Abstract
Transmissible gastroenteritis coronavirus (TGEV) contains eight overlapping genes that are expressed from a 3′-coterminal nested set of leader-containing mRNAs. To facilitate the genetic manipulation of the viral genome, genes were separated by duplication of transcription regulating sequences (TRSs) and introduction of unique restriction endonuclease sites at the 5′ end of each gene using an infectious cDNA clone. The recombinant TGEV (rTGEV) replicated in cell culture with similar efficiency to the wild-type virus and stably maintained the modifications introduced into the genome. In contrast, the rTGEV replication level in the lungs and gut of infected piglets and virulence were significantly reduced. rTGEV in which gene 7 expression was abrogated (rTGEV-Δ7) were recovered from cDNA constructs, indicating that TGEV gene 7 was a nonessential gene for virus replication. Interestingly, in vivo infections with rTGEV-Δ7 showed an additional reduction in virus replication in the lung and gut, and in virulence, indicating that TGEV gene 7 influences virus pathogenesis.
Introduction
Transmissible gastroenteritis coronavirus (TGEV) is a member of the Coronaviridae family, which, together with the Arteriviridae and Roniviridae families forms the Nidovirales order Cowley and Walker 2002, Enjuanes et al 2000a, Mayo 2002. TGEV replicates in both the villous epithelial cells of the small intestine and the lung cells of newborn piglets, resulting in a mortality of nearly 100% (Saif and Wesley, 1992). The TGEV genome contains a leader sequence at the 5′ end and a poly(A) tail at the 3′ end. Genes are arranged in the order 5′-Rep-S-3a-3b-E-M-N-7-3′ Enjuanes et al 2000b, Penzes et al 2001. The 3′ end of the majority of TGEV genes overlaps with the 5′ terminus of the next gene (Enjuanes et al., 2000b), complicating insertion of heterologous genes into the viral genome and deletion of different genes to determine whether they are essential.
The TGEV gene 7, located at the 3′ end of the genome, encodes a 78 amino acid hydrophobic protein that may play a role in membrane-associated replication complexes or in virion assembly (Tung et al., 1992). Gene 7 is a group-specific gene (de Haan et al., 2002) with homologous versions in group 1 coronaviruses such as feline infectious peritonitis virus (FCoV) and canine enteric coronavirus (CCoV) Enjuanes et al 2000a, Lai and Cavanagh 1997. In contrast, group 2 coronaviruses such as mouse hepatitis virus (MHV), bovine enteric coronavirus (BCoV), or the human coronavirus (HCoV) OC43, and group 3 coronaviruses such as avian infectious bronchitis virus (IBV), do not have a homologous gene 7 Enjuanes et al 2000a, Lai and Cavanagh 1997. Interestingly, the group 1 coronavirus HCoV-229E does not have gene 7 (Herold et al., 1993), which could indicate that this gene is nonessential for coronavirus replication, even in group 1 coronaviruses.
To study whether gene 7 is dispensable for TGEV replication, its deletion by reverse genetics would be required. We used a genomic TGEV cDNA clone assembled as a bacterial artificial chromosome (BAC) Almazán et al 2000, González et al 2002 to separate the overlapping genes by duplicating sequences at the 5′ flank of each gene and by introducing unique restriction endonuclease sites between each gene pair. Gene separation allowed the deletion of gene 7, showing that it is nonessential for virus replication. Furthermore, we show that the accumulation of modifications in gene domains where the TRSs are located and insertion of restriction sites led to the generation of a collection of recombinant TGEVs (rTGEVs) with variable virulence, including a highly attenuated recombinant. These viruses could be the basis for coronavirus vector development.
Results
Generation of rTGEV viruses with nonoverlapping genes separated by unique restriction endonuclease sites
To facilitate genetic manipulation of the viral genome, full-length cDNA clones were constructed by separating the contiguous genes and inserting unique restriction sites between each gene pair (Fig. 1A). cDNA clones with the wild-type sequence or containing one [pBAC-TGEV-PacI (P), pBAC-TGEV-MluI (M), pBAC-TGEV-FseI (F), and pBAC-TGEV-AscI (A)], two [pBAC-TGEV-FseI-PmeI (F-Pm), pBAC-TGEV-PmeI-AscI (Pm-A), and pBAC-TGEV-PacI-MluI (P-M)], three [pBAC-TGEV-FseI-PmeI-AscI (F-Pm-A)], or five [pBAC-TGEV-PacI-MluI-FseI-PmeI-AscI (RS)] restriction endonuclease sites (Fig. 1B) were transfected into BHK cells expressing the porcine amino peptidase N (pAPN) (BHK-pAPN cells). On the third day of transfection, a cytopathic effect was observed in cells transfected with each cDNA, but not in mock-treated cells. Virus production was amplified by passing the supernatants four times in cultured cells. After the fourth passage, viruses were cloned by three plaque-isolation steps, and their genomes were partially sequenced. All the rTGEV viruses conserved the modifications engineered in the cDNAs (data not shown), indicating that the ORF separation and the insertion of unique endonuclease restriction sites between genes were stably maintained in the rTGEV genomes.
