Dear editor,
Recently, an outbreak of unusual viral pneumonia in Wuhan city, China has sickened dozens of people. Preliminary studies indicated a novel coronavirus as the likely cause of the outbreak. Genetic recombination has been shown to contribute to the evolution of coronaviruses, including severe acute respiratory syndrome coronavirus (SARS-CoV) and the middle east respiratory syndrome coronavirus (MERS-CoV). Recent papers in this journal also described the involvement of recombination in viral evolution.1 , 2 Here I use ferret coronaviruses (FRCoVs) as an example to show recombination in coronaviruses.
Coronaviruses (CoVs) are enveloped, single positive-stranded RNA viruses that can infect a wide range of host species. CoV was first reported to infect ferret and associate with epizootic catarrhal enteritis in the United States in 2006, referred to ferret enteric coronavirus (FRECV).3 A fatal variant, ferret systemic coronavirus (FRSCV) caused feline infectious peritonitis-like disease in ferrets from Europe and the United States from 2002 to 2007.4 To date, FRCoVs have been detected in multiple countries, including the Netherlands, the United Kingdom, Spain, the United States, Peru and Japan.5 This study aim to assess the genetic diversity and potential role of genetic recombination in the evolutionary dynamics of FRCoVs.
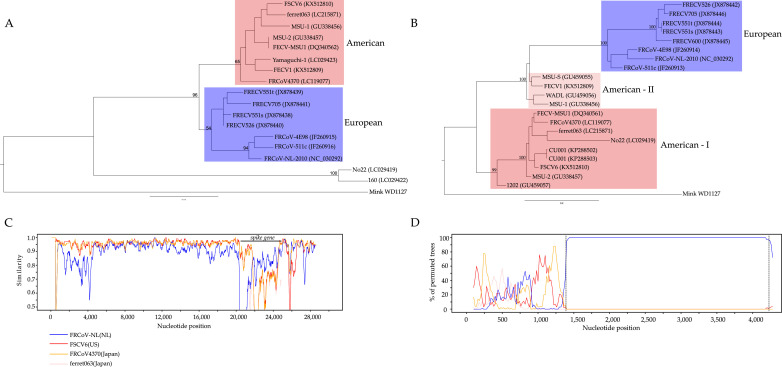
Genetic analyses were conducted with five complete genomes and 160 gene sequences of FRCoVs downloaded from the NIAID Virus Pathogen Database and Analysis Resource.6 These sequences were analysed in combination with 36 representative genomes of CoVs from other host species. Phylogenetic analysis of the complete genome confirmed the division of four genetic genera in CoVs. FRCoVs fall in alpha genera and most closely related to the CoV from mink (Fig. S1). FRCoVs from US and Japan (ferrets imported from US) were more closely related to each other than a Dutch strain. Phylogenetic tree for N gene supported two geographically dependent lineages, European and American (Fig. 1 A). The European lineage comprised FRCoVs from the Netherlands and Slovenia, while the American lineage comprised FRCoVs from US and Japan. Phylogenetic tree for RdRp gene showed that the American and Japanese strains comprised the American Lineage and are distant from the European lineage represented by the Dutch strain (Fig. S2). Different grouping was observed in the phylogenetic tree for S gene. The European lineage was consistent with that observed in the N tree, however, the American lineage was further divided into two sub-groups: American-I and American-II (Fig. 1B). The American-II sub-group comprised four FRCoVs from America, but was more closely related to European lineage rather than the American-I sub-group. Differences between the topologies of phylogenetic trees of S gene, complete genome, and N gene suggest the occurrence of potential recombination events (Fig. 1A and B).
Fig. 1.
Phylogenetic analyses for N gene (A), and S gene (B) of FRCoVs. FRCoVs from America and Europe are indicated by red and blue boxes, respectively. Two genetic groups identified for S gene of American FRCoVs are represented by American-I and American-II. Numbers at the nodes indicate bootstrap support evaluated by 500 replicates. Recombination analyses of S gene of FRCoVs (C and D). The FECV1 strain from America was used as the query and compared with four strains: FRCoV_NL_2010 (NL), FSCV6(US), ferret063(Japan), and FRCoV4370(Japan). (C) SimPlot shows the genetic distance between query sequence and each reference sequence in different parts of the genome. (D) Bootscan plot shows phylogenetic relationship between query sequence and reference sequences in different parts of the S gene. Potential region for recombination event is highlighted by dash lines.
SimPlot and Bootscan analyses of the five complete FRCoV genomes were performed to investigate the genetic variability in different parts of the genome and potential recombinations.7 The FECV1(US) strain was used as the query and compared with four strains: FRCoV_NL_2010(NL), FSCV6(US), ferret063(Japan), and FRCoV4370(Japan). Higher genetic variability was observed in the S gene, particularly at the 3′terminal end, compared to other parts of the genome (Fig. 1C). The average shared sequence identity was 91.0% for the complete genome, 93.1% for N gene, 95.0% for RdRp gene, and 73.6% for S gene, respectively. The 3′terminal end of S gene of FECV1(US) shared higher sequence identity with FRCoV_NL_2010(NL), despite the fact that FECV1(US) is more similar to FRCoVs from America in other parts of the genome. Bootscan analysis identified a potential recombination for the region between 1400 bp and 4260 bp in the S gene (Fig. 1D). Next, separate phylogenetic analyses were conducted for the recombination part and non-recombination part of the S gene. Phylogeny for the recombination part showed that American-II sub-group is closer to European lineage, whereas phylogeny for the non-recombination part showed that American-I and American-II sub-groups are both separated from European lineage (Fig. S3). Comparison of the putative transcription regulatory sequences (TRS) for these five strains showed identical core TRSs (Table 1 ). Taken together, these results suggest potential genetic recombination event at the 3′terminal end of S gene between FRCoVs from European and American lineages. However, due to relatively low sequence identity in the S gene and small number of available sequences, other possibilities cannot be excluded.
