Abstract
Great Lakes tributaries are known to deliver waterborne pathogens from a host of sources. To examine the hydrologic, land cover, and seasonal patterns of waterborne pathogens (i.e. protozoa (2), pathogenic bacteria (4) human viruses, (8) and bovine viruses (8)) eight rivers were monitored in the Great Lakes Basin over 29 months from February 2011 to June 2013. Sampling locations represented a wide variety of land cover classes from urban to agriculture to forest. A custom automated pathogen sampler was deployed at eight sampling locations which provided unattended, flow-weighted, large-volume (120–1630 L) sampling. Human and bovine viruses and pathogenic bacteria were detected by real-time qPCR in 16%, 14%, and 1.4% of 290 samples collected while protozoa were never detected. The most frequently detected pathogens were: bovine polyomavirus (11%), and human adenovirus C, D, F (9%). Human and bovine viruses were present in 16.9% and 14.8% of runoff-event samples (n = 189) resulting from precipitation and snowmelt, and 13.9% and 12.9% of low-flow samples (n = 101), respectively, indicating multiple delivery mechanisms could be influential. Data indicated human and bovine virus prevalence was different depending on land cover within the watershed. Occurrence, concentration, and flux of human viruses were greatest in samples from the three sampling locations with greater than 25% urban influence than those with less than 25% urban influence. Similarly, occurrence, concentration, and flux of bovine viruses were greatest in samples from the two sampling locations with greater than 50 cattle/km2 than those with less than 50 cattle/km2. In seasonal analysis, human and bovine viruses occurred more frequently in spring and winter seasons than during the fall and summer. Concentration, occurrence, and flux in the context of hydrologic condition, seasonality, and land use must be considered for each watershed individually to develop effective watershed management strategies for pathogen reduction.
Keywords: Human virus, Bovine virus, Land cover, Hydrology, Seasonality, Great Lakes tributaries
Graphical abstract
Highlights
-
•
Human and bovine viruses were detected in 16% and 14% of samples collected (n = 290).
-
•
Viruses occurred more frequently during winter and spring versus summer and fall.
-
•
Land cover type influenced whether human or bovine viruses occurred most often.
-
•
Hydrologic condition did not explain the variability observed in virus prevalence.
1. Introduction
Human and bovine pathogens are disease-causing microorganisms which can deteriorate groundwater and surface water resources. Fecal contamination by human and bovine pathogens, including viruses, bacteria, and protozoa, is a potential human health hazard when exposed to contaminated recreational waters (Sinclair et al., 2009), drinking water sources (Borchardt et al., 2012), wildlife (i.e. shellfish, white-tailed deer, geese, rodents, etc.) (Ley et al., 2002), crop irrigation (Bosch, 1998), and dairy production (de Oliveira et al., 2012). Various environmental factors (i.e. pH, temperature, salinity, UV light exposure, etc.) influence the fate, transport, and occurrence of human and bovine pathogens in surface water as well as other watershed-specific factors such as, land cover composition, hydrologic condition, and season.
Aquatic contamination from pathogens can vary considerably in space and time (Rutsch et al., 2008) similar to many non-point source contaminants in urban and rural runoff. To that effect, non-point sources of pathogens from human waste include leaking sanitary sewer infrastructure, landfills, degraded public and private sanitary lateral line connections or misconnections, improper sanitary sewer line connections, properly functioning and defective septic systems, land application of septic and municipal waste effluent, and stormwater drainage systems. Point sources of pathogens from human waste include municipal sanitary sewer overflows (SSO), combined sewer overflows (CSO), and treated as well as partially treated wastewater effluent, and industrial effluent (Ahmed et al., 2010, Corsi et al., 2014). Point and non-point sources of pathogens from agricultural sources related to bovine production include cattle manure in holding ponds, grazing pastures, barnyards, and agricultural practices that apply cattle manure to agricultural croplands.
Watersheds have various complex sources of human and bovine pathogens, and once released into the environment, they can be transported to surface water by way of various pathways depending on the hydrologic conditions at the time. For example, human virus occurrence during low-flow periods suggests a continuous source of sewage contamination to the watershed such as wastewater treatment effluent, exfiltration from failing wastewater infrastructure, or illicit connections of sanitary sewers and/or septic systems (Rutsch et al., 2006). In addition to the continuous sources of human viruses described above, sources during runoff periods include periods of high-flow stress on the sanitary sewage system due to increased flow volumes. These high-flow-induced sources can include sanitary sewer overflows, combined sewer overflows, and leaks in sewage conveyance infrastructure. Bovine virus occurrence during low-flow periods suggests direct cattle access to streams as a continuous source, while overland flow from barnyards, pastures, and manure application, in addition to subsurface drain tiles would be high-flow-induced sources. Watershed transport mechanisms will influence the fate of the aforementioned sources and can determine the resulting survival, occurrence, and magnitude of waterborne pathogens present in surface waters (Ferguson et al., 2003).
Given all of the potential sources, the influences on survival, and the fate and transport mechanisms within a watershed, it is challenging to properly represent variability and magnitude of pathogens in streams. Previous research has begun to address some of these challenges. For example, a diverse group of human and bovine viruses have been detected previously in watersheds with varying land use (Corsi et al., 2014). A study in Michigan sampled nine rivers for enterovirus and rotavirus, providing insight into spatial variability (Jenkins et al., 2005). Another study in California reported on human adenoviruses in urban runoff, emphasizing the influence that specific land cover class can have on pathogen presence in surface waters (Jiang, 2001).
These studies and more have made progress on understanding some of the factors that impact waterborne pathogen dynamics in streams. The challenge moving forward is to implement a study designed to address a larger proportion of these influential factors that also adequately represents pathogen occurrence and variability. Such a comprehensive monitoring program would need to include consideration of short-term (inter-event) and long-term (intra-event) hydrologic variability, seasonal and annual temporal variability, land cover, source-specific discharges (i.e. municipal wastewater effluent, CSO, etc.), and a comprehensive suite of target pathogens.
The objectives of this study were to 1) quantify multiple pathogens within four microbial categories (human viruses, bovine viruses, pathogenic bacteria and protozoa), 2) compare pathogen variability in streams due to hydrologic condition including low-flow periods, and periods of increased runoff due to rainfall and snowmelt, 3) implement a hydrologically appropriate sampling strategy that represents water from all portions of run-off event hydrographs (initial flush, rising flow, peak flow, and receding flow periods), 4) examine seasonal patterns in pathogen prevalence in streams, and 5) describe variability in pathogen prevalence in streams in relation to land cover composition. Results provide further understanding of environmental factors and inherent watershed properties which influence pathogen presence in Great Lakes tributaries and could help improve watershed management decisions aimed at minimizing human exposure to waterborne pathogens.
2. Methods
2.1. Sampling locations
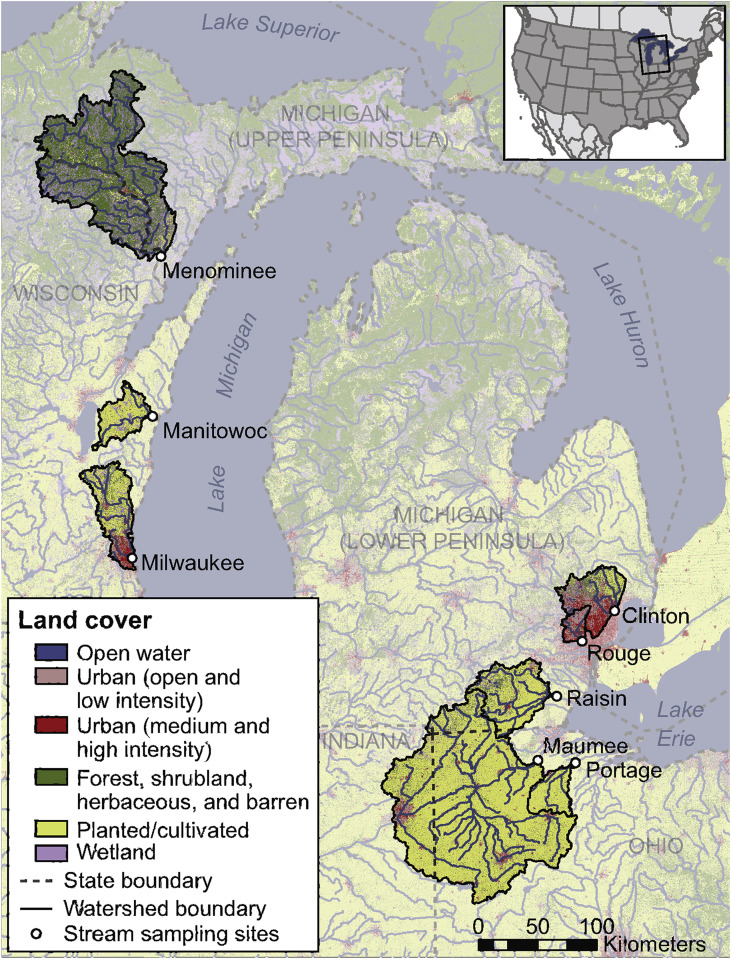
Eight Great Lakes tributaries were selected as sampling locations that represent watersheds with diverse land cover compositions from high to low urban and agricultural land cover (Fig. 1 ; Table 1 ). All land cover categories were defined by the 2011 National Land Cover Database products.