Fig. 1.
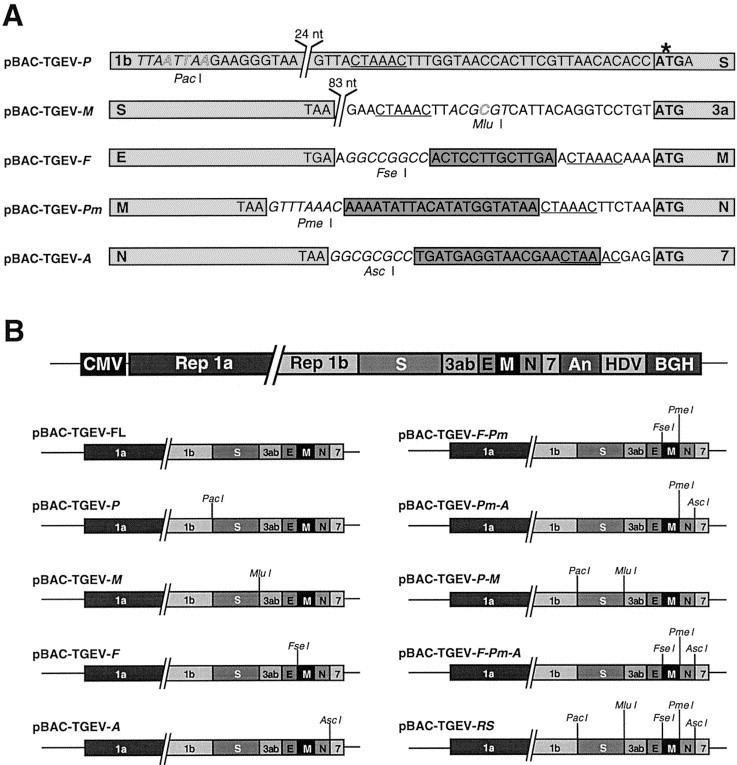
Scheme of the strategy used to separate TGEV overlapping genes. (A) Structure of the 5′ end of TGEV genes after gene separation by duplication of sequences partially overlapping with the transcription-regulating sequences and introduction of unique restriction sites. Unique endonuclease restriction sites are indicated in italics. Outlined sequences indicate the punctual mutations introduced to generate unique restriction sites. The core sequence (CS) is underlined. The ATG start codon is shown in bold. Duplicated sequences are indicated by dark boxes. TGEV genes are indicated by light boxes. 1b, replicase 1b gene; S, spike gene; E, envelope gene; M, membrane gene; N, nucleoprotein gene. *, Gene Rep 1b termination codon (TGA) and the initiation codon of gene S (ATG) partially overlap. The sequence of the 24 and 83 nt located 5′ upstream of genes S and 3a, respectively, are described in the full-length TGEV genome sequence previously reported (Penzes et al., 2001). (B) Schematic illustration of the full-length TGEV cDNA without (top bar) or with one (pBAC-TGEV-P, pBAC-TGEV-M, pBAC-TGEV-F, and pBAC-TGEV-A), two (pBAC-TGEV-F-Pm, pBAC-TGEV-Pm-A, and pBAC-TGEV-P-M), three (pBAC-TGEV-F-Pm-A), or five (pBAC-TGEV-RS) restriction endonuclease sites. CMV, cytomegalovirus immediate-early promoter; Rep, replicase; An, poly(A) tail of 24 A residues; HDV, hepatitis delta virus ribozyme; BGH, bovine growth hormone termination and polyadenylation sequences.
Cloned rTGEV viruses containing the unique restriction sites showed similar growth kinetics in cell culture to the parental rTGEV-wt after infection at both low (0.05) and high (5) m.o.i. (Fig. 2), indicating that removal of the overlapping region between TGEV genes and the insertion of endonuclease restriction sites did not significantly affect the in vitro virus replication.
Fig. 2.
