Abstract
Screening of the proteome of microdissected glutathione S-transferase placental form (GST-P) positive foci and normal-appearing liver on anionic (Q10), and cationic (CM10) surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) ProteinChip arrays demonstrated significant overexpression of cytokeratin 8 (CK8; m/z 54,020), cytokeratin 18 (CK18; m/z 47,760), microsomal cytochrome 5A (m/z 15,224) and histone type 2 H2aa3 (m/z 15,964) in the livers of rats initiated with diethylnitrosamine (DEN) followed by 10 weeks on phenobarbital (PB) at a dose of 500 ppm. Furthermore, formation of CK8 and CK18 complexes due to CK8 phosphorylation at Ser73 and Ser431 was found to be strongly associated with promotion of hepatocarcinogenesis by PB and the development of hepatocellular carcinomas. The data were confirmed by immunohistochemistry and real-time Q-PCR and profound overexpression of CK8 and CK18 (CK8/18) proteins and mRNAs were detected in several large size GST-P positive foci and liver tumors. A strong correlation between CK8/18 positive foci development and multiplicity of hepatocellular carcinomas was further observed. Moreover, elevation of CK8/18 was strongly associated with induction of cell proliferation in GST-P positive foci and tumors. In conclusion, our data imply that CK8/18 overexpression, those two cytokeratins complex formation associated with histone type 2 H2aa3 up-regulation and intermediate filament reorganization may drive neoplastic transformation of GST-P positive foci during rat hepatocarcinogenesis leading to the formation of hepatocellular carcinomas.
Keywords: Proteomics, GST-P positive foci, Hepatocarcinogenesis
Introduction
Hepatocellular carcinoma (HCC, the most common type of liver cancer) is the fifth most common cancer and the third leading cause of cancer-related mortality, with an increasing incidence worldwide (Lee et al., 2005). HCC commonly emerges on a background of chronic liver disease (Thorgeirsson and Grisham, 2002, El-Serag, 2004), with continuous rounds of necrosis and regeneration, inflammation, oxidative stress and genetic alterations, for example involving the p53 gene as suspected causative factors (Thorgeirsson and Grisham, 2002). However, the molecular mechanisms linked to lesion development and progression are still far from clear. It is known that profound changes at the cellular and subcellular level, involving RNA/DNA and protein structure and functions lead to liver cancer development and progression. Therefore, high-throughput, effective proteomic techniques targeting relevant biological molecules may provide novel insights into hepatocarcinogenesis and prognosis (Melle et al., 2007). In rat models of HCC, glutathione S-transferase placental form (GST-P) positive foci (GST-P+ foci), arise after initiation and are believed to be precursors for the later formation of tumors (Ito et al., 1998). However, only a very small proportion finally develops into hepatocellular adenomas and carcinomas, and the concrete sequence of events occurring during this transformation is unknown. Recently it has been proposed that tumor development is regulated by cancer stem cells, which have such properties as self-renewal, the ability to produce different progeny and increased expression of developmental signaling molecules (Banas et al., 2006). Identification of novel biomarkers of liver preneoplastic lesions and cancer stem cells and further elucidation of the signaling pathways that regulate their growth and survival may provide clues to novel therapeutic approaches to treat liver cancer.
It is now accepted that understanding of carcinogenesis and tumor progression on a molecular basis needs detailed studies of proteins as critical components and effector molecules of the multiple interconnected signaling pathways that drive the neoplastic phenotype. In this context, emerging proteomic analysis platforms, including mass spectrometry technologies and protein microarrays, represent powerful tools for study and understanding of cancer. It is clear that identification of biomarkers for the early diagnosis of the premalignant state should be one of the main goals in this emerging field (Alessandro et al., 2005). However, there has so far been only limited success in determining protein spectra of preneoplastic lesions mostly due to technological difficulties, and a new system is needed.
Laser-assisted microdissection is a recent technology that enables cells to be harvested from tissue sections (Banks et al., 1999). Proteins can then be extracted from the dissected cells for molecular analysis of proteins in specific cell in vivo. Although quantities of protein obtained from dissected material are generally small, it is possible to use new technology such as ProteinChip Array SELDI-TOF-MS (SELDI = surface-enhanced laser desorption ionization; BioRad, USA) specifically developed to allow profiling and identification of complex protein mixtures from a few cells (Gluckmann et al., 2007). These sensitive methods for proteome analysis have been used to discover novel biomarkers and multimarker panels relevant to diagnosis and patient stratification for Alzheimer's disease, Parkinson's disease, multiple sclerosis, schizophrenia, and HIV-induced dementia, for example (Reddy and Dalmasso, 2003).
In the present study, to analyze protein spectra of rat liver GST-P+ foci and identify new biomarkers, a combination of immunohistochemistry, laser microdissection, ProteinChip Array (SELDI), and MALDI TOF-MS/MS systems was employed.
Materials and methods
Chemicals
N-nitrosodiethylamine (DEN) was from Sakai Research Laboratory (Fukui, Japan). Phenobarbital (PB) sodium salt (CAS no. 57-30-7) (purity ≥ 98%) and other reagents were purchased from Wako Pure Chemicals Industries (Osaka, Japan) or Sigma (St. Louis, MO).
Animals
A total of 60, five-week-old male Fisher 344 rats (Charles River, Japan, Hino, Shiga, Japan) were quarantined for 1 week before the start of the experiment. They were housed in an animal facility maintained on a 12 h (7:00–19:00) light/dark cycle, at a constant temperature of 23 ± 1 °C and relative humidity of 44 ± 5%, and were given free access to tap water and food (Oriental MF pellet diet, Oriental Yeast Co., Tokyo, Japan).
Experimental design
The 60 F344 rats were divided into 2 experimental groups. After one week on basal diet all animals underwent intraperitoneal injection of DEN (100 mg/kg b.w.) dissolved in saline to initiate hepatocarcinogenesis. This was performed 3 times, once per week. After one further week on basal MF diet animals were administered diet containing PB at doses of 0 (control) or 500 ppm for 10 (10 rats per group) or 33 (20 rats per group) weeks. At experimental weeks 13 and 36 terminations were performed.
At termination animals were anesthetized with diethyl ether. At the week 13 time point, liver perfusion was performed in situ at 2 ml/min with ice-cold 1.15% KCl buffer (1.15% KCl, 1 mM EDTA, 0.25 mM PMSF) at room temperature (Kinoshita et al., 2002). Freezing fixation of liver tissue with OCT compound and isopentane was carried out for the ProteinChip Array (SELDI-TOF-MS), MALDI TOF-MS/MS (Bruker Daltonics), and analysis of mRNA expression by real-time quantitative PCR (Q-PCR). Separate portions were fixed in 10% phosphate-buffered formalin, partially frozen in liquid nitrogen and stored at − 80 °C for molecular assessment.
Immunohistochemical assessment of GST-P+ foci and histological examination of liver tumors
Immunohistochemical demonstration of GST-P+ foci and hematoxylin eosin (HE) staining for assessment of the incidence, multiplicity and histopathological classification of liver tumors were carried out at weeks 13 and 36, respectively (Kitano et al., 1998). For laser microdissection, GST-P+ foci were stained in frozen sections. The blocking step was omitted and sections were exposed to GST-P primary antibodies for 2 h at 4 °C.