Fig. 1.
Sampling locations and land cover in eight study watersheds in the Great Lakes Basin. Map comprised of various spatial datasets: state and political boundaries (Instituto Nacional de Estadística Geografía e Informática, 2006a), hydrography (Instituto Nacional de Estadística Geografía e Informática, 2006b, National Atlas of the United States, 2005), land cover, and watershed boundaries (U.S. Department of Agriculture-Natural Resources Conservation Service (2009)).
Table 1.
Physical watershed attributes, human and cattle density, land cover, and sample collection information for eight watersheds in the Great Lakes Basin. Land cover compositions for each watershed were summarized using 2011 National Land Cover Database products (Jin et al., 2013). Wastewater statistics are a combination of data from the USGS SPARROW Program and the International Joint Commission. Drainage area was derived from The Watershed Boundary Dataset (WBD) (U.S. Department of Agriculture-Natural Resources Conservation Service (2009)). [USGS, United States Geological Survey; km2, square kilometers; m3/s, cubic meters per second; WWTP, wastewater treatment plant; NA, not available].
| Sampling location | USGS station ID | Great lakes basin | Drainage area (km2) | Population density (people/km2) | Cattle density (cattle/km2) | Annual mean flow (m3/s) | WWTP flow as fraction of river discharge | Land cover percentage |
Samples collected |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Urban | Agriculture: pasture/hay | Agriculture: crops | Forest | Water/Wetland | Runoff-event periods | Low-flow periods | ||||||||
| Menominee River | 04067500 | Lake Michigan | 10179 | 38 | 3.5 | 74 | 0.00185 | 3.8 | 1.4 | 2.3 | 54.8 | 33.1 | 22 | 12 |
| Manitowoc River | 04085427 | Lake Michigan | 1362 | 164 | 143.2 | 12 | 0.00872 | 7.0 | 31.8 | 38.1 | 6.2 | 15.5 | 25 | 12 |
| Milwaukee River | 04087170 (Water Quality) 04087000, 04087142, 04087159 (Streamflow) | Lake Michigan | 2258 | 2909 | 57.2 | 22 | 0.02217 | 29.9 | 15.7 | 27.5 | 12.0 | 13.2 | 26 | 13 |
| Clinton River | 04165500 | Lake Erie | 1901 | 4097 | 2.9 | 21 | 0.06428 | 52.8 | 7.6 | 11.9 | 14.5 | 11.2 | 23 | 12 |
| River Rouge | 04166500 | Lake Erie | 484 | 6470 | 7.8 | 5 | NA | 92.0 | 0.1 | 0.0 | 4.5 | 3.1 | 23 | 13 |
| River Raisin | 04176500 | Lake Erie | 2699 | 396 | 12.8 | 25 | 0.01864 | 11.1 | 18.0 | 49.3 | 10.8 | 9.6 | 23 | 13 |
| Maumee River | 04193490 (Water Quality) 04193500 (Streamflow) | Lake Erie | 16395 | 363 | 12.4 | 182 | 0.03128 | 10.6 | 5.5 | 73.3 | 6.4 | 3.0 | 23 | 14 |
| Portage River | 04195500 | Lake Erie | 1109 | 38 | 6.6 | 15 | 0.03572 | 9.8 | 0.6 | 83.6 | 4.5 | 0.7 | 24 | 12 |
2.2. Sample collection
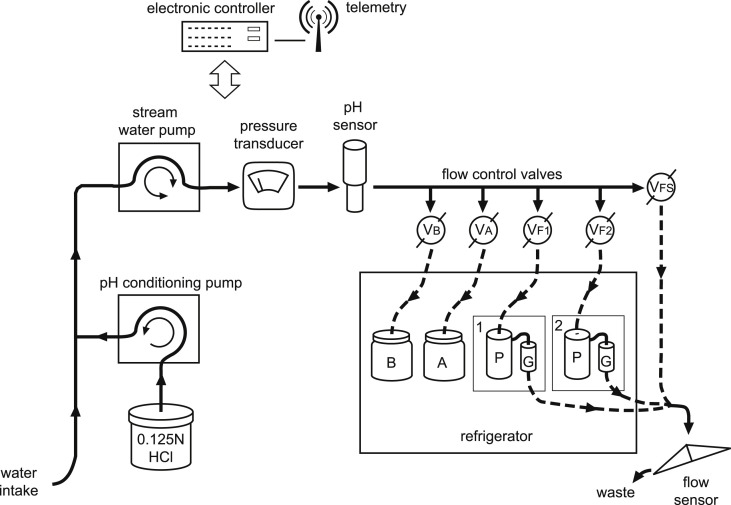
Sampling locations were monitored for waterborne pathogens over a 29 month period from February 2011 to June 2013. Flow-weighted composite samples were collected using a custom-designed automated large-volume virus sample collection and filtration system (modified from Corsi et al., 2014) (Fig. 2 ). Specific details regarding flow-weighted composite sampling using this system were previously described (Corsi et al., 2014) and details describing modifications for the current study are presented in supporting information (Text S1). Briefly, the automated subsample sequence utilized five ball valves to direct water flow between a flow sensor, two whole-water collection bottles, and two filtration cartridges (Fig. 2). The automated subsample sequence is described further in supporting information (Text S2).
Fig. 2.
Diagram of the custom automated large-volume virus sample collection and filtration system. “P” and “G” designations are for the virus filtration units and refer to a 10 μm pore size prefilter (P) and glass-wool filter (G). “A” and “B” designations are bottles for raw surface water collection. “VFS”, “VB”, “VA”, “VF1” and “VF2” designations are for the flow control valves used to direct water to the various system components: flow sensor (VFS), bottle A (VA), bottle B (VB), filtration unit 1 (VF1), and filtration unit 2 (VF2). (Modified from Corsi et al., 2014). [HCl, hydrochloric acid].
Samples were collected during low-flow and runoff-event periods with “runoff-event periods” defined as periods of increased runoff due to rainfall and snowmelt. Thresholds for runoff-event samples were set at individual sampling locations to trigger sampling when water levels increased over the most recent low-flow levels which varied temporally. Three runoff-event period samples were targeted on a quarterly/seasonal basis, and low-flow period samples were collected every other month over the 29 month sample period. Further details on field replicate and blank sample collection and results are presented in supporting information (Text S2).
Recovery controls for glass wool filtration were performed as previously (Lambertini et al., 2008). Details describing recovery control sampling and results are in supporting information (Text S3).
2.3. Laboratory methods
Pre-filters and glass wool filters were eluted immediately in the laboratory upon receipt and the eluates concentrated by polyethylene glycol following standard elution and secondary concentration procedures (Lambertini et al., 2008, Millen et al., 2012). Eluates or final concentrated sample volumes (FCSV) from a sample-paired pre-filter and glass wool filter were combined for qPCR analysis. FCSV volumes were between 0.9 mL and 57 mL (mean = 7.4 mL) which were archived at −80 °C until nucleic acid extraction. Extraction procedures were the same as those described previously (Corsi et al., 2014) except for the addition of an initial freeze-thaw step for extracting Cryptosporidium oocyst DNA (Giovanni and LeChevallier, 2005).
Real-time qPCR was performed for genes specific to eight human viruses, eight bovine viruses, four bacteria and two protozoa and is further described in supporting information (Text S4). All gene targets and references for primers and hydrolysis probes and standard curve performance parameters are listed in supporting information (Table S2).