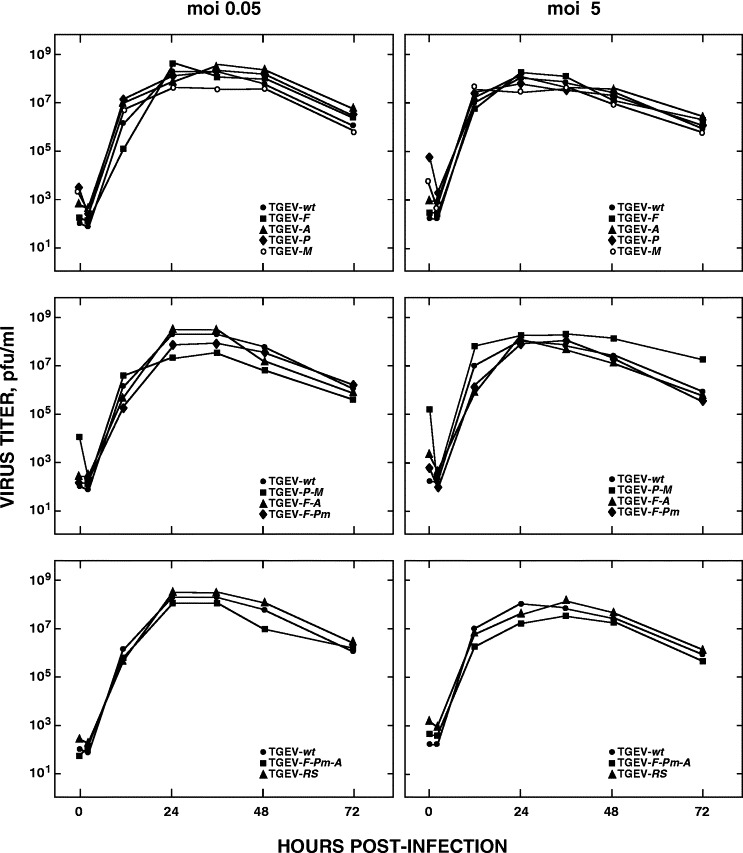
Growth kinetics of rTGEV viruses in cell culture. Growth kinetics of TGEV-wt, TGEV-F, TGEV-A, TGEV-P, TGEV-M, TGEV-P-M, TGEV-F-A, TGEV-F-Pm, TGEV-F-Pm-A, and TGEV-RS in BHK-pAPN cells after infection at low (0.05) and high (5) multiplicities of infection. Mean values of three experiments are indicated.
Rescue of infectious rTGEV-Δ7 virus from the cDNA
To analyze whether gene 7 was essential for viral growth, recombinant virus genomes with gene 7 deleted were generated from pBAC-TGEV-RS constructs, containing either the S gene from the TGEV strain PUR-C11 (Sánchez et al., 1999) able to infect both the enteric and the respiratory tracts (pBAC-TGEV-SC11-RS-Δ7) or the S gene from the strain PTV (Sánchez et al., 1999) with an exclusive respiratory tropism (pBAC-TGEV-Sptv-RS-Δ7). The rTGEV-Δ7 contained a deletion spanning 21 nucleotides upstream of the ORF 7 AUG and the first 17 nucleotides of this ORF (Fig. 3A). BHK-pAPN cells were transfected with plasmids including five restriction endonuclease sites and carrying gene 7 (pBAC-TGEV-Sptv-RS and pBAC-TGEV-SC11-RS), or without this gene (pBAC-TGEV-Sptv-RS-Δ7 and pBAC-TGEV-SC11-RS-Δ7). Virus titers were determined in supernatants throughout four additional passages in cell culture (Fig. 3B and C). Viruses were recovered from the cDNAs containing the deletion of gene 7, and viral titers increased with passage, basically to the same extent as the viruses with gene 7 (rTGEV-Sptv-RS and rTGEV-SC11-RS). As expected, no virus was recovered from the mock-transfected cultures.
Fig. 3.