Laser microdissection
For microdissection, frozen liver was embedded in TissueTek medium and sectioned on a standard cryostat (HM500-OM, Leica Microsystems GmbH, Walldorf, Germany). Ten serial frozen sections (8 μm) were cut and immediately fixed in methanol. The upper one section was stained with HE, and the remaining nine sections were used for immunohistochemistry for GST-P, performed as described above. For SELDI-TOF-MS ProteinChip analysis and protein identification by MALDI TOF-MS/MS, GST-P+ foci greater than 0.2 mm in diameter or surrounding normal-like liver areas were microdissected from cryosections using the PALM MicroBeam System with Laser Microdissection and Pressure Catapulting technology (LMPC) (P.A.L.M. Microlaser Technologies AG, Meiwa Shoji Co., Ltd., Germany) according to standard protocols (Emmert-Buck et al., 1996, Banks et al., 1999, Lehmann and Kreipe, 2006). Each LCM cap transferred about 3000 cells (∼ 5,000,000 μm2 area). To each LCM cap, 5 μl of 9 M Urea/2% CHAPS lysis buffer was added and microdissected samples were suspended. For SELDI ProteinChip Array at least 3000 cells (1 μg protein) of GST-P+ foci or normal-like liver were microdissected and subjected to analysis. For protein identification by the Peptide Mass Fingerprinting method and MALDI TOF-MS/MS, samples from approximately 75,000 cells (∼ 125,000,000 μm2 area, 30 μg protein) of GST-P+ foci and surrounding liver area of 10 rats were prepared. GST-P+ foci from the remaining two sections (2000–3000 cells) were subjected to real-time Q-PCR analysis.
SELDI ProteinChip array
Anion exchange (Q10), cation exchange (CM10) and metal ion (IMAC-Ni) SELDI ProteinChip arrays were used for protein capture (BioRad, USA). Q10 and CM10 arrays were incubated with 0.05 mol/L Tris–base (pH8) and 100 mM NaAc (pH4) binding buffers, respectively, for 5 min followed by the application of 5 μl of cell lysate to each spot, followed by binding at room temperature for 45 min. Spots were washed twice with binding buffer for 5 min and then rinsed with distilled water. All samples were run in duplicate.
SELDI-TOF-MS analysis
After the spots on the Q10 and CM10 arrays were air dried, 0.5 μl of a saturated solution of sinapinic acid (SPA) in 0.5% (v/v) trifluoroacetic acid was applied to each bait surface, allowed to air dry, and reapplied. Mass-to-charge (m/z) spectra of proteins were generated in a Protein Biology System IIc TOF-MS (PBS-IIc, BioRad, USA). External calibration was achieved using hirudin BHVK (7034 Da; BioRad), bovine cytochrome c (12,230 Da; BioRad), equine myoglobin (16,951 Da; BioRad), and bovine carbonic anhydrase (29,023 Da; BioRad) as standards. All array binding and SELDI-TOF-MS were conducted on the same day. To confirm the consistency of our assay, all samples were run in duplicate. Because estimates of the coefficient of variation (CV) based on samples of two observations could be inaccurate, we adopted the root-mean-square (RMS) method (Sauter et al., 2005). All of the duplicated spectra were compiled, and the protein peak intensities were normalized to the total ion current m/z values from 3000 to 150,000 Da using CiphergenExpress Data Manager 3.0 Software (BioRad).
To identify biomarkers predicting hepatocarcinogenesis, we compared GST-P+ foci (n = 10) with normal-appearing dissected liver area (n = 10). Logistic regression was used to compare each peak between two GST-P+ foci samples to determine the set of differentially expressed peaks. A P value of < 0.05 was considered significant.
Validation by immunohistochemistry
Differentially expressed proteins in GST-P+ foci of rats treated with DEN and DEN followed by PB were verified by immunohistochemistry. For staining in formalin-fixed paraffin embedded tissue sections, guinea pig polyclonal antibodies against CK8 and 18 (CK8/18) (dilution 1:600, Progen Biotechnik, Germany), anti-histone H2A antibodies (dilution 1:1500, Upstate Biotechnology Inc., USA), phospho-CK8 (pS73) rabbit monoclonal antibodies (dilution 1:300, Epitomics, CA), phospho-CK8 (pS23) rabbit monoclonal antibodies (dilution 1:100, Epitomics, CA) and phospho-CK8 (pS431) rabbit monoclonal antibodies (dilution 1:50, Cosmo Bio, USA) were used. In formalin-fixed sections microwave antigen retrieval was performed in citrate buffer (pH 6.0). DAB was used for antigen visualization.
Double staining for GST-P positive foci and CK8/18 was performed in formalin-fixed paraffin sections using the polyclonal guinea pig anti-rat CK8/18 antibody at 1:600 dilution and the polyclonal rabbit anti-rat GST-P primary (1:2000) (IgG, 100 μg/ml) antibody. First, immunohistochemical assessment of CK8/18 was carried out with the avidin–biotin complex method as described previously (Kinoshita et al., 2003). Thereafter sections were sequentially treated with 0.2 M glycine, pH 2.2, for 2 h to remove immune complexes and GST-P immunohistochemistry was performed. The GST-P-positive sites were demonstrated with alkaline phosphatase (Vectastain ABC-AP kit, Vector Red (SK-5100)) solution. GST-P positive foci were stained red, while brown/black staining showed a positive immunoreaction of primary antibody with CK8/18. Quantitation of CK8/18 positive foci was done using two-dimensional evaluation. The numbers and areas of foci greater than 0.2 mm in diameter, and total areas of liver sections, were measured using a color image processor (IPAP; Sumica Technos Osaka, Japan) to give values per cm2 of liver section. Quantitation of CK8/18 positive (CK8/18+) foci was done as described above for GST-P+ foci.
Double immunohistochemistry for CK8/18 and PCNA with an anti-PCNA mouse monoclonal (PC-10, IgG2a; DAKO, Kyoto, Japan; 1:500) antibody was performed with formalin-fixed sections as described above for GST-P and CK8/18 using alkaline phosphatase (Vectastain ABC-AP kit, Vector Red) solution for the immunohistochemical detection of CK8/18. First, immunohistochemistry for PCNA was performed with avidin-biotin complex method using DAB as a substrate to demonstrate sites of peroxidase binding for PCNA. Thereafter sections were sequentially treated with 0.2 M glycine, pH 2.2, for 2 h to remove immune complexes and CK8/18 immunohistochemistry was performed as described above. CK8/18-positive foci were stained red, while brown/black staining of nuclei showed a positive immunoreaction of monoclonal primary antibody with PCNA. The PCNA positive indices were estimated for CK8/18 positive areas and in background liver parenchyma as numbers of positive nuclei per 1000 cells.
RNA preparation
For RNA isolation from GST-P+ foci all solutions were prepared using diethyl pyrocarbonate-treated water. RNase free instruments and RnaseZap (Sigma, St Louis, MO) were employed. To compare gene mRNA expression in big and small foci, GST-P+ foci from 0.2 mm to 1 mm (small foci) and greater than 1 mm (big foci) in diameter were separately collected. Total RNA was isolated from microdissected GST-P+ foci in rat liver using 4 M guanidine thiocyanate, 25 mM sodium citrate with 0.5% sarcosyl buffer by the phenol–chloroform–isoamyl alcohol extraction method using glycogen as a carrier. Reverse transcription of total RNA was performed with Oligo-dT primer, and cDNA samples were stored at − 20 °C until assayed.
Validation of CK8 and CK18 mRNA expression by real-time Q-PCR
Products after PCR amplification were used for verification of CK8 and CK18 genes expression using real-time quantitative PCR. Q-PCR was performed using a TaqMan probes for 490 C. Primer sequences were designed with Primer Express software (Applied Biosystems, USA). The probes and primers were as follows: TaqMan probe TGGTGTCTGAGTCTTCTGA; primers: forward 5′ TTG AAA CCC GAG ATG GGA AA, reverse 5′ GGC CAT TCA CTT GGA CAT GAT (20 cycles of 95 °C, 59 °C and 72 °C 1 min each, product length 62 bp, for CK8 (NM_199370), TaqMan probe and primers set Rn01533363_g1 for CK18 (NM_053976.1) and TaqMan probe TGA GAC CTT CAA CAC CCC AGC CAT G, and primers: forward 5′ CCG TGA AAA GAT GAC CCA GAT C, reverse 5′ACC AGA GGC ATA CAG GGA CAA C for beta-actin, cytoplasmic (NM_007393). The cDNA product from each sample was used to perform Q-PCR according to the manufacturer's instructions with beta-actin mRNA as an internal control.