2.4. Data analysis
The summation of concentrations from individual organisms are used to compute human and bovine sum for each sample. Human and bovine sums from each sample are then used to calculate a mean sum of human and bovine virus concentration, or a percent virus occurrence for the sum of human or bovine viruses. For the purposes of computing means, a value of 0.0 was substituted for instances where viruses were not detected.
Human and cattle population (dairy and non-dairy) densities (Table 1) were calculated based on the total population within a watershed divided by the total drainage area for that watershed (Jin et al., 2013, U.S. Census Bureau Geography Division, 2010a, U.S. Census Bureau Geography Division, 2010b, U.S. Census Bureau Geography Division, 2010c, U.S. Department of Agriculture, 2015); U.S. Department of Agriculture-Natural Resources Conservation Service, 2009).
Human and bovine virus percent occurrence and mean and max concentration were computed based on three-month seasons: winter (December–February), spring (March–May), summer (June–August) and fall (September–November).
Virus flux was computed and normalized by drainage area to control for watershed size and flow variability among sampling locations. Flux provided information that was comparable among the watersheds for assessing relative impact to the Great Lakes and identifying areas that could be focused on for management efforts to have the greatest effect. Virus flux was computed as Virus flux = stream-water volume X virus concentration/(sampling duration X drainage area) (genomic copies/km2/h). Stream-water volume was computed by integrating instantaneous discharge values over the period of interest. For low-flow periods, the period of interest was the sampling period. For runoff-event periods, the period of interest began at the initial hydrograph rise, and ended when flow returned to base-flow conditions or when a subsequent runoff-event hydrograph began. When runoff-event periods included more than one sample, a summation of loadings (streamflow volume X concentration) from the individual samples were used to compute the virus flux. Beginning and ending times used for computation of flux, computed streamflow volumes, and resulting virus flux are provided in the supporting information (Table S3). U.S. Geological Survey National Water Information System (NWIS) Web Interface URL links to individual sampling locations streamflow data for the current study period are found in the supporting information (Table S4) (U.S. Geological Survey, 2016a). In addition, supporting information (Text S5) provides detailed instructions for accessing waterborne pathogen concentration data from the U.S. Geological Survey NWIS Web Interface (U.S. Geological Survey, 2016b).
Statistical significance was determined using a pairwise Wilcoxon rank sum test with corrections for multiple comparisons for concentrations (Benjamini and Hochberg, 1995). Statistical significance for pairwise comparisons of human and bovine virus occurrence among multiple factors (sampling locations, seasons, hydrologic conditions), in addition to human and ruminant specific fecal indicator bacteria, was determined by using generalized linear (binomial) models (Hothorn et al., 2008, R Core Team, 2016). In all cases, significance was evaluated at the 5% significance level.
3. Results
A total of 290 water samples were collected and analyzed for waterborne pathogens, with several of the pathogens having very low occurrence analyzed for only 162 of the 290 samples, during a 29 month period across eight sampling locations (Table 2 ). A total of 189 samples were collected during runoff-event periods and 101 samples were collected during low-flow periods. Streamflow during runoff-event and low-flow periods by sampling location are presented in supporting information (Table S6, Fig. S1). Human and bovine viruses were detected more frequently and had higher concentrations than pathogenic bacteria (p < 0.05), and protozoa were not detected during the study (Table 2). More detailed interpretations on pathogenic bacteria results are not further discussed due to their low frequency of occurrence across the eight sampling locations.
Table 2.
Mean concentrations (genomic copies/L), and frequencies of pathogens for all water samples collected from eight sampling locations on tributaries of the Great Lakes from February 2011 to June 2013. [ND, no detection].
| Pathogens | Mean Concentration (genomic copies/L) | Occurrence (%) | Samples (n = ) |
|---|---|---|---|
| Human virus | |||
| Adenovirus C, D, F | 0.5 | 9.0 | 290 |
| Adenovirus A | 0.2 | 4.1 | 290 |
| GII Norovirus | 4.0 | 2.4 | 290 |
| GI Norovirus | 0.2 | 1.0 | 290 |
| Enterovirus | 0.04 | 0.3 | 290 |
| Hepatitis-A Virus (HAV) | ND | 0.0 | 290 |
| Adenovirus B | ND | 0.0 | 290 |
| Human Rotavirus | ND | 0.0 | 162 |
| Human Sum | 4.9 | 16 | 290 |
| Pathogenic bacteria | |||
| Campylobacter jejuni | 0.1 | 0.7 | 290 |
| Salmonella enterica spp.Salmonella enterica spp. | 0.03 | 0.3 | 290 |
| Enterohemorrhagic E. coli (EHEC)Enterohemorrhagic E. coli (EHEC) | 0.02 | 0.3 | 290 |
| Mycobacterium avium subsp. Paratuberculosis | ND | 0.0 | 162 |
| Bacteria Sum | 0.2 | 1.4 | 290 |
| Bovine virus | |||
| Bovine Polyomavirus | 0.8 | 11 | 290 |
| Bovine Rotavirus A | 23.5 | 5.2 | 290 |
| Bovine Enterovirus | 1.0 | 2.1 | 290 |
| Bovine Viral Diarrhea Virus (BVDV) type 2 | 0.3 | 0.3 | 162 |
| Bovine Viral Diarrhea Virus (BVDV) type 1 | ND | 0.0 | 290 |
| Bovine Adenovirus | ND | 0.0 | 290 |
| Bovine Rotavirus C | ND | 0.0 | 162 |
| Coronavirus | ND | 0.0 | 162 |
| Bovine Sum | 26 | 14 | 290 |
| Protozoa | |||
| Giardia lamblia | ND | 0.0 | 162 |
| Cryptosporidium parvum | ND | 0.0 | 162 |
| Protozoa Sum | ND | 0.0 | 162 |
3.1. Human viruses
3.1.1. Organisms
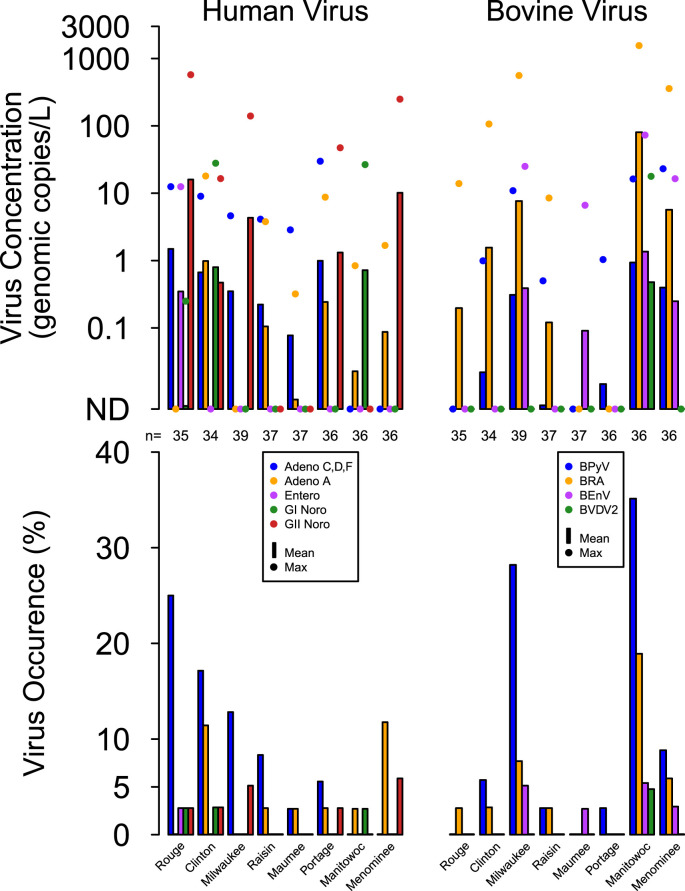
Five of the eight human viruses analyzed were detected in at least one sample (Table 2, Fig. 3 ). Adenovirus C, D, F was present most often, however, the greatest mean and maximum human virus concentrations were observed for GII norovirus. Human viruses were present most often at the River Rouge, followed by the Clinton, Milwaukee and Menominee Rivers.
Fig. 3.
