Abstract
We investigated a herpesvirus mutant that contains a single base insertion in its thymidine kinase (tk) gene yet expresses low levels of TK via a net +1 translational recoding event. Within this mutant gene, we defined a G-rich signal that is sufficient to induce recoding. Unlike other translational recoding events, downstream RNA structures or termination codons did not stimulate recoding, and paused ribosomes were not detected. Mutational analysis indicated that specific tRNAs or codon–anticodon slippage were unlikely to account for recoding. Rather, recoding efficiency correlated with the G-richness of the signal and its ability to form unusual structures. These findings identify a mechanism of translational recoding with unique features and potential implications for clinical drug resistance and other biological systems.
Introduction
Recoding is a mechanism of gene expression that permits more than one polypeptide to be translated from an otherwise monocistronic mRNA, often with important biological consequences (15, 18). Translational recoding events include shifts in reading frame during translational elongation. Normally, maintenance of the reading frame is a highly accurate process, with error rates of less than 5 × 10−5 per codon (Kurland 1992); however, specific signals and structures in mRNA can increase the probability of the elongating ribosome to change reading frame as much as 104-fold (2, 15, 18). Shifts in frame are almost always limited to one base and can occur in either the upstream, or leftward, direction (termed −1 frameshifts) or in the downstream, or rightward, direction (termed +1 frameshifts). In eukaryotes, frameshifting had been found only in mRNAs of retrotransposons, retroviruses, and certain RNA viruses until their recent discovery in certain herpesvirus and cellular mRNAs (24, 35, 38). Examples of ribosomal hopping, which have been documented in either the Escherichia coli trp gene or the T4 gene 60, have not yet been described in eukaryotes (6, 46).
To date, all examples of eukaryotic −1 frameshifting require a heptanucleotide “slippery” sequence, XXXYYYZ (where X = A, U, or G; Y = A or U; and Z = A, U, or C) and are augmented by a stimulator, either a stem–loop or pseudoknot RNA structure (2, 7, 15, 18, 26). The stimulatory structure pauses elongating ribosomes on the slippery sequence (42, 44), which is thought to facilitate slippage of the P and A site tRNAs on the mRNA from XXY–YYZ to XXX–YYY (requiring bonding between each tRNA–codon pair at at least two out of three positions). Translation then proceeds in the −1 frame. All examples of eukaryotic +1 frameshifting thus far also entail a site at which recoding occurs, and a stimulator. Frameshifting occurs either by slippage (Belcourt and Farabaugh 1990) or by a mechanism whereby specific structural features of the tRNA in the P site appear to facilitate out-of-frame binding of an aminoacyl-tRNA in the A site (16, 35, 36, 45). As is true for −1 frameshifting, stimulators of +1 frameshifting cause translational pausing. They include “hungry” codons that are decoded by rare tRNAs, termination codons, and pseudoknots (5, 16, 35). However, in the case of the yeast retrotransposon TY3, an additional distal sequence also stimulates frameshifting by an as yet unknown mechanism (15, 16).
Recently, we have described a recoding event in a herpes simplex virus (HSV) mutant that permits the expression of low levels (1%–2% of wild type) of full-length thymidine kinase (TK) despite a single base insertion in the HSV tk gene (Hwang et al. 1994). Expression of full-length TK thus requires a net +1 shift in frame, which has been shown to occur during translation (Hwang et al. 1994). The biological consequences of this recoding event include the reactivation of the mutant from latent infection upon explant of mouse trigeminal ganglia; truly TK–negative mutants do not ordinarily reactivate from such infections (10, 13, 27). This may have clinical importance, especially in immunocompromised patients such as those with AIDS, in whom reactivation of HSV from latent infection can cause severe disease. Therapy with the antiviral drug acyclovir (ACV) has been successful in the treatment of such disease; however, treatment failures associated with ACV-resistant virus are a problem (14, 23). Since the selective action of ACV entails its specific phosphorylation by HSV TK (Fyfe et al. 1978), tk mutations can confer resistance (Coen and Schaffer 1980). Indeed, the tk mutant in which recoding occurs is ACV-resistant and arose in a patient who suffered severe herpes esophagitis despite ACV therapy (Sacks et al. 1989). We hypothesize that this mutant was selected in the patient because it expresses too little TK to activate ACV effectively but enough TK to cause disease.
Here, we report the analysis of the sequences responsible for the HSV tk recoding event. The results indicate that the mechanism is unique and may depend on the ability of G-rich sequences to form unusual inter- or intramolecular structures. The results may also have implications for antiviral drug resistance and for expression of other genes.
Results
A Reporter Gene System to Analyze Net +1 Recoding In Vitro
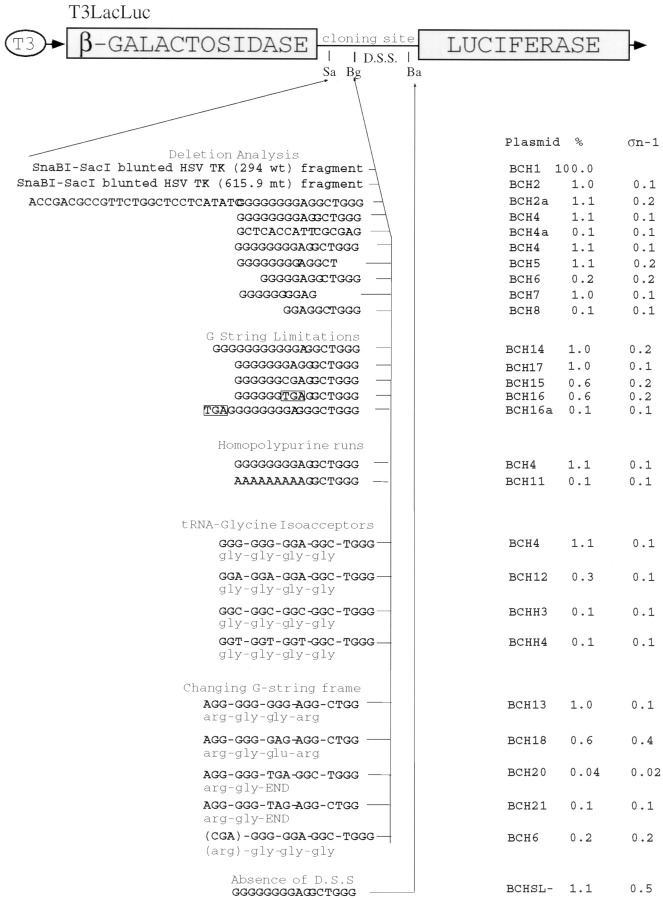
Previously, we identified a net +1 translational recoding event within the tk gene from an ACV-resistant HSV mutant (Hwang et al. 1994). Because this recoding event was not very efficient (1%–2%) and was difficult to quantify, we decided to make use of a sensitive and quantitative enzyme reporter system that has been used to investigate the role of human immunodeficiency virus (HIV) and human T cell leukemia virus II sequences in frameshifting in transfected cells (30, 37). To adapt this system for in vitro analysis, the reporter genes encoding β-galactosidase and luciferase separated by the HIV elements were placed under the control of bacteriophage T3 promoter to create T3 LucLac (Figure 1). A 207 bp SnaBI–SacI blunt-ended fragment from pTK-4, containing mutant tk sequences sufficient for recoding (Experimental Procedures), was then cloned between the reporter genes, replacing the HIV slippery sequence (Figure 1). This plasmid was called BCH2. Following transcription with T3 RNA polymerase and translation of the synthetic mRNA in reticulocyte lysates, luciferase activity from the β-galactosidase–luciferase fusion protein can be detected when a recoding event that shifts translation into the +1 frame occurs. The relative enzymatic activities of β-galactosidase and luciferase reflect the frameshifting efficiency. To normalize this efficiency, we constructed two control plasmids. Reference plasmid, BCH1, was created by cloning the corresponding wild-type tk sequences from pTK-wt (Experimental Procedures) between the reporter genes (Figure 1). This plasmid expresses an in-frame β-galactosidase–luciferase fusion. Its ratio of luciferase activity to β-galactosidase activity was set at 100%. A negative control plasmid, BCH0, was created. It contained luciferase-coding sequences in the +1 reading frame relative to β-galactosidase but lacked any HSV sequence. This allowed subtraction of background “noise” generated within the system (0.05%). For each plasmid, assays were repeated 4 to more than 30 times to obtain means and standard deviations. In these assays, BCH2 reproducibly yielded a low but significant efficiency (1%) of net +1 frameshifting (Figure 1). This efficiency will be referred to as frameshifting at “standard levels.”