Statistical analysis
The significance of differences between mean values was analyzed using the Student's t-test and Fisher's PLSD method using the StatView–J 5.0 program (Berkeley, CA). For analysis of protein expression CiphergenExpress Data Manager 3.0 Software was employed. Correlations of potential biomarkers with GST-P+ foci status and other known histopathologic factors were examined using ANOVA and multivariate logistic regression. The significance of differences in lesion incidences between groups was assessed by the χ 2 test or the Fisher's exact probability test.
Results
General observations
No significant differences among the groups were observed with regard to food and water consumption or body weight gain (data not shown). Final liver/body weight ratios were increased significantly in the group administered PB at a dose of 500 ppm for 10 weeks (4.8 ± 0.6%, P < 0.0001) and 33 weeks (5.9 ± 1.6%, P < 0.0001) following DEN initiation as compared to values for the control group (3.5 ± 0.3%).
Profiling by SELDI Q10 and CM10 ProteinChip Arrays
For the clarification of protein biomarkers predictive of GST-P+ foci developing into tumors, we compared the proteome of microdissected GST-P+ foci and normal-appearing liver area from the livers of initiation control and 500 ppm PB-administered group. With the ProteinChip and Biomarker Wizard software, sets of 80 and 60 identified peak clusters were analyzed in Q10 and CM10 arrays, respectively.
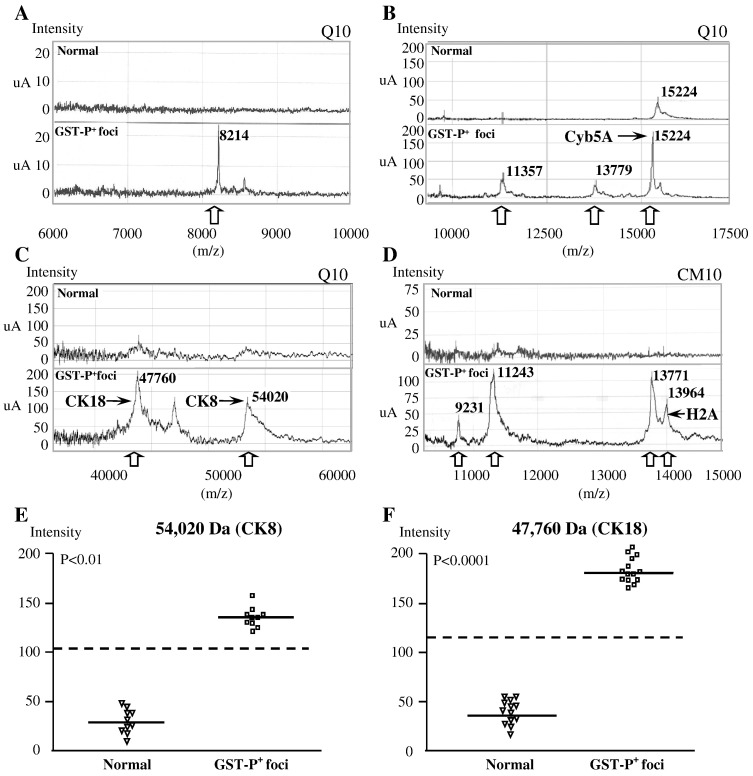
On Q10 Arrays, m/z 15,224 (P < 0.01), 47,760 (P < 0.001) and 54,020 (P < 0.0001) peaks were found to be significantly elevated in PB-promoted rat liver GST-P+ foci compared to normal-appearing liver areas (Table 1 , Figs. 1 A–C). These three proteins were identified by the PeptideMass Fingerprinting Method to be microsomal cytochrome A5 (Cyb5A; pI 4.6), cytokeratin 18 (CK18; pI 5.8) and cytokeratin 8 (CK8; pI 5.8) of rat origin, respectively. Furthermore, m/z 8214, 11,357, and 13,779 peaks were increased, albeit without significance (Table 1, Figs. 1A and B). In GST-P+ foci of DEN-initiated rats, a trend for increase was observed for m/z 8214, 11,357 and 54,020 (CK8) peaks as compared to the surrounding normal liver area (Table 1).
Table 1.
Peaks with altered expression detected by SELDI ProteinChip Array in GST-P positive foci.
| Peaks (m/z) | GST-P+ foci/DEN | Normal/DEN | P value | GST-P+ foci/DEN → PB | Normal/DEN → PB | P value |
|---|---|---|---|---|---|---|
| Q10 | ||||||
| 8214 | 36.0 ± 5.66 | 13.5 ± 6.12 | 0.06 | 21.67 ± 8.51 | 10.3 ± 6.65 | 0.21 |
| 11,357 | 48.5 ± 4.95 | 21.0 ± 11.31 | 0.09 | 64.07 ± 43.86 | 24.0 ± 22.63 | 0.17 |
| 13,780 | 23.33 ± 17.10 | 12.67 ± 6.08 | 0.34 | 32.50 ± 14.25 | 13.33 ± 7.05 | 0.1 |
| 15,190 (CYB5A) | 15.5 ± 0.71 | 11.06 ± 5.66 | 0.34 | 57.57 ± 16.49 | 19.57 ± 11.42 | 0.018 |
| 47,761 (CK18) | 42.55 ± 4.74 | 38.55 ± 3.16 | 0.43 | 181.27 ± 12.05 | 38.67 ± 5.51 | 0.00005 |
| 54,020 (CK8) | 33.35 ± 3.32 | 22.40 ± 3.92 | 0.06 | 136.27 ± 35.25 | 28.30 ± 11.05 | 0.0072 |
| CM10 | ||||||
| 6904 | 23. 1 ± 4.24 | 22.55 ± 3.61 | 0.90 | 107.65 ± 59.47 | 21.61 ± 9.33 | 0.18 |
| 8126 | 44.3 ± 16.97 | 90.1 ± 37.19 | 0.12 | 82.87 ± 30.67 | 24.47 ± 9.03 | 0.03 |
| 9231 | 15.0 ± 8.19 | 16.8 ± 11.52 | 0.82 | 97.87 ± 55.08 | 16.43 ± 8.36 | 0.03 |
| 11,243 | 79.93 ± 17.58 | 58.87 ± 10.10 | 0.15 | 130.97 ± 18.56 | 52.57 ± 25.74 | 0.01 |
| 13,771 | 85.33 ± 33.87 | 50.73 ± 27.88 | 0.24 | 138.07 ± 28.49 | 44.6 ± 10.61 | 0.01 |
| 13,964 (HistH2aa3) | 23.33 ± 17.10 | 12.67 ± 2.08 | 0.34 | 61.50 ± 10.65 | 13.33 ± 1.45 | 0.021 |
Fig. 1.
Representative findings from Q10 and CM10 ProteinChip Array showing overexpression of 8214, 11,357, 13,779, 15,224, 46,760 and 54,020 m/z peaks (Q10) and 9231, 11,243, 13,771 and 13,964 m/z peaks (CM10) in GST-P+ foci of rats treated with DEN followed by 500 ppm PB (A–D). Single marker analysis demonstrates significant up-regulation of 46,760 and 54,020 Da proteins in GST-P+ foci (E, F).