Occurrence, mean, and maximum concentrations of human and bovine viruses in water samples from eight sampling locations on tributaries of the Great Lakes from February 2011 to June 2013 (n = 290) with sampling locations ordered by highest to lowest percent urban land cover from left to right. BPyV represents bovine polyomavirus, BRA represents bovine rotavirus group A, BEnV represents Bovine enterovirus, and BVDV2 represents bovine viral diarrhea virus type 2. BVDV2 was analyzed for only 56% of samples collected at the eight sites (n = 162).
3.1.2. Hydrologic condition
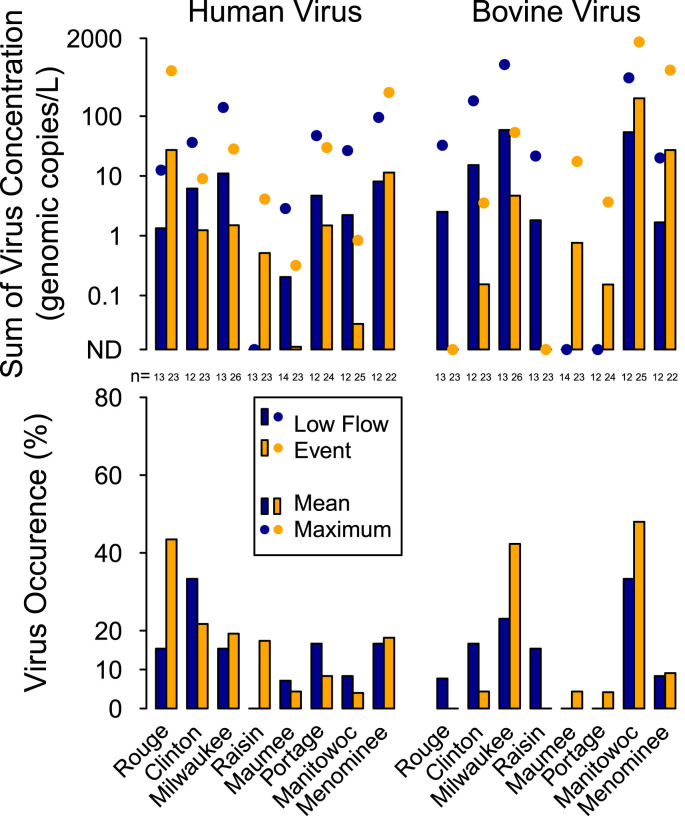
Occurrence and concentrations of human viruses were not significantly different between runoff-event and low-flow periods at individual sampling locations or when grouped by urban locations (urban land cover > 25%) and rural locations (Fig. 4 , Table S7).
Fig. 4.
Occurrence, mean concentration, and maximum concentrations of the sum of human viruses and the sum of bovine viruses during low-flow and runoff-event periods at eight sampling locations on tributaries of the Great Lakes from February 2011 to June 2013 with sampling locations ordered by highest to lowest percent urban land cover from left to right.
Flux in samples from sampling locations with greater than 25% urban influence (River Rouge, Clinton River, and Milwaukee River) were greater than those sampling locations with less than 25% urban land cover (p < 0.05; Fig. 5 ). When comparing different hydrologic conditions, however, the flux of human viruses did not indicate a consistent pattern during runoff-event or low-flow periods at any of the individual sampling locations, presumably due to low occurrence and high variability in concentrations when human viruses were present.
Fig. 5.
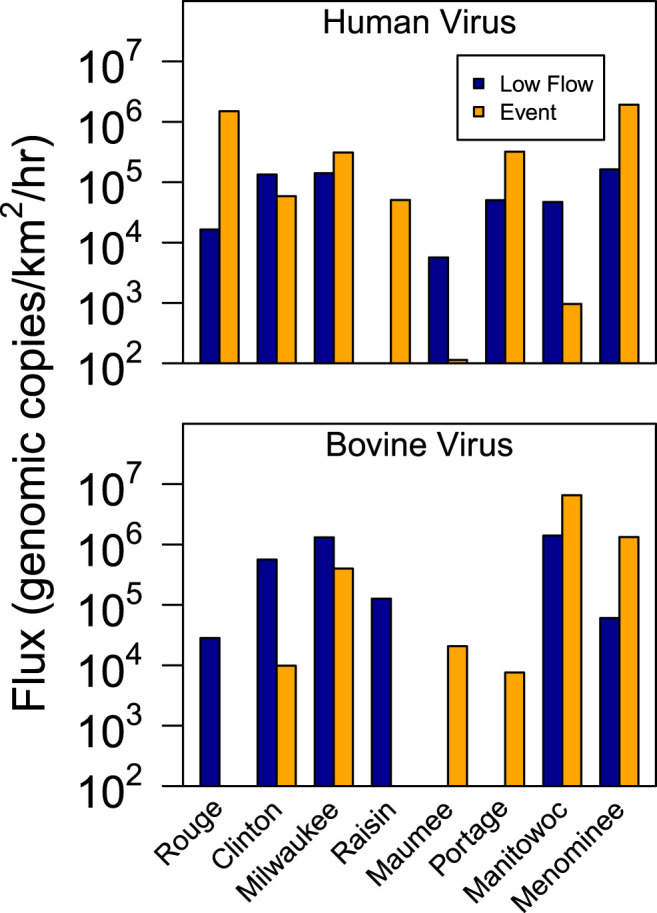
Mean flux (hourly loading per unit drainage area) of the sum of human viruses and the sum of bovine viruses during runoff-event and low-flow periods for the eight Great Lakes watersheds from February 2011 to June 2013 with sampling locations ordered by highest to lowest percent urban land cover from left to right.
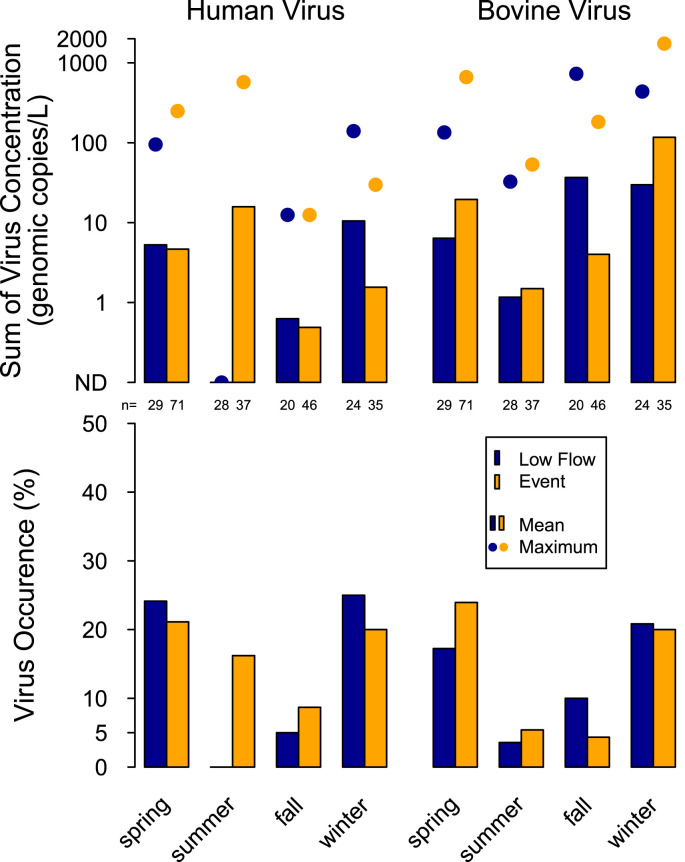
3.1.3. Season
Human viruses for all sampling locations were present in each of the four seasons with fall and summer having fewer occurrences (p < 0.05), and lower concentrations (p < 0.05) than spring and winter during low-flow and runoff-event periods (Fig. 6 ). There was no individual human virus that dominated this signal, but most of the viruses detected in more than 1% of the samples (adenovirus C, D, F, GII norovirus, and GI norovirus) contributed to this result (Fig. S2). The maximum of the sum of human virus concentrations for all sampling locations were greatest in summer, followed by spring, winter, and fall.
Fig. 6.
Occurrence, mean concentration and maximum concentrations of the sum of human viruses and the sum of bovine viruses during low-flow and event periods during different seasons at eight sampling locations in the Great Lakes watershed from February 2011 to June 2013.