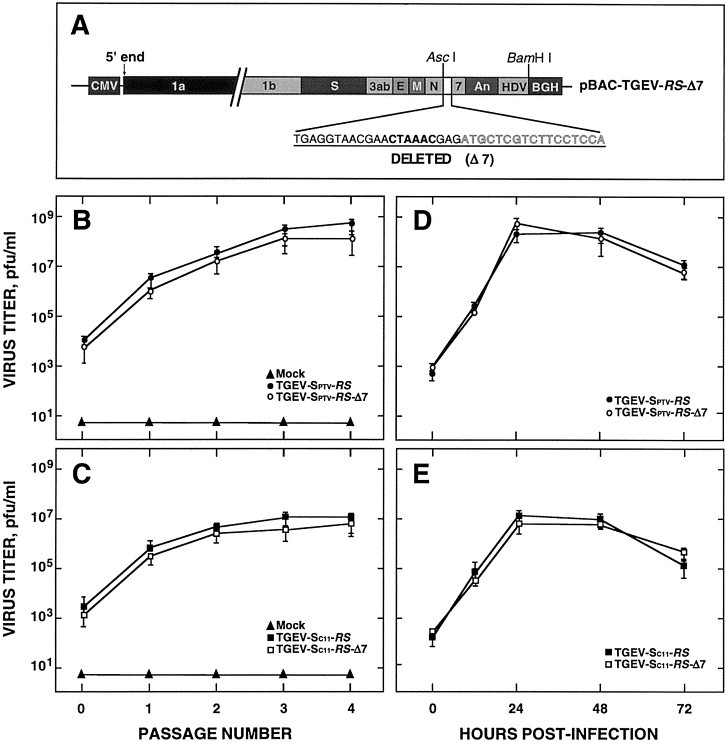
Rescue and growth kinetics of rTGEV-Δ7 from cDNA in BHK-pAPN cells. (A) Scheme of pBAC-TGEV-RS-Δ7 coding for rTGEV-RS-Δ7 virus. The deletion Δ7 virus that includes 21 nt of TRS-7 and the first 17 nt of ORF-7 (outlined letters) is underlined. The core sequence is shown in bold. Acronyms as in Fig 1. (B) and (C) Virus titers of TGEV-Sptv-RS and TGEV-Sptv-RS-Δ7 (B) and TGEV-SC11-RS and TGEV-SC11-RS-Δ7 (C) recovered from BHK-pAPN cells transfected with plasmids pBAC-TGEV-Sptv-RS, pBAC-TGEV-Sptv-RS-Δ7, pBAC-TGEV-SC11-RS, and pBAC-TGEV-SC11-RS-Δ7, respectively. The recovered virus was amplified throughout the indicated passages, and the culture supernatants were titrated on ST cells. Mock, mock transfected cells. (D) and (E) Growth kinetics of cloned TGEV-Sptv-RS-Δ7 (D) and TGEV-SC11-RS-Δ7 (E) in cell culture, after infection at an m.o.i. of 5. Aliquots were collected at the indicated times and titrated on ST cells. Error bars represent standard deviations of the mean from four experiments.
After four passages in cell cultures, the recombinant viruses were cloned by three plaque isolation steps. The cytopathic effect and plaque morphology produced by the rTGEV-Sptv-RS-Δ7 and rTGEV-SC11-RS-Δ7 were identical to those of the parental viruses containing the complete genome (data not shown). The isolate rTGEV-SC11-RS-Δ7 induced the formation of large-size plaques (3-mm-diameter), whereas the virus rTGEV-Sptv-RS-Δ7 induced small-sized plaques (1-mm-diameter). The cloned viruses with gene 7 deleted showed standard growth kinetics after infection at an m.o.i. of 5 (Fig. 3D and E). Recombinant rTGEV-Sptv-RS-Δ7 and rTGEV-SC11-RS-Δ7 generated the highest virus titers (around 6 × 108 and 107 PFU/ml, respectively) at 24 h postinfection, similar to those of the parental viruses rTGEV-Sptv-RS and rTGEV-SC11-RS. These data indicated that the protein encoded by gene 7 was not essential for TGEV replication in cell culture.
To confirm that the subgenomic mRNA (sgmRNA) 7 was not transcribed, BHK-pAPN cells were infected with rTGEV-Sptv-RS or rTGEV-Sptv-RS-Δ7 viruses. Total RNA was extracted and analyzed by Northern blot with a probe complementary to the 3′ end of the TGEV genome (Fig. 4A). The mobility and relative amount of the sgmRNAs 2, 3, 4, 5, and 6 were indistinguishable in both viruses. As expected, sgmRNA 7 was transcribed in rTGEV-Sptv-RS-infected cells, but not in cells infected with rTGEV-Sptv-RS-Δ7. Analysis of viral proteins at 16 h p.i. by Western blot showed that the amount of S, N, M, and E proteins was similar in rTGEV-Sptv-RS-Δ7-infected cells and in cells infected with the parental virus rTGEV-Sptv-RS, except for protein 7 that was not detected in rTGEV-Sptv-RS-Δ7 virus-infected cells (Fig. 4B), confirming that the partial deletion of gene 7 prevented the synthesis of protein 7. Identical results were obtained with rTGEV-SC11-RS and rTGEV-SC11-RS-Δ7 (data not shown).
Fig. 4.
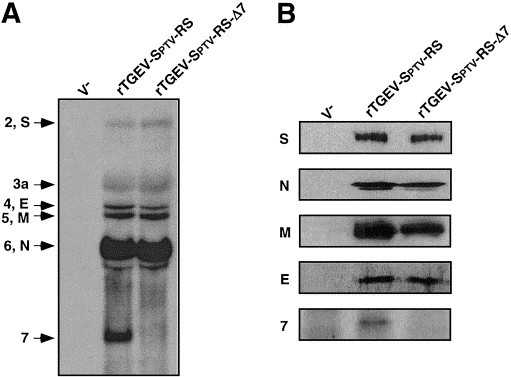
Characterization of mRNAs and proteins of rTGEV-Δ7. (A) Northern blot analysis of viral mRNAs. Total RNA was isolated from BHK-pAPN cells infected with either rTGEVSptv-RS or rTGEV-Sptv-RS-Δ7. V−, noninfected BHK-pAPN cells. The position of viral mRNAs is indicated to the left. (B) Western blot analysis of lysates from BHK-pAPN cells infected with either rTGEV-Sptv-RS or rTGEV-Sptv-RS-Δ7 viruses. Cell extracts were obtained at 16 h p.i., resolved by 5 to 20% gradient SDS–PAGE, transferred to nitrocellulose membranes, and immunoblotted with monoclonal antibodies specific for S, N, M, and E, and with an antiserum specific for a protein 7 peptide (Garwes et al., 1989).