With CM10 arrays, m/z 8126, 9231, 11,243, 13,771 and 13,964 peaks were significantly overexpressed in the GST-P+ foci of rats treated with PB (Table 1, Fig. 1D). The 13,964 Da peak (P < 0.05) was further identified as Histone type 2 H2aa3 (His H2aa3, pI 11.55). Furthermore, non-significant increases of m/z 13,771 and 13,964 peaks (His H2aa3) in the DEN initiation control group and m/z 6904 peak in PB-treated liver GST-P+ foci were detected. Our data show that the reproducibility (CV) of protein detection using SELDI-TOF-MS is acceptable (Nakagawa et al., 2006).
Single marker (univariate) analysis demonstrated that m/z 47,760 (P < 0.01) and m/z 54,020 (P < 0.00012) peaks on the Q10 Array, most profoundly overexpressed in GST-P+ foci of PB-treated rats, might be biomarkers of rat liver preneoplastic lesions associated with promotion of rat hepatocarcinogenesis by PB (Figs. 1E and F).
Validation of CK8/18 expression by IHC
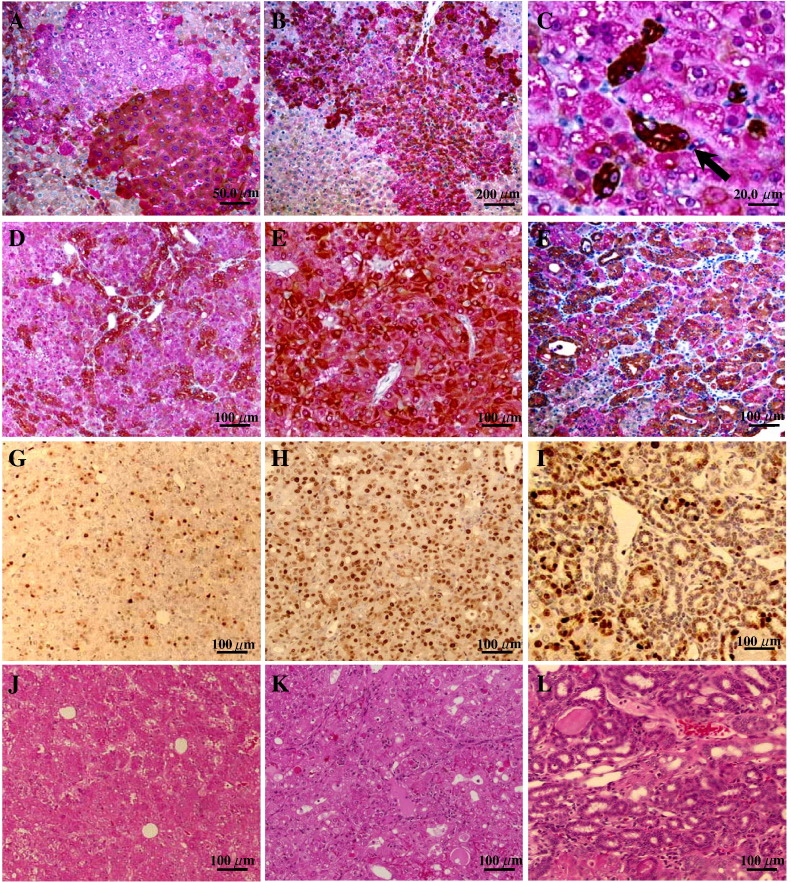
CK8/18 expression was verified in GST-P+ foci of rats treated with DEN alone and DEN followed by PB by single and double immunohistochemistry. In Fig. 2 , representative pictures of double staining for CK8/18 and GST-P for GST-P+ foci (Figs. 2A–C), and results of immunohistochemical examination of CK8/18 and GST-P, PCNA and HE staining in serial sections for hepatocellular adenomas (HAs) (Figs. 2D, G and J), HCCs (Figs. 2E, H and K) or mixed hepatocellular and chollangiocarcinomas (Figs. 2F, I and L) in rats initiated with DEN and promoted by PB are presented. Significant overexpression of CK8/18 with a homogenous staining pattern (CK8/18 positive cytoplasmic staining in almost all hepatocytes within GST-P positive foci) was detected in several large GST-P+ foci in the livers of rats administered PB (Figs. 2A and B). Furthermore, in both initiation control and PB-promoted groups some GST-P+ foci demonstrated a mosaic staining pattern. Moreover, in some GST-P+ foci, islands consisting of cells with a low nuclear/cytoplasm (N/C) ratio and very high expression of CK8/18 were observed (Fig. 2C). Elevation of CK8/18 was coordinated with GST-P overexpression in HAs, HCCs and mixed hepatocellular and chollangiocarcinomas (Figs. 2D–F). Basophilic chollangio-regions of mixed type carcinomas were highly positive for CK8/18 (Fig. 2I).
Fig. 2.
Double immunohistochemistry for GST-P (red) and CK8/18 (brown), and immunohistochemical assessment of PCNA in the livers of F344 rats treated with PB after DEN initiation. CK8/18 and GST-P double staining in rat liver preneoplastic lesions (A–C). Note strong homogenous overexpression of CK8/18 in GST-P+ foci (A and B) and several highly CK8/18+ cells with a low N/C ratio (C, arrow). CK8/18 and GST-P double staining in HAs, HCCs and mixed hepatocellular and chollangiocarcinomas, respectively (D–F), with immunohistochemical assessment of PCNA (G, H, F) and HE staining (J, K, L) in the same tumors.
Evaluation of CK8/18, GST-P+ foci and histological examination of liver tumors
Data on the effects of PB application on formation of CK8/18+ foci at week 13 and liver tumors at week 36 in experiment 1 are shown in Table 2 .
Table 2.
Quantitative data for GST-P positive foci, CK8/18+ foci and liver tumors in rats undergoing initiation with DEN, with or without subsequent PB treatment.
| No. of rats | DENa | No. of rats | DEN → PB | |
|---|---|---|---|---|
| GST-P+ foci (≥ 0.2 mm) | ||||
| Number (no./cm2) | 10 | 128.04 ± 16.51 | 10 | 161.91 ± 36.59b |
| Area (mm2/cm2) | 1.41 ± 0.54 | 3.08 ± 0.92d | ||
| CK8/18+ foci | ||||
| Number (No./cm2) | 10 | 1.82 ± 0.42 | 10 | 4.17 ± 1.41b |
| Area (mm2/cm2) | 0.26 ± 0.07 | 0.70 ± 0.15c | ||
| Liver tumors | ||||
| Incidence (%) | ||||
| Adenoma | 20 | 20(100) | 20 | 20(100) |
| HCC | 19(95) | 20(100) | ||
| Multiplicity (No./rat) | ||||
| Adenoma | 3.65 ± 1.90 | 4.10 ± 2.86 | ||
| HCC | 3.00 ± 1.95 | 9.60 ± 3.15d | ||
| Total | 6.65 ± 2.64 | 13.7 ± 3.53d | ||
aData are mean ± SD.
b–dSignificantly different from the DEN initiation control group.
bP < 0.01; cP < 0.001; dP < 0.0001.
After 10 weeks of continuous PB administration to rats initiated with DEN, significant increase in number and area of CK8/18+ foci in line with the increase of numbers and areas of GST-P+ foci was found in the group treated with PB at a dose of 500 ppm as compared to the DEN initiation control group (Table 2, Figs. 2A and B). All of the observed CK8/18+ foci were mostly large size and positivite for GST-P. On the other hand, many GST-P+ foci negative for CK8/18 were detected. The number and area of GST-P+ foci in both DEN initiation control and PB-promoted rat livers were much higher than those for CK8/18+ foci (Table 2). Histological examination of liver tumors at week 36 demonstrated a significant induction of HCC (including mixed hepatocellular and cholangiocarcinoma) (P < 0.0001) in concordance with elevation of CK8/18+ foci number and area at week 13 (Table 2). Most tumors developing in animals administered 500 ppm of PB were well differentiated HCCs.