3.2. Bovine viruses
3.2.1. Organisms
Four of the eight bovine viruses analyzed were detected at least once, and bovine polyomavirus was the most frequently detected (Table 2). The greatest maximum concentration of bovine viruses was for bovine rotavirus A at six of the eight sampling locations. Concentrations and occurrence of bovine viruses for the two sites with cattle density greater than 50 cattle/km2 (the Manitowoc River and the Milwaukee River) were greater than those with lower cattle density (p < 0.05), and the Manitowoc River was the only location where BVDV2 was detected (Fig. 3).
3.2.2. Hydrologic condition
Analysis of bovine viruses by hydrologic condition yielded the same results whether considering occurrence, concentrations, or flux. There were no statistically significant differences between results of samples collected during low-flow events as compared to those from runoff-event periods for individual sampling locations or for sampling locations grouped by high cattle density and low cattle density (Fig. 4, Fig. 5). However, consistent with evaluation of overall land cover differences, occurrence, concentrations, and flux from samples collected during only runoff-event periods were greater at the two sampling locations with the highest cattle density (p < 0.05), and the same was true for samples collected during only low-flow periods.
3.2.3. Season
Bovine viruses were present in each of the four seasons with the frequency of occurrence greater during spring and winter than during fall and summer for low-flow and runoff-event periods (p < 0.05; Fig. 6). This was the case for all of the bovine viruses that had detections. The mean sum of bovine virus concentrations for all sampling locations were greatest in winter and lowest during summer (p < 0.05), but winter was not significantly different from spring and fall.
4. Discussion
The level of human virus occurrence in the current study (5.4%–34%) was low compared to previously reported occurrence results of human viruses in river samples (30%–80%). (Corsi et al., 2014, Kishida et al., 2012, Lee et al., 2013, Rezaeinejad et al., 2014). Considering the relatively large number of human viruses analyzed compared to previous studies, the selection of organisms is not likely to be a reason for the low occurrence of human viruses observed in the current study. Numerous other factors could influence virus prevalence in a watershed and may contribute to differences from previous study results including: the prevalence of viruses in the human population during the study period, virus sources, land cover characteristics, hydrologic condition, season, and differences in sampling and analytical procedures. Given human viruses were present in all eight watersheds monitored, and each had a wide variation in proportion of areas served by municipal and septic systems, it appears all of the aforementioned factors may potentially contribute to the pathogen contamination detected in these samples.
Results for two of the human viruses illustrate very different scenarios for assessment of potential exposure to waterborne pathogens: GII norovirus was found to have the highest mean concentrations at four of the eight sampling locations, but did not occur frequently. Conversely, adenovirus C, D, F occurred relatively frequently at five of the eight sampling locations, but the mean concentrations at these five locations were all relatively low. These findings are unique when placed in the context of other previously published studies for the following reasons: 1) some studies evaluated only one human virus (Ahmed et al., 2010, Fong et al., 2010), or others considered presence/absence of viruses rather than concentration, and did not evaluate individual viruses, but evaluated “enteric viruses” as a group (Lee et al., 2013, Sidhu et al., 2012), and 2) the scenario of high concentrations and low occurrence for some organisms, and low concentrations and high occurrence rates for other organisms, was not considered in previous studies, or the pattern did not exist (Corsi et al., 2014, Kishida et al., 2012).
The three watersheds with greater than 25% urban land cover (River Rouge, Clinton River, and Milwaukee River) had an average human virus occurrence of 25%. Watersheds in the current study with less urban influence (<25%) and lower human population density (<2000 people/km2) had lower occurrence rates (p < 0.05), suggesting land cover was likely an influential factor for human virus occurrence. Human viruses have been detected previously in watersheds dominated by urban and rural land cover (Corsi et al., 2014, Sidhu et al., 2012), but a sufficient number of sampling sites with full seasonal and hydrologic condition coverage were not available to make broader statements on the influence of land cover on human virus prevalence.
The prevalence of human viruses during different hydrologic periods did not indicate differences at individual sampling locations during runoff-event periods as compared to low-flow periods whether considering concentration, occurrence, or flux. On the surface, three sampling locations (River Rouge, Milwaukee River and River Raisin) look to have greater prevalence of human viruses during runoff-event periods than during low-flow periods, and four locations (Clinton, Portage, Manitowoc and Maumee Rivers) look to have greater human virus prevalence during low-flow periods, but none of these results are statistically significant. The relatively small number of samples coupled with the low level of occurrence presents a challenge in determining statistically significant results at individual sampling locations. In further consideration of hydrologic influence, there may be watershed-specific factors that influence virus presence under different hydrologic conditions such as prevalence of viruses in the human population during the time of study (Bosch, 1998), age and condition of potential sources including septic and municipal systems (Sercu et al., 2011), precipitation characteristics (Corsi et al., 2014), snowpack, and the influence of moisture conditions in the soils adjacent to sources over different seasons (John and Rose, 2005). Even with the substantial restoration efforts which have resulted in a reduction of the number of CSOs in the River Rouge watershed in recent decades (Rouge River National Wet Weather Demonstration Project, 2014), data from the current study indicates human sewage contamination still exists during runoff-event periods in the River Rouge. This is consistent with the nature of CSO events and the potential for other infrastructure issues which lead to exfiltration of sewage during increased runoff periods (U.S. Environmental Protection Agency, 2004). Conversely, misconnected and failing septic and sanitary systems, upstream wastewater treatment plant (WWTP) discharge, and CSOs have all been identified as sources of human waste to the Clinton River watershed (Michigan Department of Environmental Quality, 2011, Michigan Department of Environmental Quality, 1995). The Clinton River has the highest percentage of wastewater effluent relative to total streamflow discharge of the eight sampling locations monitored (Table 1). Wastewater treatment often does not fully remove human viruses (Fong et al., 2010, Myrmel et al., 2006), so this can be a source of potential human virus contamination that is diluted during runoff-events, consistent with Clinton River results from the current study. An explanation for increased human virus prevalence during runoff-event periods in the River Raisin is less obvious given the rural nature of the watershed, although biosolid application of septic and municipal waste effluent may be one potential contributor (Sidhu and Toze, 2009). Land cover in the River Raisin is dominated by agriculture (67%), and sewage is largely treated by septic systems and small locality treatment systems.
Results from the Milwaukee River sampling location present a unique opportunity to compare results that used the same sampling and analytical techniques over two different time periods. In a previous study at the Milwaukee River sampling location from 2007 to 2008, occurrence of human viruses in samples (65%) was greater than the current study (16%) (Corsi et al., 2014). The largest difference between results from the two studies was the comparatively large human virus occurrence rate of 85% (n = 13) during runoff-event periods in the previous study. Examining the streamflow record at this site, mean streamflow was 29% greater during the previous study than the current study (Supporting Information; Table S8). Additionally, the sewage overflow volume released to the Milwaukee River basin was 73% greater during the previous study period compared to the current study. (Milwaukee Metropolitan Sewage District, 2016). The increased streamflow and sewage overflow volume have the potential to influence the level of human sewage contamination and could be, at least in part, an explanation for greater human virus occurrence in the previous study.
There was a higher prevalence of human viruses at the eight sampling locations during winter and spring than during summer and fall (p < 0.05), especially during low-flow conditions, a pattern which was strengthened by contributions from all human viruses with greater than 1% occurrence except adenovirus A which had similar occurrence rates throughout the year. This result could potentially be explained by four mechanisms: 1) viruses survive longer in cooler winter temperatures (Schijven and Hassanizadeh, 2000), 2) microorganism-damaging ultraviolet radiation penetrates the water less during winter due to reduced photoperiods and the presence of ice cover, and during spring due to greater turbidities from increased flows, 3) the watersheds had greater flow levels during spring than other parts of the year (Table S8), increasing stress on the wastewater conveyance infrastructure and the resulting risk of sewage exfiltration, and 4) organism (adenovirus, norovirus, rotavirus, etc.) activity and infections within human populations tend to rise and/or peak during the winter and spring seasons (Dey et al., 2013, Rohayem, 2009, Tran et al., 2010). In addition to the four mechanisms above, municipal WWTPs and/or wastewater stabilization lagoons in all studied watersheds have state regulations and/or National Pollution Discharge Elimination System (NPDES) permits that ease the disinfection requirement for WWTP effluent during the non-recreational period of October/November–April (Michigan Department of Environmental Quality (2006); Ohio Environmental Protection Agency, 2016, Wisconsin Department of Natural Resources, 1986)). Results from the current study are consistent with previous research concluding human viruses in rivers were most prevalent during the cold-weather months spanning late fall through spring. (Corsi et al., 2014, Kishida et al., 2012, Lipp et al., 2001).