Replication of rTGEV-RS and rTGEV-RS-Δ7 viruses in swine
In vivo growth of a selected set of rTGEVs containing the unique endonuclease restriction sites, and the rTGEV-RS-Δ7 viruses, was determined by infecting newborn piglets. The animals were sacrificed at 1, 2, 3, and 4 days p.i. Recombinant viruses with a modification including a restriction endonuclease site 5′ upstream of some genes, in general, showed lower titers than the wild-type virus, although there was some titer variability over the 4 days postinfection (Fig. 5A and B). Alteration in domains 5′ upstream of two or more genes did not lead to a significant decrease in virus titer in comparison with recombinants with a single modification.
Fig. 5.
In vivo growth kinetics of rTGEV viruses. Two- to three-day-old non-colostrum-deprived swine were used to study the growth kinetics of rTGEV viruses containing one (A and D) or more (B and E) endonuclease restriction sites, or partial deletion of gene 7 (C and F). Groups of 12 to 20 piglets were oronasally (2 × 108 PFU/pig) and intragastrically (3 × 108 PFU/pig) inoculated. Virus titers at the indicated number of days postinoculation were determined in the indicated tissue extracts. The whole organs were homogenized to obtain representative samples. Error bars represent standard deviations of the mean from four experiments. (G) Number of surviving piglets at different days postinoculation. Results from a representative experiment of two that gave similar results are shown. Recombinant virus titers were compared with that of the wild-type virus by the Kruskal–Wallis test and, in general, were significant (P < 0.05) between viruses with gene 7 deleted and the wild-type virus.
Interestingly, analysis of viral growth in the gut of infected piglets showed a 100- to 5000-fold reduction of recombinant viruses containing one or more restriction sites in relation to the rTGEV-wt virus (Fig. 5D and E). The rTGEV-RS, that included modifications 5′ upstream of five genes and insertion of endonuclease restriction sites between each contiguous gene, showed a titer decrease comparable with that of rTGEVs with two (TGEV-F-A) or three (TGEV-F-Pm-A) restriction endonuclease sites. These data show that modification of sequences 5′ upstream of genes affected virus replication in the gut. Recombinant viruses were isolated from the gut and sequenced. All the modifications introduced were stably maintained in the TGEV genome during in vivo infections (data not shown).
The growth in lungs of gene 7 deletion mutants (rTGEV-Sptv-RS-Δ7 and rTGEV-SC11-RS Δ7) showed around 100-fold reduction of virus titers compared with the parental viruses rTGEVSptv-RS and rTGEV-SC11-RS (Fig. 5C). In contrast, in vivo growth of rTGEV-SC11-RS-Δ7 in the gut showed slightly lower replication levels than rTGEV-SC11-RS (5 × 104 PFU/g) probably because the introduction of modifications at the 5′ upstream of five genes had already caused a significant titer reduction (Fig. 5F). As expected, the respiratory recombinants rTGEV-Sptv-RS and rTGEV-Sptv-RS-Δ7 did not grow in the gut since the S gene was derived from the respiratory PTV strain.
Piglet survival after infection by rTGEV viruses was compared with survival after infection with their virulent parental virus TGEV-PUR46-C11 (TGEV-SC11-wt). rTGEV viruses with one, two, three, or five unique restriction sites were highly attenuated (they produced mild enteritis and led to 80 to 90% survival at 5 days p.i.), except for rTGEV-P and rTGEV-M viruses, in which the restriction sites PacI and MluI were introduced by point mutations, without introducing TRS duplication. In these two recombinant viruses the survival was from 0 to 20%. Piglets infected by rTGEV-SC11-RS-Δ7 showed 100% survival, although a very mild and transitory enteritis was observed in 50% of the animals. These data indicate that gene 7 deletion further reduced virus virulence. In general, a good correlation between growth of recombinants with a single restriction endonuclease insertion and virulence was not clear. This could be due to differences in the distribution of viral antigens and inflammatory responses in pigs infected with wild-type or each mutant. Nevertheless, in viruses with two or more restriction endonuclease sites inserted there was a good correlation between virus titers in the gut and mortality.
