proteins from the murine hepatitis virus (MHV) genome have been targeted for structural analysis. The small dimeric NSP9 protein (formerly NSP6) is a homolog of the SARS NSP9, binds RNA, and is part of the replication complex (Sutton et al., 2004). Prior to methylation, no crystals of MHV NSP9 were obtained despite extensive screening efforts (native and SeMet-labeled protein were tested at different concentrations). After reductive methylation, all 11 potential methylation sites were modified and the pI of the protein was reduced from 9.5 to 8.6. Methylated forms of both native and SeMet-labeled protein crystallized readily from high salt conditions. Further crystal optimization led to structure determination by molecular replacement at 2.3 Å resolution (C.M., D.I.S., and J.M.G., unpublished data). Five lysine residues are located at interfaces, forming crystal contacts between dimers.
Discussion
A simple protocol for the modification of lysine residues has been tested on ten protein samples which had previously proved intractable to structure solution. In three cases, the structure of the methylated protein could be determined.
Mass spectrometry proves an excellent quality-control step; an incomplete reaction due to perished reagents can be readily observed as a series of ion species 14 Da apart (corresponding to addition of single methyl groups). Used with proteolysis and mass spectrometry, methylation could also be used to define candidate residues for the mutagenesis approach to surface engineering (Derewenda and Vekilov, 2006). It has been reported that methylated proteins usually display reduced solubility (Schubot and Waugh, 2004). In our test cases the concentration required for crystallization, as judged by the use of the PCT, is reduced to between two thirds and one third of that for the unmodified samples. We also observed a shift to higher apparent molecular weight as determined by the size-exclusion clean-up step, and is in line with previous reports (Kurinov et al., 2000, Schubot and Waugh, 2004). In addition, in several cases, there was a shift from higher to lower order oligomeric states (Table 1).
Another effect on the biophysical properties of the proteins upon methylation is a reduction in the pI. In the three cases where methylation led to structure determination, a measurable reduction in pI was observed. Intriguingly, these observations correlate with data previously reported for proteins of Thermotoga maritima, where plotting predicted pI and average hydropathy reveals that the majority of the proteins whose structures were solved have calculated pIs below 7.0 (Canaves et al., 2004). It is possible that this in part reflects a bias in the commercial crystallization screens currently available; however, the trend has been seen with other proteomes (Au et al., 2006). An alternative explanation for the tendency for proteins with a high pI to fail to crystallize might be that they have a high concentration of surface lysines. These side chains are likely, through entropic effects, to destabilize the crystal lattice. Conversely, the hydrophobic nature of the dimethylated lysines will favor protein-protein interactions. In line with this, well-ordered lysines are found to be involved in crystal contacts in all three of the proteins solved, and it is well known that hydrophobic interactions of methylated lysines drive the formation of certain protein complexes, notably in the interaction of histone tails with chromodomains (Flanagan et al., 2005). The change in chemistry of the free amide groups affects other biophysical properties, reducing the solubility of proteins and sometimes changing the oligomeric state. Overall, the effects of methylation on the behavior of proteins in crystallization experiments are likely to be complex; nevertheless, the significant success rate we have observed in a set of test proteins has led us to adopt methylation as a simple generic rescue tool.
Acknowledgments
The authors would like to thank Maria Harkiolaki for assistance with IEF, Robin Aplin for mass spectrometry, and Louise Bird, Kamel El-Omari, and Erika Mancini for providing reagents. C.M. is supported by a Wellcome Trust Studentship, J.M.G. by the Royal Society, and D.I.S. and the OPPF by the UK MRC and European Commission, grant numbers QLG2-CT-2002-00988 (SPINE) and LSHG-CT-2004-511960 (VIZIER).