Table 1.
Comparison of putative transcription regulatory sequences (TRS) for ORF1ab, spike, membrane, and nucleocapsid genes between five ferret coronavirus (FRCoV) strains. The core TRS is indicated in bold. Accession number and origin for each strain: FRCoV-NL-2010 (NC_030292, the Netherlands), FECV1 (KX512809, the United States), FSCV6 (KX512810, the United States), Ferret063 (LC215871, Japan), and FRCoV4370 (LC119077, Japan).
| Strain | ORF1ab | Spike | Membrane | Nucleocapsid |
|---|---|---|---|---|
| NL-2010 | TCAACTAAACGAAA | GTTACTAAACTTTG | TCAACTAAACAAAATG | AGAACTAAACTTCTATTATG |
| FECV1 | ATTACTAAACTTTG | TCAACTAAACAAAATG | AGAACTAAACTTCTATTATG | |
| FSCV6 | ATTACTAAACTTTG | TCAACTAAACAAAATG | AGAACTAAACTTTTATCATG | |
| Ferret063 | TCAACTAAACGAAA | ATTACTAAACTTTG | TCAACTAAACAAAATG | AGAACTAAACTTTTATCATG |
| 4370 | TCAACTAAACGAAA | ATTACTAAACTTTG | TCAACTAAACAAAATG | AGAACTAAACTTCTATCATG |
This study focused on the S gene, and recombination could happen in other regions of the genome. Earlier studies have identified recombinations in the 3c and envelop genes.8 , 9 The N and ORF 7b genes of two Japanese strains, No.22 and No.160, were completely different from those from other FRCoV strains.9 The phylogeny of the N gene in this study supported the observation; phylogeny of the RdRp gene also showed that these two strains are distant from other strains (Fig. 1 and Fig. S2). The sequence identities in the RdRp gene between these two strains and other FRCoVs are relatively low (87.5%). While the source of these two Japanese strains are not clear, potential recombination, including recombination with other CoVs, could have contributed to the uniqueness of their genomes.
A critical question yet to be answered is the molecular basis for pathotype switch between less pathogenic FRECV and pathogenic FRSCV. Recombination in the S gene has been suggested to associate with the transmissibility and virulence of coronaviruses.10 It is reasonable to propose that FRSCV may have evolved from FRECV through recombination.8 , 9 Although this study cannot establish a direct link between the detected recombination and change of pathogenicity, it is interesting to see that 3 out of 4 strains are FRSCV, including MSU-S, WADL, and MSU-1 (Fig. 1B). Future in vivo experiments are needed to clarify the precise biological implications of this recombination in S gene. One major limit of this study is the small number of FRCoV sequences available in the public databases. Considering the wide spread of FRCoV and extensive use of ferret as an animal model for influenza pathogenicity study, enhanced surveillance is required to monitor the spread and genetic diversity of FRCoV.
Declaration of Competing Interest
The author declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.01.016.
Appendix. Supplementary materials
References
- 1.Chen X., Guo F., Pan J., Shen X., Li Y., Irwin D.M. Rare homologous recombination in H3N2 avian influenza a viruses. J Infect. 2019 doi: 10.1016/j.jinf.2019.11.010. Elsevier. [DOI] [PubMed] [Google Scholar]
- 2.Li X., Xiao K., Zhang Z., Yang J., Wang R., Shen X. The recombination hot spots and genetic diversity of the genomes of African swine fever viruses. J Infect. 2020;80:121–142. doi: 10.1016/j.jinf.2019.08.007. Elsevier. [DOI] [PubMed] [Google Scholar]
- 3.Wise A.G., Kiupel M., Maes R.K. Molecular characterization of a novel coronavirus associated with epizootic catarrhal enteritis (ECE) in ferrets. Virology. 2006;349:164–174. doi: 10.1016/j.virol.2006.01.031. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garner M.M., Ramsell K., Morera N., Juan-Sallés C., Jiménez J., Ardiaca M. Clinicopathologic features of a systemic coronavirus-associated disease resembling feline infectious peritonitis in the domestic ferret (Mustela putorius) Vet Pathol. 2008;45:236–246. doi: 10.1354/vp.45-2-236. SAGE Publications Sage CA: Los Angeles, CA. [DOI] [PubMed] [Google Scholar]
- 5.Provacia L.B.V., Smits S.L., Martina B.E., Raj V.S., vd Doel P., Amerongen G.v. Enteric coronavirus in ferrets, the Netherlands. Emerg Infect Dis. 2011;17:1570. doi: 10.3201/eid1708.110115. Centers for Disease Control and Prevention. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V. ViPR: an open bioinformatics database and analysis resource for virology research. Nucl Acids Res. 2011;40:D593–D598. doi: 10.1093/nar/gkr859. Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol Am Soc Microbiol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamers M.M., Smits S.L., Hundie G.B., Provacia L.B., Koopmans M., Osterhaus A.D.M.E. Naturally occurring recombination in ferret coronaviruses revealed by complete genome characterization. J Gen Virol Microbiol Soc. 2016;97:2180–2186. doi: 10.1099/jgv.0.000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minami S., Kuroda Y., Terada Y., Yonemitsu K., Van Nguyen D., Kuwata R. Detection of novel ferret coronaviruses and evidence of recombination among ferret coronaviruses. Virus Genes. 2016;52:858–862. doi: 10.1007/s11262-016-1365-3. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.