Discussion
TGEV genomes with all the genes separated by unique endonuclease restriction sites have been engineered. The separation of TGEV genes implied the duplication of sequences 5′ upstream of each gene, a sequence domain involved in regulating transcription, and affected in vivo virus growth and virulence. In this article, the first demonstration that TGEV gene 7 is nonessential for virus viability is provided. In addition, it has been demonstrated that gene 7 deletion affects TGEV replication and attenuates virus virulence in piglets, its natural host. Interestingly, the introduction of one or more modifications into the TGEV genome has led to the generation of a collection of TGEV recombinants with a variable degree of virulence.
Overlapping of genes has been proposed as a mechanism by which nidoviruses preserve the genetic integrity of vital parts of their genomes (de Vries et al., 2000). Nevertheless, in coronavirus we have generated viable and stable rTGEV viruses in which genes have been separated by the insertion of unique restriction endonuclease sites and modification of the domains where the transcription-regulating sequences (TRSs) map. Maximum titers of mutant rTGEV viruses in cell culture were similar to those obtained for the parental virus, indicating that changes in the sequences between TGEV genes had little effect on viral yield. In contrast, in vivo assays showed lower TGEV mutant virus replication in the lungs and gut, and attenuation of virulence in viruses in which a TRS duplication was introduced between genes. In arteriviruses, mutants that had the overlap between ORFs 4 and 5, or between ORFs 5 and 6 removed, were also viable (de Vries et al., 2000). Taken together, these data demonstrate that gene overlapping is not an obligatory requirement for Nidovirales viability.
Viral attenuation resulting from gene separation was possibly due to modification of the TRSs affecting mRNA transcription levels. In the case of nonsegmented negative-strand RNA viruses it has also been shown that sequence alterations, such as restriction endonuclease site or heterologous gene insertion, and gene rearrangement may affect virus replication (Wertz et al., 2002).
Gene 7 is a group 1 specific gene, absent in genome of coronaviruses from groups 2 and 3 Armstrong et al 1983, Boursnell et al 1985, Kamahora et al 1989, Lapps et al 1987, Skinner and Siddell 1985. Interestingly, no mortality was observed in piglets infected with rTGEV-Δ7. Since these recombinants still replicate with titers between 103 and 105 PFU/g of tissue (Fig. 5C and F), TGEV-Δ7 mutants could be good candidates as virus vectors.
A relationship between gene 7 and virulence has also been observed in the FCoVs. The most 3′ end genes of these viruses are genes 7a and 7b. Deletions in FCoV ORF 7a, which is homologous to the TGEV ORF 7, have been reported in a natural infection of a cat population and correlated with a decrease in virulence (Kennedy et al., 2001). Similarly, mutations in FCoV ORF 7b occur in vitro and have also been correlated with loss of virus virulence (Herrewegh et al., 1995). Therefore, the most 3′ end genes of group 1 coronaviruses seem to influence in general virus pathogenesis.
Deletion of gene 7 did not affect virus replication in cell culture. Therefore, the reduction of in vivo virus replication and virulence was probably due to an effect on virus–host interaction. It has been suggested that coronavirus group-specific genes, such as gene 7, may affect host immune response (de Haan et al., 2002). It would be of interest to determine whether the immune response to a heterologous gene inserted in recombinant viruses with and without gene 7 is influenced by the presence of this gene.
In an attempt to identify gene 7 homologous protein sequences or motifs, a sequence search was performed in the databases using gene 7 sequences without success. The high hydrophobicity of TGEV protein 7 (Garwes et al., 1989) could facilitate its insertion in membranes providing a role in virus replication, since coronavirus replication complexes have been associated with membranes Denison and Sims 2001, Snijder et al 2001. Similarly, the tentative location of protein 7 within the nucleus (Garwes et al., 1989) could be taken as an indication for a possible interference with the cell cycle, similarly to coronavirus nucleoprotein that seems to interact with cell nucleolus proteins and interfere with cell cycle (Hiscox, 2002).
Therefore, genes 3a, 3b, and 7 of group 1 coronaviruses are nonessential for replication. Similarly, genes 2a/HE and 4/5a, and possibly gene E, are dispensable in MHV de Haan et al 2002, Kuo and Masters 2002. Manipulation of transcription-regulating sequences and deletion of nonessential genes, such as gene 7, will facilitate the study of the molecular basis of viral attenuation and provide an attractive approach to generate attenuated rTGEVs with high potential as virus vectors.
Materials and methods
Virus and cells
Baby hamster kidney cells (BHK-21) stably transformed with the gene coding for the porcine aminopeptidase N (BHK-pAPN) (Delmas et al., 1994) were grown in DMEM supplemented with 2% fetal calf serum and Geneticin G418 (1.5 mg/ml) as a selection agent. BHK-pAPN cells were used for all the experiments except for standard virus titrations that were performed in porcine swine testis (ST) cells (McClurkin and Norman, 1966).