Figure 1.
Analysis of Frameshifting In Vitro Using a Reporter Gene Construct
The different plasmids were transcribed and translated and the extracts assayed for luciferase and β-galactosidase. The top line shows the construct with the T3 promoter (oval) driving β-galactosidase coding sequences, which are followed by a frameshift insert and luciferase coding sequences. Below are the sequences in the different plasmids tested. Arrows show where the HSV sequences were replaced. To the right of each plasmid are the mean values and standard deviations for the percentage of luciferase activity relative to β-galactosidase activity normalized to an in-frame control (BCH1). D.S.S.: downstream structure. Sa, Bg, and Ba represent SalI, BglII, and BamHI sites, respectively (see text for cloning details). Boxes represent termination codons.
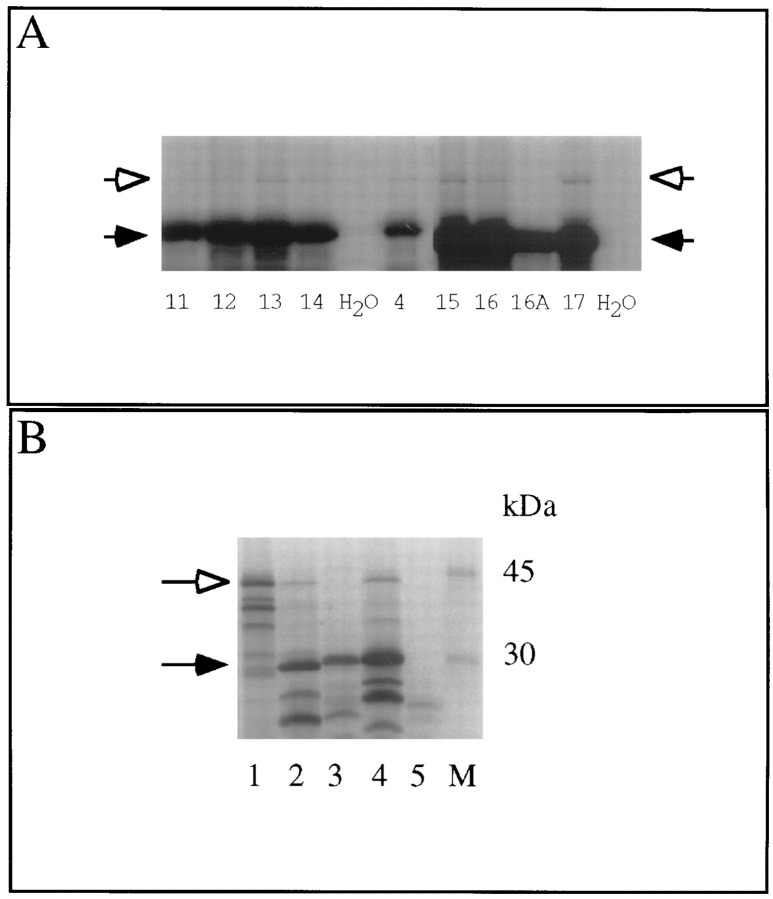
To confirm that luciferase expression was due to a net +1 frameshift and not to another translational event, (e.g., internal initiation), translations were carried out in the presence of 35S methionine and the products analyzed on SDS–polyacrylamide gels. In each case in which we obtained a corrected frameshift value (the mean minus the standard deviation) of greater than or equal to 0.2%, we could observe full-length β-galactosidase–luciferase fusion protein. (A representative sample is shown in Figure 2A). We therefore consider corrected values of greater than or equal to 0.2% as meaningful. In cases in which the corrected values fell below 0.2%, we were unable to detect fusion protein (data not shown).
Figure 2.
Translation of Wild-Type and Mutant mRNAs
(A) A representative sample of reticulocyte translation products synthesized from synthetic mRNA derived from the mutants described in Figure 1. The number in each lane corresponds to the described mutant, and the water control lanes are indicated. The white and black arrowheads represent full-length or truncated species, respectively.
(B) Translation of wild-type and mutant tk mRNAs. Lane 1, TK-wt (wild-type tk gene); lane 2, TK-4 (mutant tk gene); lane 3, TK-4a (mutant tk gene, except the recoding site sequence has been altered to correspond to that of BCH4a [Figure 1]); lane 4, TK-SL+ (mutant tk gene, except the HIV stem–loop has been inserted downstream of the recoding site, i.e., to correspond to BCH4 [Figure 1]); lane 5, water control. M indicates molecular mass standards. White and black arrowheads indicate full-length or truncated species, respectively. The frameshift products from lanes 2 and 4 are 1.4% and 1.3%, respectively, relative to wild-type levels by densitometry. The multiple TK products observed (e.g., between A and B in lane 1 and below B in lane 2) are due primarily to translational initiation at internal AUG codons owing to leaky scanning (see Irmiere et al., 1989).
A 10 Base G-Rich Sequence from the Mutant tk Gene Is Sufficient for Recoding
It seemed probable that the recoding event occurred between the first upstream termination codon in the +1 reading frame and the SacI site (Hwang et al. 1994). Therefore, HSV sequences upstream of this termination codon were deleted, creating pBCH2a. This plasmid, containing 46 nt of HSV sequence, could direct frameshifting at standard levels (Figure 1). A further 5′ deletion of 30 nt yielded plasmid BCH4. The 16 nt of HSV sequence contained within this plasmid was sufficient to promote frameshifting at standard levels. However, a control plasmid, BCH4a, which contained 16 nt of a randomly generated sequence, did not frameshift (Figure 1). To narrow the minimal signal for frameshifting, we deleted either 3 or 6 nt from the 3′ or 5′ end of the 16 nt HSV sequence, creating plasmids BCH5, BCH7, BCH6, and BCH8, respectively (Figure 1). Deletions from the 3′ end had no effect on frameshift efficiency, whereas 5′ deletions abolished frameshifting. Therefore, these results indicate that the sequence G(8)AG is sufficient to promote net +1 frameshifting at standard levels (1%). However, no recoding was detected when this motif was transplanted into a reporter system designed to detect −1 frameshifting (data not shown).
Additionally, the G(8)AGGCTGGG (BCH4) sequence in the original mutant tk gene was altered to correspond to the randomly generated (BCH4a) sequence, creating pTK-4a. While the original mutant tk gene (pTK-4) directed the expression of both a truncated product due to the insertion mutation and full-length TK, full-length TK was not detected upon transcription and translation of pTK-4a (Figure 2B). This confirms the importance of the G-rich sequence in +1 recoding in the mutant tk gene.
The HSV Recoding Event Is Not Stimulated by Downstream Structures and Occurs without Observable Ribosomal Pausing
In many examples of ribosomal recoding, the presence of a stem–loop or pseudoknot structure acts as a positive stimulator (Atkins et al. 1990). A stimulatory stem–loop structure derived from HIV sequences (Reil et al. 1993) lies 10 nt downstream of the G(8)AG motif in BCH4. However, no such structure is obvious downstream of this motif in its natural context (Hwang et al. 1994). To test whether the HIV stem–loop affects the HSV recoding event, we deleted it in BCHSL−. This deletion had no significant effect on luciferase levels produced by frameshifting (see Figure 1). Moreover, insertion of the HIV stem–loop 10 nt downstream of the G(8)AG motif in the mutant tk gene (pTK-SL+) did not alter the levels of full-length TK relative to the truncated product (Figure 2B).
As reviewed in the Introduction, stimulators of eukaryotic frameshifting are thought to act by pausing ribosomes at the recoding site. This model has been borne out in the examples examined thus far (42, 44). Given the lack of effect of the HIV stimulator, we asked whether paused ribosomes could be detected during recoding in the HSV tk system, using the translational inhibitor edeine (Somogyi et al. 1993). Briefly, ribosomes were allowed to initiate on mRNA in the presence of 35S methionine. Then, after a short time, the drug was added to prevent further initiation. This resulted in a relatively synchronous pulse of ribosomes whose products could be sampled at various times throughout the reaction. Interruption of elongation, i.e., pausing, can result in a detectable “paused” product on SDS–polyacrylamide gels.