Prediction of the development of preneoplastic lesions overexpressing CK8 and 18 into tumors
The CK8/18 protein expression and the development of CK8/18+ foci at experimental week 36 was significantly associated with development of HCCs in the rats treated with 500 ppm PB for 33 weeks after the DEN initiation (overall test, P = 0.0017 and P = 0.0005, respectively).
Overexpression of phospho-cytokeratin 8 in GST-P+ foci
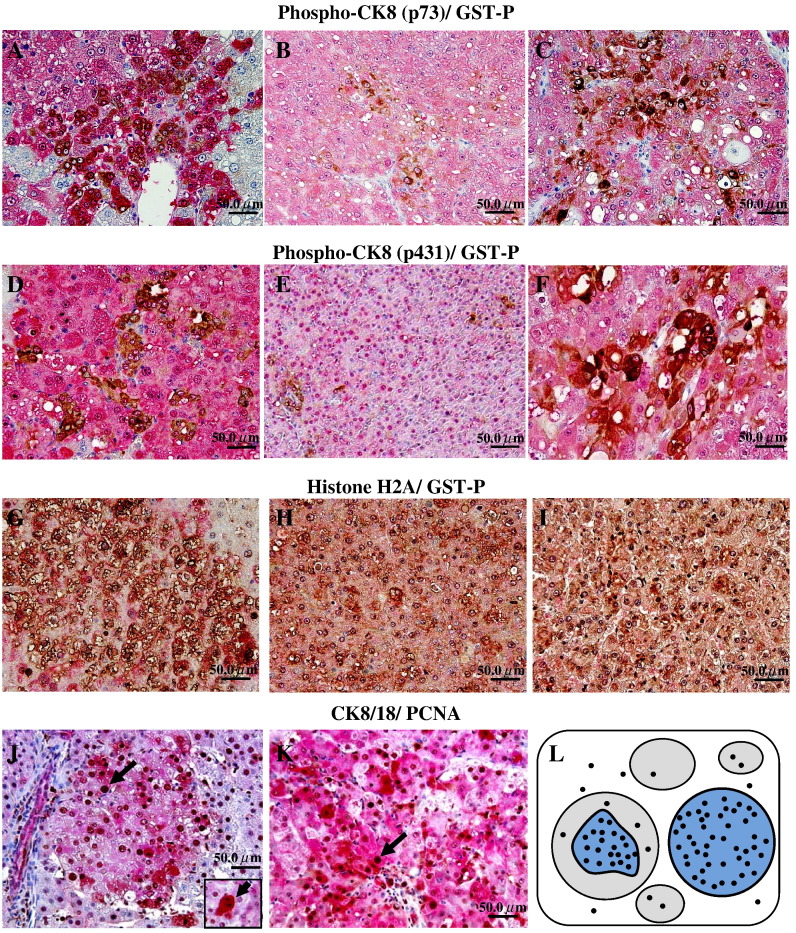
Double and single staining for GST-P and/or CK8 phosphorylated by Ser23 (p23), Ser73 (p73) or Ser431 (p431) demonstrated elevation of p73 and p431 in both GST-P+ foci and HCCs, and in some of HAs (Figs. 3 A–F). GST-P+ foci were negative for p23, but in several HCCs p23-positive regions were observed (data not shown). Cells positive for p73 were small, with a low N/C ratio (Fig. 3C). On the other hand, cells positive for p431 formed glandular structures in both GST-P+ foci and HCCs (Figs. 3D and F). HCCs with a glandular pattern were highly positive for p431. Furthermore, both p73 and p431 were up-regulated in cholangio-regions of combined hepatocellular and cholangiocarcinomas demonstrating the same pattern as observed in case of double staining for CK8/18 and GST-P (data not shown).
Fig. 3.
Double immunohistochemistry for phospho-CK8 p73 (A–C) or p431 (D–F) (brown) and GST-P (red) in GST-P+ foci, HAs and HCCs; histone H2A (brown) and GST-P (red) (G–I) and immunohistochemical assessment of CK8/18 (red) and PCNA (black) (J and K) in the livers of F344 rats treated with PB after DEN initiation. Elevation of PCNA positive cells number was observed in CK8/18+ area within totally or partially stained GST-P+ foci (L). Note high expression of CK8/18 in mitotic cells in GST-P positive focus (J, arrow).
Association of CK8/18 overexpression with induction of cell proliferation
Next, we examined whether the expression of CK8/18 might be associated with induction of cell proliferation in rat liver preneoplastic and neoplastic lesions. Double staining of CK8/18 and PCNA revealed an increase in number of positively stained cells within the areas of CK8/18+ foci in DEN control (14.9 ± 5.1/100 cells, P < 0.05) and to a great extent PB-treated rat livers (26.3 ± 7.6 per 100 cells, P < 0.01), as compared with normal-like areas (DEN: 0.41 ± 0.13 per 100 cells; DEN → PB: 0.43 ± 0.18 per 100 cells). Homogenously stained large CK8/18+ foci contained many PCNA positive nuclei (Figs. 3J–L). Furthermore, the PCNA index for CK8/18+ foci was significantly higher in the livers of rats treated with PB (P < 0.05) compared to the foci in the DEN control group (14.9 ± 5.1 per 100 cells).
Analysis by double immunohistochemistry for CK8/18 and PCNA demonstrated significant increase of cell proliferation in the area of CK8/18+ hepatocellular adenomas (35.2 ± 9.6 per 100 cells, P < 0.01) and hepatocellular and hepatochollangiocarcinomas (42.3 ± 13.8 per 100 cells, P < 0.01) (Figs. 2G–I). Furthermore, extensive staining for CK8/18 was found in hepatocytes undergoing mitotis (Fig. 3A).
Up-regulation of CK8 and CK18 mRNA in areas of GST-P+ foci
The results of real-time Q-PCR analysis of differentially expressed CK8 and CK18 in the area of GST-P+ foci and normal liver, obtained after 10 weeks of PB administration are presented in Table 3 . Reproducibility and specificity of the data were confirmed by repeating the analysis two times. In line with the SELDI ProteinChip Array results, significant overexpression of CK8 and slight elevation of CK18 mRNA, was found in the areas of microdissected (∼ 100 cell) GST-P+ foci of animals in the group treated with DEN followed by PB. Furthermore, when we compared the levels of CK8 and CK18 mRNAs in large (> 100 cell), and small (< 100 cell) foci of PB-administered rats, expression was significantly elevated in the large foci (CK8: 3010.96 ± 1700.10 (P < 0.01) and CK18:1800.08 ± 120.22 (P < 0.05)) as compared to the expression in the normal-appearing area (CK8:447.95 ± 84.85 and CK18: 15.03 ± 4.34). Observed no clear concordance between CK18 protein and mRNA expression was possibly due to the heterogeneity of microdissected GST-P positive foci in terms of CK8/18 expression and because of their restricted number used in the analysis by real-time Q-PCR.
Table 3.
mRNA expression of CK8 and CK18 in GST-P positive foci and normal-appearing liver tissue of rats.
| Gene | DEN |
DEN → PB |
||
|---|---|---|---|---|
| GST-P+ foci | Normal | GST-P+ foci | Normal | |
| CK8 | 121.86 ± 65.28 | 83.45 ± 20.22 | 1924.96 ± 1589.73 | 447.95 ± 84.85 |
| CK18 | 15.26 ± 3.01 | 15.34 ± 1.61 | 18.08 ± 8.93 | 15.03 ± 4.34 |
GST-P+ foci, GST-P positive foci; Normal, normal-appearing liver area.