Published: November 14, 2006
References
- Adams M.W.W., Dailey H.A., Delucas L.J., Luo M., Prestegard J.H., Rose J.P., Wang B.C. The Southeast Collaboratory for Structural Genomics: a high-throughput gene to structure factory. Acc. Chem. Res. 2003;36:191–198. doi: 10.1021/ar0101382. [DOI] [PubMed] [Google Scholar]
- Albeck S., Alzari P., Andreini C., Banci L., Berry I.M., Bertini I., Cambillau C., Canard B., Carter L., Cohen S., et al. SPINE bioinformatics and data-management aspects of high-throughput structural biology. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1184–1195. doi: 10.1107/S090744490602991X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au K., Berrow N.S., Blagova E., Boucher I.W., Boyle M.P., Brannigan J.A., Carter L.G., Dierks T., Folkers G., Grenha R., et al. Application of high-throughput technologies to a structural proteomics-type analysis of Baccilus anthracis. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1267–1275. doi: 10.1107/S0907444906033555. [DOI] [PubMed] [Google Scholar]
- Brown J., Walter T.S., Carter L., Abrescia N.G.A., Aricescu A.R., Batuwangala T.D., Bird L.E., Brown N., Chamberlain P.P., Davis S.J., et al. A procedure for setting up high-throughput nanolitre crystallization experiments. II. Crystallization results. J. Appl. Crystallogr. 2003;36:315–318. [Google Scholar]
- Canaves J.M., Page R., Wilson I.A., Stevens R.C. Protein biophysical properties that correlate with crystallization success in Thermotoga maritima: maximum clustering strategy for structural genomics. J. Mol. Biol. 2004;344:977–991. doi: 10.1016/j.jmb.2004.09.076. [DOI] [PubMed] [Google Scholar]
- Derewenda Z.S., Vekilov P.G. Entropy and surface engineering in protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 2006;62:116–124. doi: 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. An RNA cap (nucleoside-2'-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf R.M., Ren J., Garman E.F., Somers D.O., Ross C.K., Jones E.Y., Stammers D.K., Stuart D.I. Continuous and discontinuous changes in the unit cell of HIV-1 reverse transcriptase crystals on dehydration. Acta Crystallogr. D Biol. Crystallogr. 1998;54:938–953. doi: 10.1107/s0907444998004284. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M., Hamer R., Sussman J.L., Silman I., Trudgian D., Yang Z.R., Prilusky J. Honing the in silico toolkit for detecting protein disorder. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1260–1266. doi: 10.1107/S0907444906033580. [DOI] [PubMed] [Google Scholar]
- Flanagan J.F., Mi L.Z., Chruszcz M., Cymborowski M., Clines K.L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Fogg M.J., Alzari P., Bahar M., Bertini I., Betton J.-M., Burmeister W.P., Cambillau C., Canard B., Carrondo M., Coll M., et al. Application of the use of high-throughput technologies to the determination of protein structures of bacterial and viral pathogens. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1196–1207. doi: 10.1107/S0907444906030915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerlof A., Brown J., Coutard B., Egloff M.-P., Enguita F.J., Fogg M.J., Gilbert R.J.C., Groves M.R., Haouz A., Nettleship J.E., et al. The impact of protein characterization in structural proteomics. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1125–1136. doi: 10.1107/S0907444906030307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras B., Edeling M.A., Byriel K.A., Jones A., Raina S., Martin J.L. Dehydration converts DsbG crystal diffraction from low to high resolution. Structure. 2003;11:139–145. doi: 10.1016/s0969-2126(03)00005-4. [DOI] [PubMed] [Google Scholar]
- Iwata S., Ostermeier C., Ludwig B., Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Kubota M., Matsuura Y. Crystallization and improvement of crystal quality for X-ray diffraction of maltooligosyl trehalose synthase by reductive methylation of lysine residues. Acta Crystallogr. D Biol. Crystallogr. 1999;55:931–933. doi: 10.1107/s0907444999002115. [DOI] [PubMed] [Google Scholar]
- Kurinov I.V., Mao C., Irvin J.D., Uckun F.M. X-ray crystallographic analysis of pokeweed antiviral protein-II after reductive methylation of lysine residues. Biochem. Biophys. Res. Commun. 2000;275:549–552. doi: 10.1006/bbrc.2000.3329. [DOI] [PubMed] [Google Scholar]