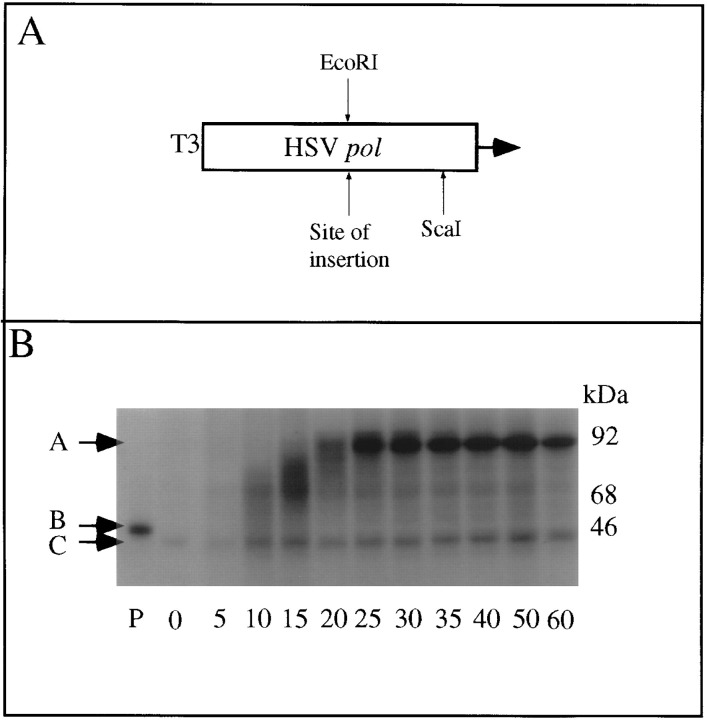
To facilitate the analysis, we cloned the EcoRV–SacI fragment of pTK-4 and pTK-4a (i.e., frameshift-competent BCH4 and frameshift-incompetent BCH4a-containing sequences, respectively; Figure 1) into the HSV DNA polymerase (pol) open reading frame under the control of the T3 promoter, forming plasmids pBH19 and pBH21, respectively (Figure 3A). pBH19 and pBH21 were linearized with either ScaI, which cuts downstream of protein coding sequences, or with EcoRI. EcoRI cuts just 3′ to the G-rich sequence; thus, transcription and translation resulted in a labeled product of the same size as the potential paused product, approximately 44 kDa (Figure 3B, lane P). ScaI-digested DNA was transcribed, the synthetic RNA translated (with edeine added following initiation), and the products electrophoresed through SDS–polyacrylamide gels. Paused products of the expected size were not detected (Figure 3B; data not shown [the product below and clearly resolved from the 44 kDa marker was due to methonine labeling of an endogenous reticulocyte polypeptide]). In contrast, a control plasmid, pPS1a (Somogyi et al. 1993; a gift of I. Brierley), which contains a coronavirus pseudoknot, produced a paused product of the expected size, 42 kDa (data not shown). However, as the sensitivity of these assays was limited, we cannot totally discount the possibility that a very low level pause occurs at the HSV recoding site. Nevertheless, in contrast to results obtained in other translational recoding systems, there was no observable detectable pause associated with the HSV recoding event, nor was recoding stimulated by a downstream structure believed to act by pausing ribosomes.
Figure 3.
Schematic of Plasmids and Labeled Products
(A) Schematic showing construction of pausing construct pBH19 and pBH21.
(B) Time courses of translation of pBH19/ScaI–derived mRNAs in reticulocyte lysates. No paused products of the expected size were detected. The black arrows A, B, and C indicate the position of full-length product, the expected position of a G-string–induced ribosomal pause (prepared by translating BH19/EcoRI–derived mRNA), and the background-labeled rabbit reticulocyte 42 kDa protein, respectively.
Recoding Requires and Initiates within a G-String
As the sequence defined by deletion analysis was highly G-rich, we wished to assess the importance of monotonous runs of G's (G-strings) in net +1 frameshifting. Replacing the G(8)AG motif of BCH4 with A(8)AG abolished frameshifting (Figure 1), indicating a requirement for G's rather than for any purines. We lengthened the run of G's from 8 in BCH4 to 11 (BCH14) or shortened it to 7 (BCH17; Figure 1). The frameshift efficiencies were unchanged. However, mutation of the seventh G in pBCH4 to C, creating a run of six G's, reduced frameshift efficiencies by approximately 50% (BCH15; Figure 1). As BCH6, which contained a string of five G's, did not frameshift at detectable levels, it would appear that the threshold for G-string–induced frameshifting in our reporter system is six residues.
These results suggest that the recoding event occurs as ribosomes encounter the run of G's. Thus far, however, all attempts to determine the site of recoding by protein sequencing have failed, primarily due to low frameshift efficiency (B. C. H. and S. Matusfuji, unpublished data). Therefore, we decided to use site-directed mutagenesis to address this question. In an initial attempt to define the site of frameshifting, we mutated the C at position 7 in BCH15 to a T, yielding plasmid BCH16, i.e., GGG-GGG-CGA-GGC-TGG-G to GGG-GGG-TGA-GGC-TGG-G. (Bold characters represent the differences; T's indicated in the DNA sequence become U's in the RNA). This mutation resulted in a termination codon (underlined) in-frame with β-galactosidase. This termination codon did not measurably affect frameshifting, since luciferase expression of BCH16 and BCH15 were comparable (Figure 1). This indicates that ribosomes enter the luciferase reading frame without recognizing the termination codon. (Also, the termination codon, which is known to stimulate +1 frameshifting in other systems, did not stimulate the HSV recoding event.) Luciferase expression from pBCH16a, TGA-GGG-GGG-GGA-GGC-TGG-G, which has a TGA codon inserted in the β-galactosidase reading frame immediately upstream of the G(8) motif, was not detectable (Figure 1). The results from BCH 16 and BCH16A, taken together, imply that the frameshift at least initiates between the termination codons; that is, on the G-string.
Recoding Is Associated with G-Richness Rather Than with Specific tRNAs
We next asked whether the recoding event is mediated either by specific tRNAs or by a structural element within the mRNA or both. Certain +1 frameshifts are dependent upon a specific tRNA occupying the ribosomal P site and a rare tRNA or termination codon occupying the ribosomal A site. The hungry codon induces a ribosomal pause, allowing recoding (5, 16, 34, 36, 45).
The BCH4 sequence, GGG-GGG-GGA-GGC-TGG-G, is decoded as GLY-GLY-GLY-GLY-TRP. The likely site of recoding, underlined, is decoded by two tRNAGLY CCC species, whereas the next two codons, GGA and GGC, are decoded by tRNAGLY UCC and tRNAGLY GCC. None of these is a rare tRNA (Gupta et al. 1980) and consequently should not induce a pause to facilitate tRNA/mRNA realignment. Nevertheless, we wished to investigate the effect that different glycine tRNA isoacceptors might have on frameshifting. (Glycine tRNAs decode the sequence GGX, where X = G, A, C, or UGupta et al. 1980). We mutated the third and sixth G of the recoding site (bold characters above) to A, C, or T (plasmids BCH12, BCHH3, and BCHH4, respectively). Analysis of these constructs revealed that BCH12 (GGA-GGA-GGA-GGC-TGG-G) expressed low but detectable levels of luciferase, whereas BCHH3 (GGC-GGC-GGA-GGC-TGG-G) and BCHH4 (GGT-GGT-GGA-GGC-TGG-G) did not (Figure 1). These data suggest that if specific tRNAs are involved in the recoding event, they would have to be tRNAGLY CCC decoding GGG or tRNAGLY UCC decoding GGA. Importantly, the results with BCH12 indicate that a tRNAGLY UCC slippage model is unlikely because tRNA does not form a middle base pair with the next +1 codon, GAG; the predicted C:A pair has been shown to inhibit frameshifting 80- to 240-fold in a +1 frameshift system that utilizes slippage (Curran 1993).