The sampling technique used for the current study was different from many previous studies. The current study sampled for 24 h during low-flow periods and throughout the hydrograph during runoff-event periods, and samples were collected over longer periods than the more traditional “grab” sampling techniques of many previous studies (pumping and filtering large volumes of water within 1–3 h) (Fong et al., 2010, Sidhu et al., 2012). This cannot; however, be the sole explanation of low occurrence and concentration in the current study given the results from the 2007 to 2008 study from the Milwaukee River described above. Identical sampling techniques and laboratory techniques resulted in much lower occurrence and magnitude of human viruses in the current study as compared to the previous study at the same sampling location.
Bovine viruses were most prevalent in watersheds with the highest cattle density, but were also present, at a lower frequency, in the two most urban watersheds, the Clinton River (bovine rotavirus A and bovine polyomavirus) and the River Rouge (bovine rotavirus A). The Clinton River does have some agricultural land cover that could account for this, but the River Rouge consists primarily of urban land cover. It is possible that the bovine rotavirus A detected in urban watersheds originates from the human-bovine reassortant vaccine amplified by bovine rotavirus A primers (Matthijnssens et al., 2010). The vaccine is administered orally and may be present in WWTP effluent and leaking sanitary/septic systems. In an analysis of virus presence as compared to human-specific and ruminant-specific bacteria, bovine rotavirus A does not increase with human-specific bacteria, but does increase with ruminant-specific bacteria. These findings suggest the human-bovine reassortant vaccine is not a substantial contributor to bovine rotavirus A detected in this study.
All eight sampling locations had one or more bovine viruses present during the current study. Similar to the human viruses, results for two of the bovine viruses illustrate different scenarios for assessment of potential exposures. Bovine rotavirus A had the highest mean concentrations at six of the eight sampling locations, but the occurrence of rotavirus A was low relative to the other bovine viruses present. Conversely, bovine polyomavirus was present most often at five of the eight sampling locations, but the mean concentrations at those locations were all relatively low. Similar to the human viruses, this finding is unique for bovine viruses because: 1) most studies evaluated only one bovine virus (Ahmed et al., 2010, Jiménez-Clavero et al., 2005), 2) others only considered presence/absence of viruses (Fong et al., 2005), and 3) the scenario of high concentrations and low occurrence for some organisms, and low concentrations and high occurrence rates for other organisms did not exist (Corsi et al., 2014).
In previous studies of rivers that had cattle farming in their watersheds, approximately 50% of river samples were positive for bovine viruses (Corsi et al., 2014, de Oliveira et al., 2012, Fong et al., 2005, Hundesa et al., 2010, Jiménez-Clavero et al., 2005, Ley et al., 2002) with the exception of two studies: the Maroochy River, Australia (10% occurrence; Ahmed et al., 2010) and in runoff from Brazilian dairy farms (30% occurrence; de Oliveira et al., 2012). Bovine virus occurrence in the current study was lower than previous studies with 14% of all samples positive for at least one bovine virus. Considering only the two watersheds with the greatest cattle density (the Milwaukee River with 57 cattle/km2 and the Manitowoc River with 143 cattle/km2), results are similar to those from previous studies with occurrence rates of 36% and 43% respectively. All other sampling locations had less than 13 cattle/km2 (U.S. Department of Agriculture (2015)) and less than 18% occurrence of bovine viruses.
Given the result there were bovine viruses present during runoff-event and low-flow periods, and hydrologic condition did not explain variability of occurrence, concentrations, or flux, suggests that sources during both hydrologic conditions are influential in overall prevalence of bovine viruses. In addition, the importance of land use (high compared to low cattle density) with respect to bovine virus prevalence is apparent in each of the different methods of evaluation presented in the present study. The presence of bovine viruses during runoff-event periods and low-flow periods indicates the possibility of different delivery mechanisms from the source to the stream that become influential during the different hydrologic conditions. Presence of bovine viruses during runoff-event periods is likely a result of overland runoff in barnyard and pasture settings as well as runoff from crop land after manure spreading operations. Potential sources of bovine viruses during low-flow periods include instances where cattle have direct access or are located adjacent to the stream, and situations where applied land manure from cattle manure spreading operations can flow to the stream through tile drainage or the shallow groundwater system. These sources and transport mechanisms are consistent with those identified in previous research on livestock-related waterborne pathogens (Crane et al., 1983, Givens et al., 2016, Jamieson et al., 2002, Wilkes et al., 2014).
Similar to human viruses, and consistent with previous research in the Milwaukee River watershed (Corsi et al., 2014) and other previously published research (Fong et al., 2005), bovine virus concentration and occurrence was greater during spring and winter than other seasons. This was not only true for the sum of all bovine viruses, but for all bovine viruses with detections. This result is likely influenced by several factors including: prevalence of viruses in the bovine herd population, greater virus survival in cooler winter temperatures, ice cover, shorter photoperiod, winter manure spreading, and increased soil-moisture content, which enhances the efficiency of hydrologic pathways from sources to receiving streams (Bosch et al., 2006, Mawdsley et al., 1995, Stuntebeck et al., 2011, Xagoraraki et al., 2014). In addition, this result is consistent with results from research on other agricultural contaminants such as suspended solids and phosphorus which are recognized to be greater during winter and spring periods (Danz et al., 2010, Stuntebeck et al., 2011).
Flux computed for the current study provided relatively unique information for pathogen studies performed in rivers. Considering microorganism concentrations can vary substantially during short time periods and different phases of a hydrograph or during low-flow periods (Templar et al., 2016), a technique to characterize concentrations over relevant time periods is necessary to compare results among different hydrologic conditions and watersheds. The flow-compositing technique described herein allowed for determination of mean pathogen concentrations throughout entire runoff-event periods or over extended low-flow periods that included the full light cycle. This type of characterization provided a way to minimize uncertainty in determination of concentrations and flux computations that arise from less intensive sampling techniques (e.g. grab sampling). Using these data, and normalizing by drainage area, provided an equal measure for comparison among watersheds to allow stakeholders proper data to make informed management decisions on the most effective ways to reduce pathogen delivery to receiving water bodies.
5. Conclusions
-
•
Occurrence and concentration of human and bovine viruses appear to have similarities in implications for assessing the potential of waterborne pathogen exposure; organisms with high mean concentrations were found infrequently (human GII norovirus and bovine rotavirus A), and organisms with higher occurrence frequency were found to have lower mean concentrations (human adenovirus C, D, F and bovine polyomavirus).
-
•
Sampling locations with greater than 25% urban land cover and cattle densities exceeding 50 cattle/km2 had the highest human and bovine virus occurrence, concentration, and flux, respectively, suggesting virus prevalence was dependent on land cover within the watershed.
-
•
Human and bovine virus prevalence across all eight sampling locations exhibited similar seasonal response during low-flow and runoff-event periods with elevated prevalence during winter and spring as compared to summer and fall, emphasizing the potential influence of colder water temperature, sunlight exposure and watershed conditions on virus survival.
-
•
Hydrologic condition, whether runoff-events or low-flow periods, did not explain variability observed in human and bovine virus occurrence, concentration, or flux which suggests contrasting delivery mechanisms from the source to the stream become influential during the different hydrologic conditions.
-
•
Waterborne pathogen prevalence observed across the eight Great Lakes watersheds appears to be influenced by virus sources, hydrology, land cover, and seasonal characteristics, but the exact influence varied by sampling location, presumably due to the source-specific differences among watersheds.
-
•
Concentration, occurrence, and flux in the context of hydrologic condition, seasonality, and land use must be considered for each watershed individually to develop effective watershed management strategies for pathogen reduction.