Virus titers were compared by the Kruskal–Wallis test (Motulsky, 1995) and, when significant (P < 0.05), it was indicated. rTGEV viruses were generated from pBAC-TGEV constructs containing the S gene from the virulent TGEV strain PUR-C11 (SC11) as described Almazán et al 2000, González et al 2002. Viruses containing the S gene from the attenuated strain PTV (Sptv) were derived from the corresponding pBAC TGEV vectors with gene SC11 by replacing this gene by that (Sptv) of the respiratory strain.
Construction of modified rTGEV genomes
Two different approaches were followed to introduce into pBAC-TGEV unique restriction endonuclease sites separating each gene of TGEV genome (Fig. 1A), leading to pBAC-TGEV-RS. The first approach was the introduction of punctual mutations in the TGEV genome to generate the restriction sites PacI (between genes Rep 1b and S) and MluI (between genes S and 3a) (Fig. 1B). Due to overlapping in the TGEV genome, the second approach involved the duplication of 13, 22, and 19 nucleotides from the 5′ transcription-regulating sequences of genes M, N, and 7 (TRS-M, TRS-N, and TRS-7) after the restriction sites FseI, PmeI, and AscI, respectively (Fig. 1A and B). Point mutations, duplications, and insertion of restriction endonuclease sites were generated by overlapping PCR amplification from the TGEV genome as described (Ortego et al., 2002). The assembly of the full-length pBAC-TGEV constructs was performed as previously reported Almazán et al 2000, González et al 2002.
To generate the deletion Δ7, oligonucleotide primers 5′-Asc-7.17-VS (5′-GAGGCGCGCCTGCTGTATTTATTACAG-3′), including the restriction site AscI (underlined) and the deletion of 21 nucleotides from TRS-7 plus the first 17 nucleotides of ORF 7, and BGH34-RS (5′-CAGATGGCTGGCAACTAGAAGGC-3′) were used to generate a PCR product comprising from nt 28,094 to nt 28,764 of the TGEV cDNA clone. PCR product was digested with AscI and BamHI and cloned into the AscI-BamHI-digested pBAC-TGEV-SC11-RS and pBAC-TGEV-Sptv-RS, generating pBAC-TGEV-SC11-RS-Δ7 and pBAC-TGEV-Sptv-RS-Δ7, respectively.
Transfection and recovery of infectious TGEV from cDNA clones
BHK-pAPN cells were grown to 60% confluence on 35-mm-diameter plates and transfected with 10 μg of cDNA plasmid and 15 μg of Lipofectin (Life Technologies, GIBCO) according to the manufacturer’s specifications. Recovery and amplification of viruses were performed as described (Ortego et al., 2002).
Western and Northern blot analysis
Cell lysates were analyzed by 5 to 20% gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Proteins were transferred to a nitrocellulose membrane and analyzed as described (Ortego et al., 2002), using mAbs specific for S (5B.H1), N (3D.C10), M (9D.B4), and E (V27) proteins (Ortego et al., 2002), and a swine polyclonal antibody specific for TGEV protein 7 (provided by P. Britton, Compton, UK). Total RNA was extracted by using an Ultraspec RNA isolation system (Biotecx) according to the manufacturer’s instructions and analyzed by Northern blotting as described (Ortego et al., 2002).
In vivo growth kinetics of rTGEV viruses
Two- to three-day-old non-colostrum-deprived swine, from crossing Large White and Belgium Landrace, were used to study in vivo growth kinetics of rTGEV, as described (Sanchez et al., 1999). Piglets were obtained from sows seronegative for TGEV, as tested by radioimmunoassay.
Acknowledgements
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT), La Consejería de Educación y Cultura de la Comunidad de Madrid, Fort Dodge Veterinaria, and the European Union (Frame V, Key Action 2, Control of Infectious Disease Projects QLRT-1999-00002, QLRT-1999-30739, and QLRT-2000-00874). I.S. received postdoctoral fellowships from the Community of Madrid and the European Union (Frame V, Key Action 2, Control of Infectious Disease Projects).