To address whether G-rich sequences might be involved rather than these two specific tRNAs, we placed the G-strings in different reading frames. We first mutated the BCH4 sequence, GGG-GGG-GGA- GGC-TGG-G, to AGG-GGG-GGG-AGG-CTG-G, creating BCH13 (Figure 1). This mutant, decoded by ARG-GLY-GLY-ARG-LEU, frameshifted at standard (1%) levels (Figure 1), indicating that when two Gly GGG codons are bounded by G residues, they direct recoding as well as do eight or more G residues comprising three glycine codons. This finding was underscored and extended by the construction and expression of BCH18. The seventh nucleotide in the G-string of BCH13 was mutated to an A. While this conserves the purine richness of the sequence, it shortens the G-string to 6 nt. BCH18 (AGG-GGG-GAG-AGG-CTG-G), decoded by ARG-GLY-GLU-ARG-LEU (only one Gly codon, GGG), expressed luciferase at just slightly lower levels than BCH15 and BCH16, whereas BCH6 (GGG-GGA-GGC-TGG-G), decoded by GLY-GLY-GLY-TRP (one GGG and one GGA codon), did not frameshift (Figure 1). However, we would not expect BCH6 to frameshift if the recoding event were a −2 slip, because tRNAGLY CCC is able to slip either +1 or −2 in mutant BCH15, BCH16, and BCH18 but only +1 in mutant BCH6 (tRNAGLY CCC does not form a middle base pair with the −2 codon GAG in BCH6; Figure 1). Therefore, we constructed two additional mutants, BCH20 (AGG-GGG-TGA-GGC-TGG-G) and BCH21 (AGG-GGG-TAG-AGG-CTG-G), in which tRNAGLY CCC would be able to slip −2 or +1 equally well. These mutants included a UAG or UGA termination codon (underlined), which when downstream of a recoding site can act as stimulators of programmed frameshifting (35, 46). Neither mutant frameshifted (Figure 1). These results imply that neither specific glycine tRNAs nor tRNA slippage mechanisms are likely to be responsible for the recoding event. Rather, the G-richness of the mRNA sequence is important.
Correlation of the Ability of G-Strings to Form Unusual Structures with Recoding
A plethora of reports indicates that G-rich sequences (e.g., telomeres) are able to form unusual structures in vitro via noncanonical base pairing between G's, e.g., Hoogsteen base pairing, to form two- or four-stranded (G-quartet) structures (reviewed byWilliamson 1994). These structures generally are stabilized by monovalent ions (Williamson 1994). We hypothesized that the G-string affected the local RNA architecture to cause net +1 recoding.
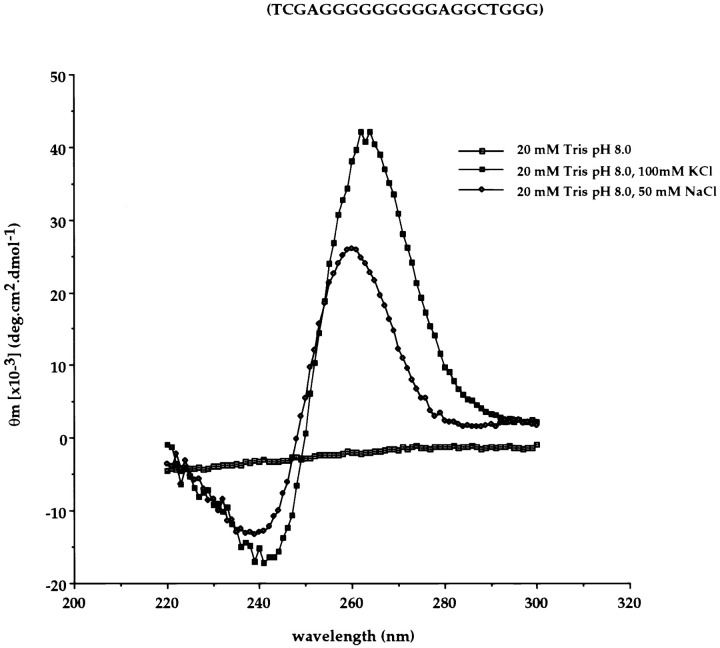
As DNA G-strings behave similarly to RNA G-strings in formation of these structures (41, 43, 49), we used the DNA oligonucleotides from our mutagenesis experiment to address this hypothesis. First, we subjected the oligonucleotide used to create BCH4 to circular dichroism experiments. The oligo (2 μM) was dissolved in either 20 mM Tris (pH 8.0) alone or 100 mM KCl or 50 mM NaCl, denatured, and then cooled before recording wavelengths on a spectrophotometer. Molecular rotation at specific wavelengths can be measured because G-quartets are optically active in polarized light. The resulting wavelength scans revealed a minimum at 240 nm and a maximum at 260 nm in the presence of K+ or Na+ ions but not in the absence of salt (Figure 4). This pattern is indicative of parallel quadraplex formation (Balagurumoorthy et al. 1992). Interestingly, addition of 50 mM NaCl to a pTK-4 RNA translation mixture, although lowering overall translation, resulted in a 2–3-fold increase in TK frameshifting (data not shown).
Figure 4.
Circular Dichroism Wavelength Scans of Oligonucleotide A3
Sequence shown at top. The oligonucleotide (2 μM) was denatured by boiling for 15 min, then cooled on ice for 30 min before recording wavelengths on an Aviv 62DS spectrophotometer. Wavelength scans were recorded at 1 nm intervals (10 s averaging time), and three scans were averaged. Minima at 240 nm and maxima at 260 nm are characteristic of parallel quadraplexes.
In a second approach, end-labeled oligonucleotides were electrophoresed through native 20% polyacrylamide gels as described in Experimental Procedures. Figure 5A shows that the control oligonucleotide used to create plasmid BCH4a or oligonucleotides in which the G(8) sequence of BCH4 (Figure 1) was changed to either A(8), C(8), or T(8), migrated mainly as homogeneous species of the expected size. However, G-rich oligonucleotides, including those used to create plasmids BCH4, BCH12, BCH13, and BCH14 exhibited three different mobilities; faster- and slower-moving species were observed as well as the species of expected size (Figure 5A). These results suggest that the G-rich oligonucleotides fold into compact (fast-migrating) and complex (slow-migrating) structures as a result of G–G base pairing. The slower-migrating species (Figure 5A) were more responsive to sequence alterations than the faster-moving species. Dilution experiments suggested that the faster-moving species were monomeric (data not shown). Further, repetition of the experiment in the presence of 50 mM NaCl yielded similar results (data not shown).
Figure 5.
Correlation between Frameshifting and Structure Formation
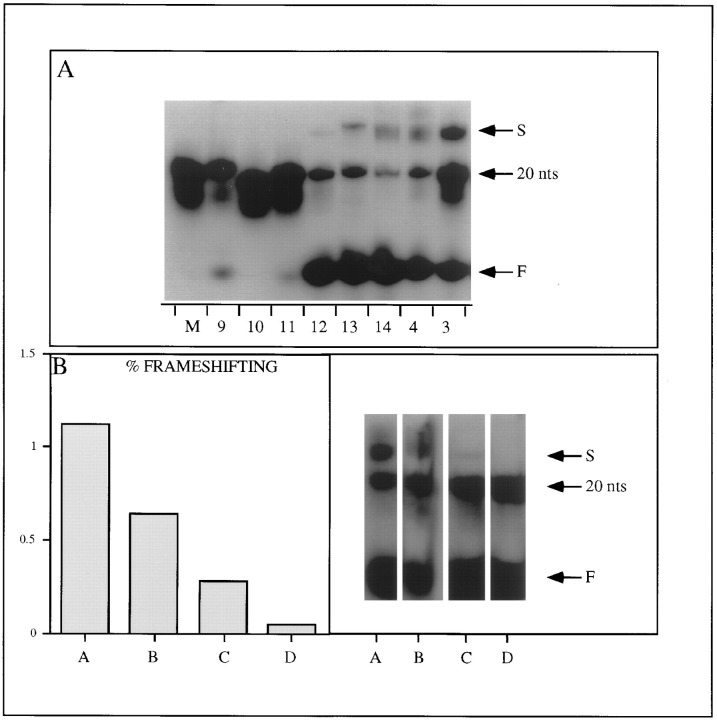
(A) Autoradiograms of nondenaturing 20% polyacrylamide gels. 32P-labeled DNA samples (200 fmol) were denatured at 95°C for 15 min, ice-cooled, and mixed with marker dyes and electrophoresed through a nondenaturing gel. All gels were run at 4°C. Oligonucleotides used: (M) tcgaGCTCACCATTCGCGAG; (9) tcgaTTTTTTTTAGGCTGGG; (10) tcgaCCCCCCCCAGGCTGGG; (11) tcgaAAAAAAAAAGGCTGGG (BCH11); (12) tcgaGGAGGAGGAGGCTGGG (BCH12); (13) tcgaAGGGGGGGGAGGCTGG (BCH13); (14) tcgaGGGGGGGGGGGAGGCTGGG (BCH14); (4) tcgaGGGGGGGGAGGCTGGG (BCH4); (3) tcgaGGGGGGGAGGCTGGG (BCH3). Faster (compact)- and slower (complexes)-moving species are indicated by letters F and S, respectively. The position of a marker 20 mer is also indicated.