Acknowledgments
Support of this research was provided by the Great Lakes Restoration Initiative, contract number: DW-014-92453901. Special thanks to Ben Siebers for the graphical abstract art he created for the manuscript. Thanks to colleagues in the Michigan, Ohio and Wisconsin U.S. Geological Survey Water Science Centers and U.S. Department of Agriculture, Agricultural Research Service station for their contributions. Any use of trade, product or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.watres.2017.01.060.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ahmed W., Goonetilleke A., Gardner T. Human and bovine adenoviruses for the detection of source-specific fecal pollution in coastal waters in Australia. Water Res. 2010;44:4662–4673. doi: 10.1016/j.watres.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Borchardt M.A., Spencer S.K., Kieke B.A., Lambertini E., Loge F.J. Viruses in nondisinfected drinking water from municipal wells and community incidence of acute gastrointestinal illness. Environ. Health Perspect. 2012;120:1272–1279. doi: 10.1289/ehp.1104499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A. Human enteric viruses in the water environment: a minireview. Int. Microbiol. 1998;1:191–196. [PubMed] [Google Scholar]
- Bosch A., Pinto R.M., Abad F.X. Springer; 2006. Survival and Transport of Enteric Viruses in the Environment, in: Viruses in Foods; pp. 151–187. [Google Scholar]
- Corsi S.R., Borchardt M.A., Spencer S.K., Hughes P.E., Baldwin A.K. Human and bovine viruses in the Milwaukee River watershed: hydrologically relevant representation and relations with environmental variables. Sci. Total Environ. 2014;490:849–860. doi: 10.1016/j.scitotenv.2014.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane S.R., Moore J.A., Grismer M.E., Miner J.R. Bacterial pollution from agricultural sources: a review. Trans. ASAE. 1983;26:858–0866. [Google Scholar]
- Danz M.E., Corsi S.R., Graczyk D.J., Bannerman R.T. U.S. Geological Survey; 2010. Characterization of Suspended Solids and Total Phosphorus Loadings from Small Watersheds in Wisconsin (No. 2010–5039) [Google Scholar]
- de Oliveira L.K., Fleck J.D., Comerlato J., Kluge M., Bergamaschi B., Fabres R.B., da Luz R.B., da Silva J.V., dos S., Rodrigues M.T., Genro J.L., Staggemeier R., Baldasso N., Spilki F.R. Enteric viruses in water samples from Brazilian dairy farms. Agric. Water Manag. 2012;111:34–39. doi: 10.1016/j.agwat.2012.05.001. [DOI] [Google Scholar]
- Dey S.K., Hoq I., Okitsu S., Hayakawa S., Ushijima H. Prevalence, seasonality, and peak age of infection of enteric adenoviruses in Japan, 1995–2009. Epidemiol. Infect. 2013;141:958–960. doi: 10.1017/S0950268812001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson C., Husman A.M., de R., Altavilla N., Deere D., Ashbolt N. Fate and transport of surface water pathogens in watersheds. Crit. Rev. Environ. Sci. Technol. 2003;33:299–361. doi: 10.1080/10643380390814497. [DOI] [Google Scholar]
- Fong T.-T., Griffin D.W., Lipp E.K. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl. Environ. Microbiol. 2005;71:2070–2078. doi: 10.1128/AEM.71.4.2070-2078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.-T., Phanikumar M.S., Xagoraraki I., Rose J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010;76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanni G.D.D., LeChevallier M.W. Quantitative-pcr assessment of Cryptosporidium parvum cell culture infection. Appl. Environ. Microbiol. 2005;71:1495–1500. doi: 10.1128/AEM.71.3.1495-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens C.E., Kolpin D.W., Borchardt M.A., Duris J.W., Moorman T.B., Spencer S.K. Detection of hepatitis E virus and other livestock-related pathogens in Iowa streams. Sci. Total Environ. 2016;566–567:1042–1051. doi: 10.1016/j.scitotenv.2016.05.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T., Bretz F., Westfall P. Simultaneous inference in general parametric models. Biom. J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Hundesa A., Bofill-Mas S., Maluquer de Motes C., Rodriguez-Manzano J., Bach A., Casas M., Girones R. Development of a quantitative PCR assay for the quantitation of bovine polyomavirus as a microbial source-tracking tool. J. Virol. Methods. 2010;163:385–389. doi: 10.1016/j.jviromet.2009.10.029. [DOI] [PubMed] [Google Scholar]
- Instituto Nacional de Estadística Geografía e Informática . Instituto Nacional de Estadística, Geografía e Informática, Aguascalientes, Aguascalientes; Mexico;U.S. Geological Survey, Reston, Virginia, USA: 2006. North American Atlas - Political Boundaries: Government of Canada, Ottawa, Ontario, Canada; pp. 1.1–1.9.. The Atlas of Canada, U.S. Geological Survey. [Google Scholar]
- Instituto Nacional de Estadística Geografía e Informática . Instituto Nacional de Estadística, Geografía e Informática, Aguascalientes, Aguascalientes; Mexico;U.S. Geological Survey, Reston, Virginia, USA: 2006. North American Atlas - Hydrography: Government of Canada, Ottawa, Ontario, Canada. The Atlas of Canada, U.S. Geological Survey. [Google Scholar]
- Jamieson R.C., Gordon R.J., Sharples K.E., Stratton G.W., Madani A. Movement and persistence of fecal bacteria in agricultural soils and subsurface drainage water: a review. Can. Biosyst. Eng. 2002;44:1.1–1.9. [Google Scholar]
- Jenkins T.M., Scott T.M., Morgan M.R., Rose J.B. Occurrence of alternative fecal indicators and enteric viruses in Michigan rivers. J. Gt. Lakes Res. 2005;31:22–31. doi: 10.1016/S0380-1330(05)70235-5. [DOI] [Google Scholar]
- Jiang S.C. Vibrio cholerae in recreational beach waters and tributaries of Southern California. Hydrobiologia. 2001;460:157–164. doi: 10.1023/A:1013152407425. [DOI] [Google Scholar]
- Jiménez-Clavero M.A., Escribano-Romero E., Mansilla C., Gómez N., Córdoba L., Roblas N., Ponz F., Ley V., Sáiz J.-C. Survey of bovine enterovirus in biological and environmental samples by a highly sensitive real-time reverse transcription-PCR. Appl. Environ. Microbiol. 2005;71:3536–3543. doi: 10.1128/AEM.71.7.3536-3543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Yang L., Danielson P., Homer C., Fry J., Xian G. A comprehensive change detection method for updating the national land cover database to circa 2011. Remote Sens. Environ. 2013;132:159–175. [Google Scholar]
- John D.E., Rose J.B. Review of factors affecting microbial survival in groundwater. Environ. Sci. Technol. 2005;39:7345–7356. doi: 10.1021/es047995w. [DOI] [PubMed] [Google Scholar]
- Kishida N., Morita H., Haramoto E., Asami M., Akiba M. One-year weekly survey of noroviruses and enteric adenoviruses in the Tone River water in Tokyo metropolitan area. Jpn. Water Res. 2012;46:2905–2910. doi: 10.1016/j.watres.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Lambertini E., Spencer S.K., Bertz P.D., Loge F.J., Kieke B.A., Borchardt M.A. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 2008;74:2990–2996. doi: 10.1128/AEM.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.-C., Jheong W.-H., Kim M., Choi D.H., Baik K.-H. A 5-year survey (2007–2011) of enteric viruses in Korean aquatic environments and the use of coliforms as viral indicators. Microbiol. Immunol. 2013;57:46–53. doi: 10.1111/j.1348-0421.2012.00515.x. [DOI] [PubMed] [Google Scholar]
- Ley V., Higgins J., Fayer R. Bovine enteroviruses as indicators of fecal contamination. Appl. Environ. Microbiol. 2002;68:3455–3461. doi: 10.1128/AEM.68.7.3455-3461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp E.K., Kurz R., Vincent R., Rodriguez-Palacios C., Farrah S.R., Rose J.B. The effects of seasonal variability and weather on microbial fecal pollution and enteric pathogens in a subtropical estuary. Estuaries. 2001;24:266–276. doi: 10.2307/1352950. [DOI] [Google Scholar]