Discussion
High-throughput, proteomic techniques targeting biological molecules should provide novel insights into hepatocarcinogenesis and prognosis. This is the first report characterizing protein profiles of GST-P+ foci using ProteinChip technology (SELDI), able to detect minute amounts of proteins and moreover to analyze complex patterns. Screening of proteomes on anionic (Q10), and cationic (CM10) ProteinChip arrays here demonstrated significant overexpression of intermediate filament members CK8 and CK18, microsomal cytochrome 5A and histone type 2 H2aa3 in the livers of rats initiated with DEN and promoted with PB. Furthermore, from the CM10 array results, the m/z 8126, 9231, 11,243, and 13,771 peaks were significantly overexpressed in GST-P+ foci of rats treated with PB. Trends for increase of m/z 8214, 11,357 peaks and CK8 on the Q10 array and m/z 13,771 and His H2aa3 on the CM10 array were also observed in the DEN initiation control group. Single marker analysis showed CK8 and CK18 to be profoundly overexpressed in GST-P+ foci of PB-treated rats, thus presenting as promising biomarkers. The data of immunohistochemistry and real-time Q-PCR supported the results of SELDI ProteinChip array, demonstrating concordance between induction of CK8 and CK18 proteins and their mRNAs. Furthermore, a correlation of CK8/18+ foci development and multiplicity of HCCs was observed. The results thus extended earlier findings from proteomic profiling in microdissected HCC tissue (Melle et al., 2004).
In line with our results, overexpression of CK8/18 in human HCCs has been demonstrated by immunohistochemistry previously (Johnson et al., 1988, Athanassiadou et al., 2007). Furthermore, proteomes of human HCCs were recently examined using SELDI ProteinChip and LC–MS/MS techniques (Melle et al., 2004, Melle et al., 2007, Chaerkady and Pandey, 2008). Surprisingly, the latest results for microdissected human HCCs using iTRAQ quantification in combination with LC–MS/MS are controversial regarding expression of CK8 and CK18 (Chaerkady and Pandey, 2008). However, in this study the MS/MS data were not supported by Western Blot or the immunohistochemical examination. Furthermore, the controversial results might be due to the specificity of protein spectrum of HCC used for the examination.
CK8 and CK18 are known to be distributed in cytoplasmic filament networks and as bands associated with the plasma membrane (Abe and Oshima, 1990). A number of in vitro experiments and transgenic mouse model studies have shown that CK8/18 carry out essential functions in protecting hepatocytes from a variety of stresses such as toxins (e.g., griseofulvin, acetaminophen and cadmium) (Omary et al., 2002, Lau and Chiu, 2007). CK8 and CK18 mutations are thus reported as risk factors for developing liver diseases of multiple etiologies, such as biliary atresia and viral hepatitis (Ku et al., 2003). One specific mutation of CK8 (G61C) was found to be a genetic risk factor for the development of cryptogenic liver cirrhosis (Schoniger-Hekele et al., 2006). A CK8 mutation was also found in another series of cryptogenic cirrhosis cases in association with particular autoimmune features (Ku et al., 2001). In humans, CK18 mutation can also predispose to cryptogenic cirrhosis (Ku et al., 1997, Ku et al., 2001). Furthermore, transgenic mice expressing a CK18 conserved arginine mutant were shown to be susceptible to chronic hepatitis, with increased levels of soluble phosphoglycokeratins (Ku et al., 1995). It has been further demonstrated that cells transfected with human CK8 and CK18 exhibit enhanced migratory and invasive potential (Chu et al., 1993). Therefore, keratins may play important roles in migration, through interactions with the extracellular environment and influencing cell shape (Chu et al., 1993).
In the present study, CK8 phosphorylation at Ser73 and Ser431 and overexpression of phosphorylated forms in GST-P+ foci was found to be strongly associated with promotion of hepatocarcinogenesis by PB and the development of hepatocellular carcinomas. It has been previously reported that CK8 and CK18 are able to form complexes due to CK8 phosphorylation (Toivola et al., 1998, Ku et al., 2002, Nousiainen et al., 2006). Dynamic phosphorylation is one of the mechanisms regulating more than 20 keratin type I and II intermediate filament proteins in epithelial cells. The major type II keratin in “simple type” glandular epithelia is CK8. Biochemical and mutational approaches have been used to localize the two major in vivo phosphorylation sites of human CK8 to the head (Ser23) and tail (Ser-431) domains. CK8 Ser73 occurs within a relatively conserved type II keratin motif ((68)NQSLLSPL) and becomes phosphorylated in cultured cells and organs during mitosis, cell stress, and apoptosis (Nousiainen et al., 2006). Furthermore, Ser73 phosphorylation is reported to play an important role in keratin filament reorganization (Toivola et al., 1998, Ku et al., 2002). CK8 was also suggested to be a new cytoplasmic target for c-Jun N-terminal kinase (JNK) in Fas receptor-mediated signaling, and has been shown to be exclusively phosphorylated in vitro by p38 mitogen-activated protein kinase (MAPK) (He et al., 2002, Ku et al., 2002). In another study, it was demonstrated that shear stress, but not stretch, causes disassembly of keratin intermediate filaments in lung alveolar epithelial cells and that this disassembly is regulated by protein kinase C delta (PKCδ)-mediated phosphorylation of CK8 Ser73 (Ridge et al., 2005). The functional significance of these phosphorylations could relate to regulation of JNK signaling, p38 MAPK or PKCδ signaling and/or regulation of keratin dynamics (He et al., 2002, Ku et al., 2002). On the other hand, CK8 Ser431 phosphorylation was found to occur after EGF stimulation and during mitotic arrest and is likely to be mediated by MAPK and cdc2 kinase (Ku and Omary, 1997). A monoclonal antibody that specifically recognizes phosphoserine 431-CK8 manifests increased reactivity with CK8 and recognizes reorganized CK8/18 filaments after EGF stimulation. In vivo, the specific serine sites 73 and 431 of CK8 phosphorylated by p38, influence Mallory body formation (Nan et al., 2006). On the other hand, CK18, when phosphorylated, plays a role in filament reorganization and together with CK8, is involved in interleukin-6 (IL-6)-mediated barrier protection (Wang et al., 2007).
Our results indicated that elevation of CK8/18 is strongly associated with induction of cell proliferation in GST-P+ foci and tumors. Recently, CK8 and CK18 were reported to bind to the cytoplasmic domain of tumor necrosis factor receptor 2 (TNFR2) and moderate tumor necrosis factor (TNF)-induced JNK intracellular signaling and NFkappa B activation. This moderation of the effects of TNF, a cytokine produced by macrophages and T lymphocytes affecting cellular proliferation, differentiation, survival, and cell death, may be the fundamental function of CK8 and CK18 common to liver regeneration, hepatotoxin sensitivity, and the diagnostic, persistent expression of these cytokeratins in many carcinomas (Caulin et al., 2000). Kitano et al. (1998) have reported that development of transforming growth factor-α (TGF-α) positive foci in the livers of rats treated with PB is basically similar to that of GST-P+ foci (Kitano et al., 1998). Thus, the elevation of cell proliferation detected by double immunohistochemical staining in areas of CK8/18+ foci suggests a correspondence with the previously observed effect of PB administration on formation of TGF-α positive foci. Rise of the PCNA labeling index within the CK8/18+ areas observed in our study 10 weeks post treatment with 500 ppm dose of PB is generally in line with published results regarding hepatocarcinogenesis (Kolaja et al., 1996).