(B) The left panel shows a bar graph of mean percentage frameshifting (see Figure 1) of (A) BCH4, tcgaGGGGGGGGAGGCTGGG; (B) BCH15, tcgaGGGGGGCGAGGCTGGG (C) BCH12, tcgaGGAGGAGGAGGCTGGG; (D) BCHH3 and BCHH4 (the oligonucleotide used to create these mutant was synthesized as a mixture, i.e., tcgaGGRGGRGGAGGCTGGG, where R equals pyrimidine). The right panel shows an autoradiogram of a polyacrylamide gel of the corresponding 32P-labeled oligonucleotides, run as described in (A). As above, F and S indicate the faster-moving and slower-moving species, respectively.
To investigate this further, we compared oligonucleotide complex formation of BCH4 with other G-rich oligonucleotide sequences that directed different levels of frameshifting, i.e., BCHH3 and BCHH4 (background), BCH12 (0.3%), BCH15 (0.6%), and BCH4 (1.0%; Figure 5B). Labeled oligonucleotides were electrophoresed through polyacrylamide gels as described previously. BCHH3 and BCHH4 sequences did not form the slower-migrating complex (S; Figure 5B) or frameshift, whereas BCH12 sequences both complexed and frameshifted weakly and BCH15 sequences both complexed and frameshifted at an intermediate level relative to BCH4 sequences (S; Figure 5B). Thus, there is an excellent correlation between the efficiency of recoding and the ability of the corresponding oligonucleotides to form unusual structures.
Discussion
During expression of the tk gene of an HSV drug–resistant mutant, recoding occurs to shift translation into the +1 reading frame (Hwang et al. 1994; this study). We have found that a specific G-rich signal embedded within the tk mRNA corrupts the translational machinery. The ability of sequences to induce recoding correlates well with their ability to form unusual RNA structures. We discuss below in what way this recoding event differs from previously described translational frameshifts, possible mechanisms to explain the correlation between RNA structure and recoding, and the potential relevance of our results to herpesviruses and other biological systems.
Our mutational analysis is consistent with net +1 recoding occurring within the G-string (Figure 1; data not shown) but is not definitive. Unfortunately, due to the low efficiency of recoding, it has not yet been possible to determine by protein sequencing whether the net +1 recoding event is a +1 frameshift or a −2 frameshift (B. C. H. and S. Matusfuji, unpublished data). Nevertheless, for the sake of simplicity and brevity, we will assume in the ensuing discussion that recoding occurs within the G-string as a +1 frameshift.
The HSV Recoding Event Differs from Previously Described Frameshifts: Failure to Detect Pausing or Stimulation
Current models to explain translational frameshifting entail two elements: first, a recoding site at which frameshifting occurs; and second, a stimulatory element that increases the frameshift efficiency by pausing ribosomes at the recoding site (2, 15, 18). Mechanisms that have been invoked to explain the actual recoding event at the frameshifting site, which both entail a kinetically slow step that is more favorable when ribosomes are paused, are, first, slippage; and second, specific peptidyl tRNAs, which are thought to facilitate out-of-frame binding of tRNAs at the A site (reviewed byFarabaugh 1996). The efficiency of frameshifting is generally considered to depend on the extent of pausing, and, in particular, weakly functioning recoding sites are most affected by the presence of stimulators. We failed to detect paused ribosomes at the HSV tk recoding site (Figure 3B), suggesting that the HSV frameshift does not involve a kinetically slower second step. Given the low efficiency of the tk frameshift, it could be argued that the pause was too short for us to detect. Nevertheless, if pausing were important for the tk frameshift, one would expect that stimulators would greatly increase frameshift efficiency. Instead, neither a downstream RNA structure nor termination codons increased frameshift efficiency. Thus, it appears that the tk frameshift operates via a mechanism that does not entail a kinetically slow step that is enhanced by ribosomal pausing.
Does Slippage Account for the tk Frameshift?
Conventional +1 frameshifting utilizing slippage is exemplified by the frameshift in the overlap between the TYA and TYB genes in the retrotransposon ty1 (5, 15). The frameshift requires a 7 nt sequence, CUU AGG C, and occurs by slippage of the P-site tRNA from CUU to UUA. This slip is stimulated by a translational pause induced by the slowly decoded hungry codon AGG in the A site. The frameshift requires no other factors.
If the HSV frameshift were conventional, then its P-site codons could be GGG or GGA. Given either of these P-site codons, we analyzed several A-site codons that could permit +1 slippage while maintaining base pairing at at least two out of three codon–anticodon positions (UAG [BCH21], UGA [BCH20], GGA [gly; BCH6], GGG [gly; BCH13], and GAG [glu; BCH18]) when placed downstream of a single GGG or GGA codon. Of these, UAG and UGA are nonsense codons and are, in general, more slowly decoded than sense codons. (These nonsense codons operated efficiently in the antizyme frameshift system, which, like the HSV frameshift system, has been characterized in a mammalian systemMatsufuji et al. 1995). GGA is an intermediately used codon, and GGG and GAG are very commonly used (“well-fed”) codons (Haas et al. 1996). Thus, if the HSV frameshift were conventional, we would expect higher frameshift efficiencies with A-site codons UAG and UGA, intermediate frameshift efficiencies with GGA codons, and lower frameshift efficiencies with GGG and GAG codons. This was not observed. Instead, only the well-fed A-site codons promoted frameshifting, whereas slowly decoded A-site codons did not (e.g., compare BCH18 and BCH21). Thus, the HSV frameshift is not conventional.
Moreover, it has been argued that tRNAs should not slip on G-strings because of the predicted strength of the mRNA–tRNA interactions (Jacks et al. 1988). Indeed, replacement of A- or U-rich codons with G- or C-rich triplets reduced frameshifting in −1 frameshift systems that utilize slippage (7, 26), and replacement of third position U's in the in-phase triplet decreased frameshifting in a +1 system, suggesting that the weakness of wobble pairs facilitates +1 slippage (Curran 1993). One might imagine a scenario whereby the mRNA G-string interacts, for example, with rRNA (see below) in a way that might promote tRNA slippage. For example, if the G-string formed non–Watson-Crick base pairs with rRNA, that would weaken the hydrogen bonds between the O6 positions of G's in the codon and C's in the anticodon. It would be tempting to speculate that the weakening of these bonds could add to the slipperiness of the mRNA–tRNA complex. However, for this scenario to be viable, we would expect that an A string would also promote net +1 frameshifting, since A–U base pairs have even less energy than the weakened G–C base pairs imagined above. This did not occur (BCH 11; Figure 1). Moreover, this model would predict that an RNA structure or termination codon that enhanced the probability of the second, kinetically slower, slip would increase the efficiency of frameshifting, which did not occur. Thus, it is difficult to consider slippage on G-strings as a viable explanation for our results.
Furthermore, although tRNAGLY UCC will form base pairs at positions one and three with the +1 codon, GAG in BCH12, the U:G wobble base pair is relatively weak at codon position 3 while the clashing middle base pair (A:C) in the slipped codon–anticodon complex should result in destabilization. Grosjean and co-workers (1978) demonstrated that centrally positioned C:A or A:C pairs abolished synthetic anticodon–codon complexes. Such mismatches decrease frameshifting more than 80-fold in the +1 E. coli RF2 system (Curran 1993), compared with the only 3-fold decrease in the HSV tk frameshift (BCH12, Figure 1). Thus, the ability of BCH12 to frameshift is very difficult to explain by either conventional slippage or by the unconventional mechanism considered above. Taken together, our results argue that tRNA slippage is highly unlikely to account for the HSV recoding event.