References
- Almazán F., González J.M., Pénzes Z., Izeta A., Calvo E., Plana-Durán J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J., Smeekens S., Rottier P. Sequence of the nucleocapsid gene from murine coronavirus MHV-A59. Nucleic Acids Res. 1983;11:883–891. doi: 10.1093/nar/11.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E.G., Binns M.M., Foulds I.J., Brown T.D.K. Sequences of the nucleocapsid genes from two strains of avian infectious bronchitis virus. J. Gen. Virol. 1985;66:573–580. doi: 10.1099/0022-1317-66-3-573. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Walker P.J. The complete genome sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 2002;147:1977–1987. doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Masters P.S., Shen S., Weiss S., Rottier P.J.M. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A.A.F., Glaser A.L., Raamsman M.J.B., de Haan C.A.M., Sarnataro S., Godeke G.J., Rottier P.J.M. Genetic manipulation of equine arteritis virus using full-length cDNA clones: separation of overlapping genes and expression of a foreign epitope. Virology. 2000;270:84–97. doi: 10.1006/viro.2000.0245. [DOI] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Sjöström H., Noren O., Laude H. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 1994;342:293–298. doi: 10.1007/978-1-4615-2996-5_45. [DOI] [PubMed] [Google Scholar]
- Denison M.R., Sims A.C. MHV-A59 gene 1 proteins are associated with two distinct membrane populations. Adv. Exp. Med. Biol. 2001;494:655–661. doi: 10.1007/978-1-4615-1325-4_97. [DOI] [PubMed] [Google Scholar]
- Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P., Rottier P., Siddell S.G., Spaan W.J.M., Taguchi F., Talbot P. Coronaviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carsten E.B., Estes M.K., Lemon S.M., McGeoch D.J., Maniloff J., Mayo M.A., Pringle C.R., Wickner R.B., editors. Virus Taxonomy. Classification and Nomenclature of Viruses, Academic Press; New York: 2000. pp. 835–849. [Google Scholar]
- Enjuanes L., Spaan W., Snijder E., Cavanagh D. Nidovirales. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carsten E.B., Estes M.K., Lemon S.M., McGeoch D.J., Maniloff J., Mayo M.A., Pringle C.R., Wickner R.B., editors. Virus Taxonomy. Classification and Nomenclature of Viruses, Academic Press; New York: 2000. pp. 827–834. [Google Scholar]
- Garwes D.J., Stewart F., Britton P. The polypeptide of Mr 14000 of porcine transmissible gastroenteritis virus: gene assignment and intracellular location. J. Gen. Virol. 1989;70:2495–2499. doi: 10.1099/0022-1317-70-9-2495. [DOI] [PubMed] [Google Scholar]
- González J.M., Penzes Z., Almazán F., Calvo E., Enjuanes L. Stabilization of a full-length infectious cDNA clone of transmissible gastroenteritis coronavirus by the insertion of an intron. J. Virol. 2002;76:4655–4661. doi: 10.1128/JVI.76.9.4655-4661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Raabe T., Schelle-Prinz B., Siddell S.G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Vennema H., Horzinek M.C., Rottier P.J.M., Groot P.J. The molecular genetics of feline coronavirus comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology. 1995;212:622–631. doi: 10.1006/viro.1995.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A. The nucleolus—a gateway to viral infection? Arch. Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamahora T., Soe L.H., Lai M.M.C. Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res. 1989;12:1–9. doi: 10.1016/0168-1702(89)90048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M., Boedeker N., Gibbs P., Kania S. Deletions in the 7a ORF of feline coronavirus associated with an epidemic of feline infectious peritonitis. Vet. Microbiol. 2001;81:227–234. doi: 10.1016/S0378-1135(01)00354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76:4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapps W., Hogue B.G., Brian D.A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo M.A. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 2002;147:1655–1656. doi: 10.1007/s007050200039. [DOI] [PubMed] [Google Scholar]
- McClurkin A.W., Norman J.O. Studies on transmissible gastroenteritis of swine. II. Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can. J. Comp. Med. Vet. Sci. 1966;30:190–198. [PMC free article] [PubMed] [Google Scholar]
- Motulsky, H. (1995). Comparing two paired groups: paired t and Wilcoxon tests, in: Intuitive Biostatistics, Oxford Univ. Press, New York, pp. 225–229
- Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z., González J.M., Calvo E., Izeta A., Smerdou C., Mendez A., Sánchez C.M., Sola I., Almazán F., Enjuanes L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes. 2001;23:105–118. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J., Wesley R.D. Transmissible gastroenteritis. In: Leman A.D., Straw B.E., Mengeling W.L., D’Allaire S., Taylor D.J., editors. Diseases of Swine. Seventh ed. Wolfe Publishing Ltd; Ames, IA: 1992. pp. 362–386. [Google Scholar]
- Sánchez C.M., Izeta A., Sánchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Durán J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M.A., Siddell S.G. Coding sequence of coronavirus MHV-JHM mRNA 4. J. Gen. Virol. 1985;66:593–596. doi: 10.1099/0022-1317-66-3-593. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van Tol H., Roos N., Pedersen K.W. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 2001;82:985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- Tung F.Y.T., Abraham S., Sethna M., Hung S.L., Sethna P., Hogue B.G., Brian D.A. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz G.W., Moudy R., Ball L.A. Adding genes to the RNA genome of vesicular stomatitis virus: positional effects on stability of expression. J. Virol. 2002;76:7642–7650. doi: 10.1128/JVI.76.15.7642-7650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