Histones are basic nuclear proteins that are responsible for the nucleosome structure of chromosomal fibers in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in nucleosomes. The linker histone H1 gene is intronless and encodes a member of the histone H2A family. Histone cluster 2, H2aa3 (Hist2H2aa3) elevation was previously reported in human serous ovarian carcinoma and severe acute respiratory syndrome (Donninger et al., 2004). Furthermore, overexpression of histones was recently reported in human HCCs (Ho et al., 2008). Moreover, it was revealed that histone 3 co-immunoprecipitated with CK18 in HCCs. It was therefore suggested that modulation of the cytoskeleton might disturb the organization of the nucleoskeleton which might further cause instability and fragility of nuclei, possibly exposing the histone and co-immunoprecipitating it with CK18. Thus, the overexpression of Hist2H2aa3 observed in the present study might be related to modulation of CK18 and play an important role in hepatocarcinogenesis.
We previously reported significant elevation of 8-hydroxy-2′-deoxyguanosine (8-OHdG), a marker of oxidative DNA damage potentially involved in carcinogenesis in various experimental models, in the livers of rats administered DEN promoted by PB. This was presumed due to increase of intracellular OH• and enhanced protein and activity levels of cytochrome P450 isoenzymes CYP2B1/2 and CYP3A2 (Kinoshita et al., 2003). While expression of those P450 isoenzymes is generally lacking in GST-P+ foci, we here found elevation of liver microsomal cytochrome b5 (Cyb5A), a membrane-bound hemoprotein which functions as an electron carrier for several membrane-bound oxygenases. Cyb5A interacts with NADH-cytochrome b5 reductase and has been recently shown to be required for some cytochrome P-450-catalyzed reactions (Shimakata et al., 1972).
The present study demonstrated that CK8/18 is an indicator for the likehood of rat liver GST-P+ foci developing into HCCs. Our data imply that CK8/18 overexpression, those two cytokeratins complex formation associated with up-regulation of Hist2H2aa3 and intermediate filament reorganization may drive neoplastic transformation of GST-P+ foci during rat hepatocarcinogenesis leading to the formation of HCCs. Coupling laser assisted microdissection and SELDI ProteinChip Array technology provides tremendous opportunities to identify cell and tumor specific proteins and understand molecular events underlying liver preneoplasia development, and furthermore, might become a powerful tool for the early diagnosis of cancer.
Acknowledgments
We thank Kaori Touma and Rie Onodera for their technical assistance, and Yukiko Iura for her help during the preparation of this manuscript.
This research was supported by grants from the Ministry of Health, Labor and Welfare of Japan and the Japan Science and Technology Corporation.
References
- Abe M., Oshima R.G. A single human keratin 18 gene is expressed in diverse epithelial cells of transgenic mice. J. Cell. Biol. 1990;111:1197–1206. doi: 10.1083/jcb.111.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessandro R., Fontana S., Kohn E., De Leo G. Proteomic strategies and their application in cancer research. Tumori. 2005;91:447–455. doi: 10.1177/030089160509100601. [DOI] [PubMed] [Google Scholar]
- Athanassiadou P., Psyhoyiou H., Grapsa D., Gonidi M., Ketikoglou I., Patsouris E. Cytokeratin 8 and 18 expression in imprint smears of chronic viral hepatitis, autoimmune hepatitis and hepatocellular carcinoma. A preliminary study. Acta Cytol. 2007;51:61–65. doi: 10.1159/000325684. [DOI] [PubMed] [Google Scholar]
- Banas A., Quinn G., Yamamoto Y., Teratani T., Ochiya T. “Stem cells into liver”—basic research and potential clinical applications. Adv. Exp. Med. Biol. 2006;585:3–17. doi: 10.1007/978-0-387-34133-0_1. [DOI] [PubMed] [Google Scholar]
- Banks R.E., Dunn M.J., Forbes M.A., Stanley A., Pappin D., Naven T., Gough M., Harnden P., Selby P.J. The potential use of laser capture microdissection to selectively obtain distinct populations of cells for proteomic analysis—preliminary findings. Electrophoresis. 1999;20:689–700. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<689::AID-ELPS689>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Caulin C., Ware C.F., Magin T.M., Oshima R.G. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J. Cell Biol. 2000;149:17–22. doi: 10.1083/jcb.149.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaerkady R., Pandey A. Applications of proteomics to lab diagnosis. Annu. Rev. Pathol. 2008;3:485–498. doi: 10.1146/annurev.pathmechdis.3.121806.151419. [DOI] [PubMed] [Google Scholar]
- Chu Y.W., Runyan R.B., Oshima R.G., Hendrix M.J. Expression of complete keratin filaments in mouse L cells augments cell migration and invasion. Proc. Natl. Acad. Sci. U. S. A. 1993;90:4261–4265. doi: 10.1073/pnas.90.9.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donninger H., Bonome T., Radonovich M., Pise-Masison C.A., Brady J., Shih J.H., Barrett J.C., Birrer M.J. Whole genome expression profiling of advance stage papillary serous ovarian cancer reveals activated pathways. Oncogene. 2004;23:8065–8077. doi: 10.1038/sj.onc.1207959. [DOI] [PubMed] [Google Scholar]
- El-Serag H.B. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck M.R., Bonner R.F., Smith P.D., Chuaqui R.F., Zhuang Z., Goldstein S.R., Weiss R.A., Liotta L.A. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Gluckmann M., Fella K., Waidelich D., Merkel D., Kruft V., Kramer P.J., Walter Y., Hellmann J., Karas M., Kroger M. Prevalidation of potential protein biomarkers in toxicology using iTRAQ reagent technology. Proteomics. 2007;7:1564–1574. doi: 10.1002/pmic.200600836. [DOI] [PubMed] [Google Scholar]
- He T., Stepulak A., Holmstrom T.H., Omary M.B., Eriksson J.E. The intermediate filament protein keratin 8 is a novel cytoplasmic substrate for c-Jun N-terminal kinase. J. Biol. Chem. 2002;277:10767–10774. doi: 10.1074/jbc.M111436200. [DOI] [PubMed] [Google Scholar]
- Ho C.C., Cheng C.C., Liu Y.H., Pei R.J., Hsu Y.H., Yeh K.T., Ho L.C., Tsai M.C., Lai Y.S. Possible relation between histone 3 and cytokeratin 18 in human hepatocellular carcinoma. In Vivo. 2008;22:457–462. [PubMed] [Google Scholar]
- Ito N., Imaida K., Hirose M., Shirai T. Medium-term bioassays for carcinogenicity of chemical mixtures. Environ. Health Perspect. 1998;106(Suppl. 6):1331–1336. doi: 10.1289/ehp.98106s61331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.E., Herndier B.G., Medeiros L.J., Warnke R.A., Rouse R.V. The diagnostic utility of the keratin profiles of hepatocellular carcinoma and cholangiocarcinoma. Am. J. Surg. Pathol. 1988;12:187–197. doi: 10.1097/00000478-198803000-00004. [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Wanibuchi H., Imaoka S., Ogawa M., Masuda C., Morimura K., Funae Y., Fukushima S. Formation of 8-hydroxydeoxyguanosine and cell-cycle arrest in the rat liver via generation of oxidative stress by phenobarbital: association with expression profiles of p21(WAF1/Cip1), cyclin D1 and Ogg1. Carcinogenesis. 2002;23:341–349. doi: 10.1093/carcin/23.2.341. [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Wanibuchi H., Morimura K., Wei M., Shen J., Imaoka S., Funae Y., Fukushima S. Phenobarbital at low dose exerts hormesis in rat hepatocarcinogenesis by reducing oxidative DNA damage, altering cell proliferation, apoptosis and gene expression. Carcinogenesis. 2003;24:1389–1399. doi: 10.1093/carcin/bgg079. [DOI] [PubMed] [Google Scholar]