Is the HSV Recoding Event a Consequence of Peptidyl tRNA–Induced Out-of-Frame Binding?
A second mechanism for frameshifting involves specific peptidyl tRNAs, which are thought to facilitate out-of-frame binding of tRNAs at the A site (16, 35, 36, 45). In our case, this would implicate tRNAGLY CCC and tRNAGLY UCC as responsible for the frameshift when they occupy the P site. In yeast, GGG codons in the P site can induce frameshifting but only when ribosomal pausing is increased by a stimulator, in this case a hungry codon (Vimaladithan and Farabaugh 1994). However, this explanation is not sufficient, since BCH6 (in which either tRNAGLY CCC or tRNAGLY UCC can occupy a P site), and BCH20 and BCH21 (in which tRNAGLY CCC would occupy the P site, and a stimulator, in this case a termination codon, would occupy the A site), fail to frameshift (Figure 1). These results also argue against the possibility that a subpopulation of glycine tRNAs contain 4 nt anticodons and therefore function as +1 frameshift suppressors.
In summary, the tk recoding event appears to function with the same efficiency with or without a stimulator and in the absence of detectable pausing (Figure 1 Figure 2 Figure 3). It does not behave as if it utilizes slippage or peptidyl-tRNA–induced misalignment. Thus, the tk recoding event appears to be truly novel.
Possible Mechanisms for the tk Recoding Event
The efficiency of the tk recoding event correlates with the G-richness of the signal and its ability to form unusual RNA structures. Such structures depend on the ability of G residues to pair with each other, e.g., via non–Watson-Crick interactions (Williamson 1994). Therefore, we suggest that G-string structure mediates recoding within the ribosome via Hoogsteen or other non–Watson-Crick interactions.
We envision three possible models to account for this. In the first model (intramolecular), the G-string in the mRNA forms a structure in the ribosome. Formation of an intramolecular structure in the ribosome has been proposed to account for the T4 gene 60 ribosomal hop (22, 47). In the intramolecular model for the tk frameshift, one of the nucleotides within the G-string would bulge out so that it does not pair with a tRNA, either in the P site or in the A site. Thus, in the ribosome, the two tRNAs bound to the mRNA would form an RNA helix in which an extra nucleotide between the tRNAs is excluded from base pairing. However, as pointed out by Farabaugh et al. 1993, who considered a similar model to explain the TY3 +1 frameshift, it is more likely that such an excluded base would still stack between the paired nucleotides on either side. This has been observed in several RNA structures (1, 28). Thus, in this model, some kind of folding of the mRNA on itself would be required to stabilize a bulge. We have obtained preliminary evidence that the G-string is contained within a structure resistant to RNase T1, consistent with this idea (data not shown).
In our second model (intermolecular), ribosome–mRNA interactions play a role in the occlusion of one nucleotide. Given the correlation between frameshifting and the ability of the G-strings to form unusual structures, an appealing possibility would be non–Watson-Crick interactions between the viral mRNA G-string and G-rich element within mammalian rRNAs. There already are precedents for rRNA–mRNA interactions affecting the efficiency of frameshifting in the E. coli release factor 2 and dnaX genes (32, 47). However, in these cases, the interactions are Watson-Crick and do not involve the recoding signal per se.
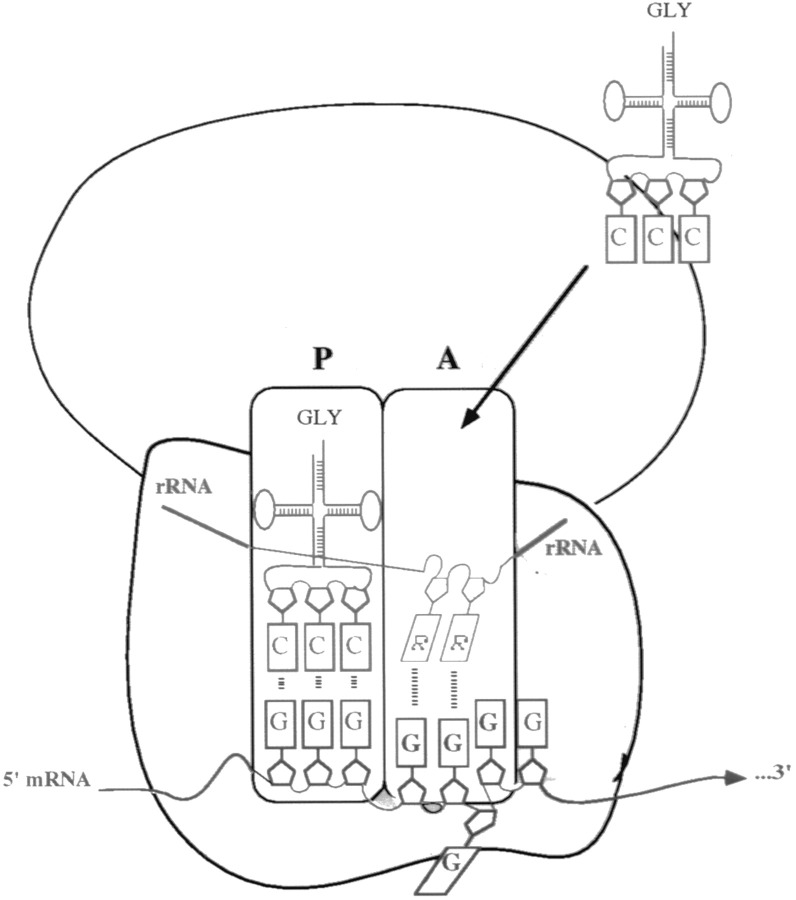
A version of this model is cartooned in Figure 6. Purines within rRNA at the ribosomal A site interact via noncanonical base pairs on the major groove side of a GGG codon within the G-string. Other potential rRNA–mRNA interactions are omitted from the cartoon for simplicity. The incoming aminoacyl-tRNA interacts with the same GGG codon via Watson-Crick base pairs. The result is a small pseudohelix (tRNA–mRNA and mRNA–rRNA) with a nucleotide bulged out. Supporting evidence for bulging nucleotides stabilized by non–Watson-Crick base pairs comes from studies on the HIV Rev Responsive Element RNA bound to Rev peptide (Battiste et al. 1994). In this structure, purine–purine base pairs form within an internal loop of a helix to create a quasi-continous helix with the concomitant bulging of bases (Battiste et al. 1994).
Figure 6.
A Cartoon Model of +1 Ribosomal Recoding Due to a Bulged Base
G–G (or G–A) base-pairing through an rRNA–mRNA interaction permits a base to loop out into solution, thereby correcting the reading frame for the incoming aminoacyl tRNA (tRNA-GLY). The ribosome P and A sites are indicated. R equals purine. Watson-Crick base-pairing is indicated by shorter dashed lines, whereas non–Watson-Crick base-pairing is indicated by longer dashed lines. Non–Watson-Crick base pairs could also form in the ribosomal P site but are not shown for simplicity.
In our third model, a non–Watson-Crick base-paired structure forms within the ribosome, distorting the ribosomal A site, thus favoring binding of tRNA to the mRNA in the +1 frame. This structure could result from either intramolecular base pairing or intermolecular base pairing between mRNA and rRNA. In this model, no bulge in the mRNA is required. However, for this model to fit our data, the distortion of the A site would have to be a relatively fast step kinetically; otherwise, frameshifting would be expected to be increased by pausing.
Each of these models makes specific predictions that can be tested. Furthermore, any of these models provides a new example for a role of non–Watson-Crick base pairs in biology, in this case, translational recoding.
Implications for Herpesviruses and Other Systems
Our results indicate that G-strings are sufficient to induce net +1 recoding in vitro. Furthermore, the degree of frameshift efficiency (approximately 1%) from tk sequences measured in reticulocyte lysates matches TK activity quantitated in TK mutant–infected cells using new assays developed in our laboratory (S.-H. Chen, B. C. H., and D. M. C., unpublished data). Encouragingly, preliminary results indicate that some of our constructs placed under the control of the SV40 promoter recapitulate this phenomenon in vivo (H. K., B. C. H., D. M. C., and H. H., unpublished data).