- Lawson D.M., Artymiuk P.J., Yewdall S.J., Smith J.M.A., Livingstone J.C., Treffry A., Luzzago A., Levi S., Arosio P., Cesareni G., et al. Solving the structure of human H-ferritin by genetically engineering intermolecular crystal contacts. Nature. 1991;349:541–544. doi: 10.1038/349541a0. [DOI] [PubMed] [Google Scholar]
- Lesley S.A., Kuhn P., Godzik A., Deacon A.M., Mathews I., Kreusch A., Spraggon G., Klock H.E., McMullan D., Shin T., et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. USA. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longenecker K.L., Garrard S.M., Sheffield P.J., Derewenda Z.S. Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI. Acta Crystallogr. D Biol. Crystallogr. 2001;57:679–688. doi: 10.1107/s0907444901003122. [DOI] [PubMed] [Google Scholar]
- Mateja A., Devedjiev Y., Krowarsch D., Longenecker K., Dauter Z., Otlewski J., Derewenda Z.S. The impact of Glu → Ala and Glu → Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1983–1991. doi: 10.1107/s090744490201394x. [DOI] [PubMed] [Google Scholar]
- McElroy H.E., Sisson G.W., Schoettlin W.E., Aust R.M., Villafranca J.E. Studies on engineering crystallizability by mutation of surface residues of human thymidylate synthase. J. Cryst. Growth. 1992;122:265–272. [Google Scholar]
- Means G.E., Feeney R.E. Reductive alkylation of amino groups in proteins. Biochemistry. 1968;7:2192–2201. doi: 10.1021/bi00846a023. [DOI] [PubMed] [Google Scholar]
- Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J., Grimes J.M., Stuart D.I. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 1997;276:171–179. [PubMed] [Google Scholar]
- Rayment I., Rypniewski W.R., Schmidtbase K., Smith R., Tomchick D.R., Benning M.M., Winkelmann D.A., Wesenberg G., Holden H.M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Ren J., Esnouf R., Garman E., Somers D., Ross C., Kirby I., Keeling J., Darby G., Jones Y., Stuart D., et al. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat. Struct. Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- Ren J.S., Sainsbury S., Berrow N.S., Alderton D., Nettleship J.E., Stammers D.K., Saunders N.J., Owens R.J. Crystal structure of nitrogen regulatory protein IIA(Ntr) from Neisseria meningitidis. BMC Struct. Biol. 2005;10:5–13. doi: 10.1186/1472-6807-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice R.H., Means G.E. Radioactive labeling of proteins in-vitro. J. Biol. Chem. 1971;246:831–832. [PubMed] [Google Scholar]
- Rypniewski W.R., Holden H.M., Rayment I. Structural consequences of reductive methylation of lysine residues in hen egg-white lysozyme: an X-ray analysis at 1.8-Å resolution. Biochemistry. 1993;32:9851–9858. doi: 10.1021/bi00088a041. [DOI] [PubMed] [Google Scholar]
- Saxena A.K., Singh K., Su H.P., Klein M.M., Stowers A.W., Saul A.J., Long C.A., Garboczi D.N. The essential mosquito-stage P25 and P28 proteins from Plasmodium form tile-like triangular prisms. Nat. Struct. Mol. Biol. 2006;13:90–91. doi: 10.1038/nsmb1024. [DOI] [PubMed] [Google Scholar]
- Schubot F.D., Waugh D.S. A pivotal role for reductive methylation in the de novo crystallization of a ternary complex composed of Yersinia pestis virulence factors YopN, SycN and YSc. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1981–1986. doi: 10.1107/S0907444904023005. [DOI] [PubMed] [Google Scholar]
- Siebold C., Berrow N., Walter T.S., Harlos K., Owens R.J., Stuart D.I., Terman J.R., Kolodkin A.L., Pasterkamp R.J., Jones E.Y. High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule. Proc. Natl. Acad. Sci. USA. 2005;102:16836–16841. doi: 10.1073/pnas.0504997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroh J.G., Loulakis P., Lanzetti A.J., Xie J.L. LC-mass spectrometry analysis of N- and C-terminal boundary sequences of polypeptide fragments by limited proteolysis. J. Am. Soc. Mass Spectrom. 2005;16:38–45. doi: 10.1016/j.jasms.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter T.S., Diprose J., Brown J., Pickford M., Owens R.J., Stuart D.I., Harlos K. A procedure for setting up high-throughput nanolitre crystallization experiments. I. Protocol design and validation. J. Appl. Crystallogr. 2003;36:308–314. [Google Scholar]
- Walter T.S., Diprose J.M., Mayo C.J., Siebold C., Pickford M.G., Carter L., Sutton G.C., Berrow N.S., Brown J., Berry I.M., et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.R., Thomson R., McNeil P., Esnouf R.M. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]