- Kitano M., Ichihara T., Matsuda T., Wanibuchi H., Tamano S., Hagiwara A., Imaoka S., Funae Y., Shirai T., Fukushima S. Presence of a threshold for promoting effects of phenobarbital on diethylnitrosamine-induced hepatic foci in the rat. Carcinogenesis. 1998;19:1475–1480. doi: 10.1093/carcin/19.8.1475. [DOI] [PubMed] [Google Scholar]
- Kolaja K.L., Stevenson D.E., Walborg E.F., Jr., Klaunig J.E. Dose dependence of phenobarbital promotion of preneoplastic hepatic lesions in F344 rats and B6C3F1 mice: effects on DNA synthesis and apoptosis. Carcinogenesis. 1996;17:947–954. doi: 10.1093/carcin/17.5.947. [DOI] [PubMed] [Google Scholar]
- Ku N.O., Azhar S., Omary M.B. Keratin 8 phosphorylation by p38 kinase regulates cellular keratin filament reorganization: modulation by a keratin 1-like disease causing mutation. J. Biol. Chem. 2002;277:10775–10782. doi: 10.1074/jbc.M107623200. [DOI] [PubMed] [Google Scholar]
- Ku N.O., Omary M.B. Phosphorylation of human keratin 8 in vivo at conserved head domain serine 23 and at epidermal growth factor-stimulated tail domain serine 431. J. Biol. Chem. 1997;272:7556–7564. doi: 10.1074/jbc.272.11.7556. [DOI] [PubMed] [Google Scholar]
- Ku N.O., Michie S., Oshima R.G., Omary M.B. Chronic hepatitis, hepatocyte fragility, and increased soluble phosphoglycokeratins in transgenic mice expressing a keratin 18 conserved arginine mutant. J. Cell Biol. 1995;131:1303–1314. doi: 10.1083/jcb.131.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N.O., Wright T.L., Terrault N.A., Gish R., Omary M.B. Mutation of human keratin 18 in association with cryptogenic cirrhosis. J. Clin. Invest. 1997;99:19–23. doi: 10.1172/JCI119127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N.O., Darling J.M., Krams S.M., Esquivel C.O., Keeffe E.B., Sibley R.K., Lee Y.M., Wright T.L., Omary M.B. Keratin 8 and 18 mutations are risk factors for developing liver disease of multiple etiologies. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6063–6068. doi: 10.1073/pnas.0936165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N.O., Gish R., Wright T.L., Omary M.B. Keratin 8 mutations in patients with cryptogenic liver disease. N. Engl. J. Med. 2001;344:1580–1587. doi: 10.1056/NEJM200105243442103. [DOI] [PubMed] [Google Scholar]
- Lau A.T., Chiu J.F. The possible role of cytokeratin 8 in cadmium-induced adaptation and carcinogenesis. Cancer Res. 2007;67:2107–2113. doi: 10.1158/0008-5472.CAN-06-3771. [DOI] [PubMed] [Google Scholar]
- Lee I.N., Chen C.H., Sheu J.C., Lee H.S., Huang G.T., Yu C.Y., Lu F.J., Chow L.P. Identification of human hepatocellular carcinoma-related biomarkers by two-dimensional difference gel electrophoresis and mass spectrometry. J. Proteome. Res. 2005;4:2062–2069. doi: 10.1021/pr0502018. [DOI] [PubMed] [Google Scholar]
- Lehmann U., Kreipe H. Laser-assisted microdissection and isolation of DNA and RNA. Methods Mol. Med. 2006;120:65–75. doi: 10.1385/1-59259-969-9:65. [DOI] [PubMed] [Google Scholar]
- Melle C., Ernst G., Scheibner O., Kaufmann R., Schimmel B., Bleul A., Settmacher U., Hommann M., Claussen U., von Eggeling F. Identification of specific protein markers in microdissected hepatocellular carcinoma. J. Proteome Res. 2007;6:306–315. doi: 10.1021/pr060439b. [DOI] [PubMed] [Google Scholar]
- Melle C., Kaufmann R., Hommann M., Bleul A., Driesch D., Ernst G., von Eggeling F. Proteomic profiling in microdissected hepatocellular carcinoma tissue using ProteinChip technology. Int. J. Oncol. 2004;24:885–891. [PubMed] [Google Scholar]
- Nakagawa T., Huang S.K., Martinez S.R., Tran A.N., Elashoff D., Ye X., Turner R.R., Giuliano A.E., Hoon D.S. Proteomic profiling of primary breast cancer predicts axillary lymph node metastasis. Cancer Res. 2006;66:11825–11830. doi: 10.1158/0008-5472.CAN-06-2337. [DOI] [PubMed] [Google Scholar]
- Nan L., Dedes J., French B.A., Bardag-Gorce F., Li J., Wu Y., French S.W. Mallory body (cytokeratin aggresomes) formation is prevented in vitro by p38 inhibitor. Exp. Mol. Pathol. 2006;80:228–240. doi: 10.1016/j.yexmp.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Nousiainen M., Sillje H.H., Sauer G., Nigg E.A., Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary M.B., Ku N.O., Toivola D.M. Keratins: guardians of the liver. Hepatology. 2002;35:251–257. doi: 10.1053/jhep.2002.31165. [DOI] [PubMed] [Google Scholar]
- Reddy G., Dalmasso E.A. SELDI ProteinChip(R) array technology: protein-based predictive medicine and drug discovery applications. J. Biomed. Biotechnol. 2003;2003:237–241. doi: 10.1155/S1110724303210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge K.M., Linz L., Flitney F.W., Kuczmarski E.R., Chou Y.H., Omary M.B., Sznajder J.I., Goldman R.D. Keratin 8 phosphorylation by protein kinase C delta regulates shear stress-mediated disassembly of keratin intermediate filaments in alveolar epithelial cells. J. Biol. Chem. 2005;280:30400–30405. doi: 10.1074/jbc.M504239200. [DOI] [PubMed] [Google Scholar]
- Sauter E.R., Shan S., Hewett J.E., Speckman P., Du Bois G.C. Proteomic analysis of nipple aspirate fluid using SELDI-TOF-MS. Int. J. Cancer. 2005;114:791–796. doi: 10.1002/ijc.20742. [DOI] [PubMed] [Google Scholar]
- Schoniger-Hekele M., Petermann D., Muller C. Mutation of keratin 8 in patients with liver disease. J. Gastroenterol. Hepatol. 2006;21:1466–1469. doi: 10.1111/j.1440-1746.2006.04392.x. [DOI] [PubMed] [Google Scholar]
- Shimakata T., Mihara K., Sato R. Reconstitution of hepatic microsomal stearoyl-coenzyme A desaturase system from solubilized components. J. Biochem. 1972;72:1163–1174. doi: 10.1093/oxfordjournals.jbchem.a130004. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson S.S., Grisham J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- Toivola D.M., Omary M.B., Ku N.O., Peltola O., Baribault H., Eriksson J.E. Protein phosphatase inhibition in normal and keratin 8/18 assembly-incompetent mouse strains supports a functional role of keratin intermediate filaments in preserving hepatocyte integrity. Hepatology. 1998;28:116–128. doi: 10.1002/hep.510280117. [DOI] [PubMed] [Google Scholar]
- Wang L., Srinivasan S., Theiss A.L., Merlin D., Sitaraman S.V. Interleukin-6 induces keratin expression in intestinal epithelial cells: potential role of keratin-8 in interleukin-6-induced barrier function alterations. J. Biol. Chem. 2007;282:8219–8227. doi: 10.1074/jbc.M604068200. [DOI] [PubMed] [Google Scholar]