An obvious question is whether recoding events mediated by G-strings could be occurring in genes other than the mutant HSV tk gene that we have studied. One possibility is the wild-type tk gene. This gene contains the motif G(7)AG, which, like the mutant form G(8)AG, is sufficient for standard levels of frameshifting in our reporter system (Figure 1). This raises the possibility that the wild-type tk gene normally expresses low levels of a previously undetected polypeptide. This polypeptide would retain the ATP-binding site of TK but would lack the nucleoside binding site and other conserved residues (Brown et al. 1995). The question of whether this polypeptide is expressed and, if so, whether it has any function, is under investigation.
Generally, long G-strings are not found in eukaryotic coding sequences, suggesting that there is selection against these motifs. Searches of the database for the motif G(8)AG revealed two occurrences within herpes genome sequences (types 1 and 2) and one example in a cellular gene. We hypothesize that low level net +1 frameshifts will be detected from other genes containing this sequence motif. Our results indicate that shorter G-strings (e.g., G6), within purine-rich contexts, promote net +1 recoding, although at lower levels (Figure 1). This implies that biologically relevant G-string–mediated translational leakiness may occur more frequently than would otherwise be expected, since genes containing G(6) sequences are more common than genes containing G(8) sequences.
HSV genes are very G-C–rich and contain higher numbers of guanine repeats than their cellular counterparts. For example, HSV-1 tk genes contain one G(6) and one G(7) string, and HSV-2 tk genes contain two G(6) and one G(7) string. It has been suggested that the high mutation frequency in the tk gene of HSV may be a consequence of these guanine repeats (Kit et al. 1987), and, indeed, Hwang and Chen 1995 have obtained evidence that frameshift mutations can occur frequently in the G(7) string. If this is so, why would a virus retain sequences that accumulate mutations? Perhaps the virus can tolerate mutations in these sequences because it generates so many wild-type copies per infected cell. Nevertheless, one speculation is that the G-string sequences have been retained because they permit the expression of alternate polypeptides. Regardless, a consequence of our results is that the translational machinery can partly compensate for mutations in the G-string, permitting low level “ribosomal rescue” of the mutations and expression of some of the normal gene products. In the case of the drug-resistant mutant we have studied, it may be that this low level of TK expression was insufficient to activate ACV effectively but was sufficient for pathogenesis in a human, leading to selection of the mutant in the infected patient. Given that the G(7) string is a mutational hotspot, we predict that other ACV-resistant mutants associated with human disease will contain the same mutation. Study of other drug-resistant HSV tk mutants may identify different signals that allow relaxation of the constraints involved in reading frame maintenance.
Experimental Procedures
Plasmid Construction
pT3LacLuc was created by cloning the T3 promoter into the HindIII site of pBgalluc-1 (Reil et al. 1993). The BCH-1, BCH3–BCH19 plasmids were constructed by cloning synthetic oligonucleotides containing specific HSV sequence into the BglII and SalI sites of pT3LacLuc. BCH1 and BCH2 were constructed by cloning the SnaBI–SacI fragment from pTK-wt or pTK-4 into T4 DNA polymerase blunt-ended BglII and SalI sites of pT3LacLuc. (Plasmids TK-wt and TK-4 were described previously as pBH15 and pBH13 by Hwang et al. [1994]). Plasmid BCHSL− was created by digesting BCH4 with BglII and BamHI and religating. Oligonucleotides (TCGAG and GATCC) were cloned into SalI-BglII–digested pT3LacLuc to create BCH0. This construct, lacking HSV sequences, has luciferase in the +1 frame relative to β-galactosidase. Plasmids pBH13 and pBH15 have been previously described (Hwang et al. 1994). pTK-4a (laboratory plasmid BH17) was constructed by digesting pBH13 with SphI and SacI. The intervening 51 nt were replaced with identical oligonucleotide sequences, except that the G(8)AGGCTGGG motif was changed to GCTCACCATTCGCGAG. Insertion of a duplex of synthetic oligonucleotides with sequences 5′ GGCCTTCCTACAAGGGAAGGCCAGGGAGCT and 5′ CCCTGGCCTTCCCTTGTAGGAAGGCCAGCT into the SacI site of pBH13 created pTK-SL+ (laboratory plasmid BH13SL+). This plasmid contains the HIV stem–loop 10 nt downstream of the G-string.
Plasmids pBH19 and pBH21, for pausing experiments, were created by digesting pBH13 or pBH17 with EcoRV and SacI. The fragment were T4 DNA polymerase blunt-ended, gel-eluted, and cloned into the EcoRI (T4 DNA polymerase blunt-ended)–EcoRV sites of p911 (Digard et al. 1993). All plasmid constructs were verified by DNA sequencing.
In Vitro Transcription and Translation
In vitro transcription reactions were carried out as described previously (Hwang et al. 1994). Product RNA was recovered by phenol-chloroform extraction and ethanol precipitation in the presence of 2 M ammonium acetate. The RNA pellet was dissolved in water and checked for integrity by electrophoresis on 1% agarose gels containing 0.1% SDS.
In ribosomal frameshift assays, serial dilutions of purified RNAs were translated in the rabbit reticulocyte lysate system as previously described (Hwang et al. 1994). Frameshift efficiencies were calculated from β-galactosidase, and luciferase assays were performed exactly as described (33, 40) and frameshift efficiencies calculated as described in Results.
In ribosomal pausing assays, translations were carried out essentially as described by Somogyi et al. 1993. The translational inhibitor, edeine (5 μM final concentration), was added 5 min after the start of the reaction in order to obtain synchronous initiation. Aliquots (3 μl) were withdrawn at specific intervals, mixed with an equal volume of pancreatic RNase A (100 μg/ml) in 10 mM EDTA, and incubated at room temperature for 10 min. Laemmli's buffer (12.5% glycerol, 2% bromophenol blue, 25 mM Tris [pH 6.8], 100 mM dithiothreitol, and 2% SDS) was added to the samples prior to electrophoresis through SDS–12.5% (wt/vol) polyacryamide gels. The products were analyzed by autoradiography of dried gels.
Circular Dichroism
Samples (2 μM) were prepared by heating at 95°C for 15 min and cooled to room temperature, and circular dichroism spectra were recorded on an Aviv 62DS spectrophotometer (made available by Professor Stephen Harrison). Wavelength scans were recorded at 1 nm intervals (10 s averaging time), and three scans were averaged.
Polyacrylamide Gel Electrophoresis of Oligonucleotides
Native gel electrophoresis was performed in 20% polyacrylamide gels using 0.5 × TBE (0.45 M Tris–borate [pH 8.3], 1 mM EDTA buffer at 4°C [7.5 V cm−1]). Gels run with added salt contained salt in the gel as well as the running buffer. DNA samples were in 5 μl of TE plus salt at the same concentration as the gel and prior to electrophoresis were heated to 95°C for 20 min, cooled to room temperature, and mixed with 1 μl of 30% glycerol-containing marker dyes. Gels were dried and visualized by autoradiography.
Acknowledgements
We thank Paul Digard, Martha Kramer, Louane Hann, and Ian Brierley for their comments on the manuscript; Steve Harrison for allowing us to use his circular dichroism equipment; Charles Hwang and Steven Sacks for collaborating on earlier aspects of this project; and Senya Matsufuji for help on protein sequencing attempts. We also thank the Drug Synthesis and Chemistry Branch of the National Cancer Institute for the drug edeine. This work was supported by National Institutes of Health grant AI26126 to D. M. C. and by the German Ministry of Science and Technology.