- Matthijnssens J., Joelsson D.B., Warakomski D.J., Zhou T., Mathis P.K., van Maanen M.-H., Ranheim T.S., Ciarlet M. Molecular and biological characterization of the 5 human-bovine rotavirus (WC3)-based reassortant strains of the pentavalent rotavirus vaccine. RotaTeq. Virol. 2010;403:111–127. doi: 10.1016/j.virol.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Mawdsley J.L., Bardgett R.D., Merry R.J., Pain B.F., Theodorou M.K. Pathogens in livestock waste, their potential for movement through soil and environmental pollution. Appl. Soil Ecol. 1995;2:1–15. doi: 10.1016/0929-1393(94)00039-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michigan Department of Environmental Quality . 2011. Stage 2 Remedial Action Plan Clinton River Area of Concern [WWW Document]http://www.crwc.org/wp-content/uploads/Clinton-River-Stage-2-RAP-2011-_2_.pdf URL. [Google Scholar]
- Michigan Department of Environmental Quality . Michigan Administrative Code. Michigan Department of Environmental Quality, Lansing, MI. 2006. Part 4 water quality standards, rule 62, microorganisms; pp. 1–62. [Google Scholar]
- Michigan Department of Environmental Quality . 1995. Clinton River Watershed Remedial and Preventive Action Plan: 1995 Update [WWW Document]http://www.epa.gov/glnpo/aoc/clintonriver/pdfs/1995_Clinton%20River%20RAP%20update.pdf URL. [Google Scholar]
- Millen H.T., Gonnering J.C., Berg R.K., Spencer S.K., Jokela W.E., Pearce J.M., Borchardt J.S., Borchardt M.A. Glass wool filters for concentrating waterborne viruses and agricultural zoonotic pathogens. J. Vis. Exp. 2012 doi: 10.3791/3930. JoVE e3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milwaukee Metropolitan Sewage District . 2016. SEWAGE OVERFLOWS-(MMSD) Milwaukee Metropolitan Sewerage District [WWW Document]http://www.mmsd.com/wastewatertreatment/overflows URL. (Accessed 09 July 2016) [Google Scholar]
- Myrmel M., RimstadE E., Berg E., Grinde B. Enteric viruses in inlet and outlet samples from sewage treatment plants. J. Water Health. 2006;4:197–209. [PubMed] [Google Scholar]
- National Atlas of the United States . National Atlas of the United States; Reston, VA: 2005. National Atlas of the United States, County Boundaries of the United States, 2001, National Atlas of the United States. [Google Scholar]
- Ohio Environmental Protection Agency . Ohio Administrative Code. Ohio Environmental Protection Agency. 2016. O.A.C. 3745-1-07 (B)(4), water use designations and statewide criteria; pp. 1–32. Columbus, OH. [Google Scholar]
- R Core Team . 2016. R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. [Google Scholar]
- Rezaeinejad S., Vergara G.G.R.V., Woo C.H., Lim T.T., Sobsey M.D., Gin K.Y.H. Surveillance of enteric viruses and coliphages in a tropical urban catchment. Water Res. 2014;58:122–131. doi: 10.1016/j.watres.2014.03.051. [DOI] [PubMed] [Google Scholar]
- Rohayem J. Norovirus seasonality and the potential impact of climate change. Clin. Microbiol. Infect. 2009;15:524–527. doi: 10.1111/j.1469-0691.2009.02846.x. [DOI] [PubMed] [Google Scholar]
- Rouge River National Wet Weather Demonstration Project . 2014. Rouge River Restoration Summary: Wayne County Rouge River National Wet Weather Demonstration Project 1992-2014 [WWW Document]http://www.rougeriver.com/pdfs/overview/rouge-summary-1992-2014.pdf URL. [Google Scholar]
- Rutsch M., Rieckermann J., Cullmann J., Ellis J.B., Vollertsen J., Krebs P. Towards a better understanding of sewer exfiltration. Water Res. 2008;42:2385–2394. doi: 10.1016/j.watres.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Rutsch M., Rieckermann J., Krebs P. Quantification of sewer leakage: a review. Water Sci. Technol. 2006;54:135. doi: 10.2166/wst.2006.616. [DOI] [PubMed] [Google Scholar]
- Schijven J.F., Hassanizadeh S.M. Removal of viruses by soil passage: overview of modeling, processes, and parameters. Crit. Rev. Environ. Sci. Technol. 2000;30:49–127. doi: 10.1080/10643380091184174. [DOI] [Google Scholar]
- Sercu B., Van De Werfhorst L.C., Murray J.L.S., Holden P.A. Sewage exfiltration as a source of storm drain contamination during dry weather in urban watersheds. Environ. Sci. Technol. 2011;45:7151–7157. doi: 10.1021/es200981k. [DOI] [PubMed] [Google Scholar]
- Sidhu J.P.S., Hodgers L., Ahmed W., Chong M.N., Toze S. Prevalence of human pathogens and indicators in stormwater runoff in Brisbane, Australia. Water Res. 2012;46:6652–6660. doi: 10.1016/j.watres.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Sidhu J.P.S., Toze S.G. Human pathogens and their indicators in biosolids: a literature review. Environ. Int. 2009;35:187–201. doi: 10.1016/j.envint.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Sinclair R.G., Jones E.L., Gerba C.P. Viruses in recreational water-borne disease outbreaks: a review. J. Appl. Microbiol. 2009;107:1769–1780. doi: 10.1111/j.1365-2672.2009.04367.x. [DOI] [PubMed] [Google Scholar]
- Stuntebeck T.D., Kominskey M.J., Peppler M.C., Owens D.W., Frame D.R. U.S. Geological Survey; 2011. Precipitation-runoff Relations and Water-quality Characteristics at Edge-of-field Stations, Discovery Farms and Pioneer Farm, Wisconsin, 2003-8 (No. 2011–5008) [Google Scholar]
- Templar H.A., Dila D.K., Bootsma M.J., Corsi S.R., McLellan S.L. Quantification of human-associated fecal indicators reveal sewage from urban watersheds as a source of pollution to Lake Michigan. Water Res. 2016;100:556–567. doi: 10.1016/j.watres.2016.05.056. [DOI] [PubMed] [Google Scholar]
- Tran A., Talmud D., Lejeune B., Jovenin N., Renois F., Payan C., Leveque N., Andreoletti L. Prevalence of rotavirus, adenovirus, norovirus, and astrovirus infections and coinfections among hospitalized children in northern France. J. Clin. Microbiol. 2010;48:1943–1946. doi: 10.1128/JCM.02181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Census Bureau Geography Division . 2010. 2010 State-based Census Block TIGER/Line Shapefile (Michigan) [Google Scholar]
- U.S. Census Bureau Geography Division . 2010. 2010 State-based Census Block TIGER/Line Shapefile (Ohio) [Google Scholar]
- U.S. Census Bureau Geography Division . 2010. 2010 State-based Census Block TIGER/Line Shapefile (Wisconsin) [Google Scholar]
- U.S. Department of Agriculture, 2015. 2012 Census of Agriculture [WWW Document]. URL http://www.agcensus.usda.gov/Publications/2012/ 80(12), 3708-3720.
- U.S. Department of Agriculture-Natural Resources Conservation Service, U. S. Geological Survey, U.S Environmental Protection Agency, 2009. The Watershed Boundary Dataset (WBD) (Vector Digital Data). (Fort Worth, Texas).80(12), 3708-3720.
- U.S. Environmental Protection Agency . 2004. Report to Congress: Impacts and Control of CSOs and SSOs (No. EPA 833-R-04-001) (Office of Water, Washington, D.C) [Google Scholar]
- U.S. Geological Survey . 2016. Surface Water Data for the Nation: USGS Surface-water Annual Statistics [WWW Document]http://waterdata.usgs.gov/nwis/sw USGS Surf.-Water Annu. Stat. Wis. URL. [Google Scholar]
- U.S. Geological Survey . 2016. USGS Water-quality Data for the Nation: Web Interface [WWW Document]http://waterdata.usgs.gov/nwis/qw URL. (Accessed 10 November 2016) [Google Scholar]
- Wilkes G., Brassard J., Edge T.A., Gannon V., Gottschall N., Jokinen C.C., Jones T.H., Khan I.U.H., Marti R., Sunohara M.D., Topp E., Lapen D.R. Long-term monitoring of waterborne pathogens and microbial source tracking markers in paired-agricultural watersheds under controlled and conventional tile drainage management. Appl. Environ. Microbiol. 2014;80(12):3708–3720. doi: 10.1128/AEM.00254-14. AEM.00254–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisconsin Department of Natural Resources . Wisconsin Department of Natural Resources; Madison, WI: 1986. Chapter NR 210, Sewage Treatment Works, in: Wisconsin Administrative Code. 57–60.5. [Google Scholar]
- Xagoraraki I., Yin Z., Svambayev Z. Fate of viruses in water systems. J. Environ. Eng. 2014;140:04014020. doi: 10.1061/(ASCE)EE.1943-7870.0000827. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.