References
- 1.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principals of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 2.Atkins J.F, Weiss R.B, Gesteland R.F. Ribosome gymnastics: degree of difficulty 9.5, style 10.0. Cell. 1990;62:413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balagurumoorthy P, Brahmachari S.M, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucl. Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battiste J.L, Tan R, Frankel A.D, Williamson J.R. Binding of an HIV Rev peptide to Rev response element RNA induces formation of purine–purine base pairs. Biochemistry. 1994;33:2741–2747. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- 5.Belcourt M.F, Farabaugh P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nt minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benhar I, Engleberg K.H. Frameshifting in the expression of E. coli. 1993;trpR(gene occurs by the bypassing of a segment of its coding sequence. Cell 72):121–130. doi: 10.1016/0092-8674(93)90056-v. [DOI] [PubMed] [Google Scholar]
- 7.Brierley I, Jenner A.J, Inglis S.C. Mutational analysis of the “slippery sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown D.G, Visse R, Sandhu G, Davies A, Rizkallah P.J, Melitz C, Summers W.C, Sanderson M.R. Crystal structure of the thymidine kinase from herpes simplex virus type-1 in complex with deoxythymidine and Ganciclovir. Nature Struct. Biol. 1995;2:876–881. doi: 10.1038/nsb1095-876. [DOI] [PubMed] [Google Scholar]
- 9.Coen D.M, Schaffer P.A. Two distinct loci confer resistance to acyloguanosine in herpes simplex virus type-1. Proc. Natl. Acad. Sci. USA. 1980;77:2265–2269. doi: 10.1073/pnas.77.4.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coen D.M, Kosz-Vnenchak M, Jacobson J.G, Leib D.A, Bogard C.L, Schaffer P.A, Tyler K.L, Knipe D.M. Thymidine kinase–negative herpes simplex mutant establish latency in mouse trigeminal ganglia but do not reactivate. Proc. Natl. Acad. Sci. USA. 1989;86:4735–4739. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran J.F. Analysis of effects of tRNA: message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucl. Acids Res. 1993;21:1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Digard P, Bebrin W.R, Weisshart K, Coen D.M. The extreme C-terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J. Virol. 1993;67:398–406. doi: 10.1128/jvi.67.1.398-406.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efstatiou S, Kemp S, Darby G, Minson A.C. The role of herpes simplex virus type-1 thymidine kinase in pathogenesis. J. Gen. Virol. 1989;70:869–879. doi: 10.1099/0022-1317-70-4-869. [DOI] [PubMed] [Google Scholar]
- 14.Englund J.A, Zimmerman M.E, Swierkosz E.M, Goodman J.L, Scholl D.R, Balfour H.H. Herpes simplex virus resistant to acyclovir: a study in a tertiary care center. Ann. Intern. Med. 1990;112:416–422. doi: 10.7326/0003-4819-76-3-112-6-416. [DOI] [PubMed] [Google Scholar]
- 15.Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farabaugh P.J, Zhao H, Vimaladithan A. A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fyfe J.A, Keller P.M, Furman P, Miller R.L, Elion G.B. Thymidine kinase from herpes simplex virus phosphorylates the new antiviral compound, 9-(2-hydroxyethoxymethy)guanine. J. Biol. Chem. 1978;253:8721–8727. [PubMed] [Google Scholar]
- 18.Gesteland R.F, Weiss R.B, Atkins J.F. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- 19.Grosjean H.J, de Henau S, Crothers D.M. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. USA. 1978;75:610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta R.C, Roe B.A, Randerath K. Sequence of human glycine transfer ribonucleic acid (anticodon CCC): determination by a newly developed thin-layer readout sequencing technique and comparison with other glycine transfer ribonucleic acids. Biochemistry. 1980;19:1699–1705. doi: 10.1021/bi00549a028. [DOI] [PubMed] [Google Scholar]
- 21.Haas J, Park E.-C, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 22.Herbst K.L, Nichols L.M, Gesteland R.F, Weiss R.B. A mutation in ribosomal protein L9 affects ribosomal hopping during translation of gene 60 from bacteriphage T4. Proc. Natl. Acad. Sci. USA. 1994;91:12525–12529. doi: 10.1073/pnas.91.26.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch M.S, Schooley R.T. Resistance to antiviral drugs: the end of innocence. N. Engl. J. Med. 1989;320:313–314. doi: 10.1056/NEJM198902023200510. [DOI] [PubMed] [Google Scholar]
- 24.Hwang C.B.C, Horsburgh B.C, Pelosi E, Roberts S, Digard P, Coen D.M. A net +1 frameshift permits synthesis of a thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc. Natl. Acad. Sci. USA. 1994;91:5461–5465. doi: 10.1073/pnas.91.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang C.B.C, Chen H.J. An altered spectrum of herpes simplex virus mutations mediated by an antimutator DNA polymerase. Gene. 1995;152:191–193. doi: 10.1016/0378-1119(94)00712-2. [DOI] [PubMed] [Google Scholar]
- 26.Jacks T, Madhani H.D, Masiarz F.R, Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobson J.G, Ruffner K.L, Kosz-Vnenchak M, Hwang C.B.C, Wobbe K, Knipe D.M, Coen D.M. Herpes simplex virus thymidine kinase and specific stages of latency in murine trigeminal ganglia. J. Virol. 1993;67:6903–6908. doi: 10.1128/jvi.67.11.6903-6908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalnik M.W, Norman D.G, Swann P.F, Patel D.J. Conformation of adenosine bulge–containing deoxytridecanucleotide duplexes in solution. J. Biol. Chem. 1989;264:3702–3712. [PubMed] [Google Scholar]
- 29.Kit S, Sheppard M, Ichimura H, Nusinoff-Lehrman S, Ellis M.N, Fyfe J.A, Otsuka H. Nucleotide sequence changes in thymidine kinase gene of herpes simplex virus type -2 clones from an isolate of a patient treated with acyclovir. Antimicrob. Agents Chemother. 1987;31:1483–1490. doi: 10.1128/aac.31.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollmus H, Honigman A, Panet A, Hauser H. The sequences of and distance between two cis-acting signals determine the efficiency of ribosomal frameshifting in human immunodeficiency virus type I and human T-cell leukaemia virus type II in vivo. J. Virol. 1994;68:6087–6091. doi: 10.1128/jvi.68.9.6087-6091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurland C.G. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 32.Larsen B, Wills N.M, Gesteland R.F, Atkins J.F. rRNA–mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeBowitz J.H, Coburn C.M, Beverley S.M. Simultaneous transient expression assays of the trypanosomid parasite Leishmania using β-galactosidase and β-glucuronidase as reporter enzymes. Gene. 1991;103:119–123. doi: 10.1016/0378-1119(91)90402-w. [DOI] [PubMed] [Google Scholar]
- 34.Lindsley D, Gallant J. On the directional specificity of ribosome frameshifting at a “hungry” codon. Proc. Natl. Acad. Sci. USA. 1993;90:5469–5473. doi: 10.1073/pnas.90.12.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins J.F, Gesteland R.F, Hayashi S. Autoregulatory frameshifting in decoding mammalian orthinine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pande S, Vimaladithan A.H.Z, Farabaugh P.J. Pulling the ribosome out of frame by +1 at a programmed frameshift site by cognate binding of aminoacly-tRNA. Mol. Cell. Biol. 1995;15:298–304. doi: 10.1128/mcb.15.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reil H, Kollmus H, Weidle U.H, Hauser H. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J. Virol. 1993;67:5579–5584. doi: 10.1128/jvi.67.9.5579-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rom E, Kahana C. Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frameshifting. Proc. Natl. Acad. Sci. USA. 1994;91:3959–3963. doi: 10.1073/pnas.91.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sacks S.L, Wanklin R.J, Reece D.E, Hicks K.A, Tyler K.L, Coen D.M. Progressive esophagitis from acyclovir-resistant herpes simplex: clinical roles for DNA polymerase mutant and viral heterogeneity? Ann. Intern. Med. 1989;111:893–899. doi: 10.7326/0003-4819-111-11-893. [DOI] [PubMed] [Google Scholar]
- 40.Schenborn E.T, Goiffen V. Note. Promega Notes. 1993;41:11. [Google Scholar]
- 41.Smith F.W, Feigon J. Quadraplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 42.Somogyi P, Jenner A.J, Brierley I, Inglis S.C. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sundquist W.I, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 44.Tu C, Tzeng T.-H, Bruenn J.A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl. Acad. Sci. USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vimaladithan A, Farabaugh P.J. Special peptidyl-tRNA molecules can promote translational frameshifting without slippage. Mol. Cell. Biol. 1994;14:8107–8116. doi: 10.1128/mcb.14.12.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss R.B, Dunn D.M, Atkins J.F, Gesteland R.F. Slippery runs, shifty stops, backward steps and forward hops: −2, −1, +1, +2, +5 and +6 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- 47.Weiss R.B, Dunn D.M, Atkins J.F, Gesteland R.F. Ribosomal frameshifting from −2 to +50 nucleotides. Prog. Nucl. Acids Res. Mol. Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- 48.Williamson J.R. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 49.Zimmerman S.B, Cohen G.H, Davies D.R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975;92:181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]