Abstract
A quantum dot-based lateral flow immunoassay system (QD-LFIAS) was developed to simultaneously detect both influenza A virus subtypes H5 and H9. Water-soluble carboxyl-functionalized quantum dots (QDs) were used as fluorescent tags. The QDs were conjugated to specific influenza A virus subtype H5 and H9 antibodies via an amide bond. When influenza A virus subtype H5 or H9 was added to the QD-LFIAS, the QD-labeled antibodies specifically bound to the H5 or H9 subtype viruses and were then captured by the coating antibodies at test line 1 or 2 to form a sandwich complex. This complex produced a bright fluorescent band in response to 365 nm ultraviolet excitation. The intensity of fluorescence can be detected using an inexpensive, low-maintenance instrument, and the virus concentration directly correlates with the fluorescence intensity. The detection limit of the QD-LFIAS for influenza A virus subtype H5 was 0.016 HAU, and the detection limit of the QD-LFIAS for influenza A virus subtype H9 was 0.25 HAU. The specificity and reproducibility were good. The simple analysis step and objective results that can be obtained within 15 min indicate that this QD-LFIAS is a highly efficient test that can be used to monitor and prevent both Influenza A virus subtypes H5 and H9.
Keywords: Influenza A virus, Immunoassay, Quantum dots
Highlights
-
•
A QD-LFIAS was developed using quantum dots as fluorescent tags.
-
•
This QD-LFIAS can detect both H5 and H9 virus in a single test.
-
•
QD-LFIAS is ultrasensitive and can yield objective results in 15 min.
-
•
This QD-LFIAS is efficiency for both H5 and H9 virusmonitoring and prevention.
1. Introduction
Influenza can occur in pandemics and localized outbreaks. Avian influenza viruses, which are known as highly pathogenic avian influenza (HPAI) virus, cause serious illness and death in domestic poultry. Outbreaks of HPAI have led to human infections, some of which resulted in deaths (Chan, 2002, Yuan et al., 2013). Influenza A viruses can be subdivided into several subtypes based on the antigenic nature of the surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) (Hinshaw et al., 1982). Human infections with influenza A virus subtypes H5, H7 and H9 cause severe respiratory disease (Chan, 2002, Koopmans et al., 2004, Lin et al., 2000). For example, H5 subtype causes infection and mortality rates of up to 60% (Guzelian et al., 1996, Xiong et al., 2013), whereas the H9 subtype (mainly the H9N2 subtype) has been reported in domestic fowl and wild birds in various regions and is considered to be one of the most likely candidates to cause a new influenza pandemic in humans (Yuan et al., 2013).
The HPAI virus is continually being isolated from live-bird markets in multiple countries and consequently a serious threat to individual and global health (Pepin et al., 2013). Therefore, the epidemiology of this virus needs to be studied, and potential sources of exposure to the pathogen need to be identified. To date, different types of methods have been used to detect influenza A viruses. These methods are categorized based on the type of detection target: nucleic acid-based detection, virus isolation and identification, antigen detection, antibody detection, etc. (Alberini et al., 2009; Chen et al., 2008; Ho et al., 2009; Moore et al., 2010; Payungporn et al., 2006; Xie et al., 2006; Yang et al., 2008) The most commonly used detection methods are nucleic acid-based detection, such as reverse-transcription PCR, Real-time PCR, RT-LAMP, antibody detection and antigen detection (Antarasena et al., 2007; Deng et al., 2011; He et al., 2007; Kang et al., 2010; Khurana et al., 2011; Shahsavandi et al., 2011; Yea et al., 2010; Zhao et al., 2010). However, most of these methods require specialized equipment and highly trained operators, and these detection processes are time consuming, thereby making them impractical for point-of-care (POCT) detection (Yager et al. 2008). Thus, a sensitive, portable and rapid technology needs to be developed for influenza A virus (including different subtypes) surveillance and prevention.
Quantum dots (QDs) have been broadly used for biological labeling and imaging due to their unique optical properties, i.e., a broad excitation spectrum and a narrow and symmetric emission peak (Lu et al., 2011, Medintz et al., 2005, Zhou et al., 2011b). Therefore, QDs have been successfully utilized for rapid diagnosis. Specifically, QD immune-labeling, which is highly sensitive, quantitative, and reproducible technology that features a wide linear detecting range, has been developed to detect pathogens, characteristic proteins and even nucleic acids (Li et al., 2012; Liu et al., 2013; Sapountzi et al., 2015; Shen et al., 2011b; Shen et al., 2012; Zhou et al., 2011a).
In this study, we developed a QDs-based lateral flow immunoassay system (QD-LFIAS) to simultaneously detect influenza A virus subtypes H5 and H9. We synthesized water-soluble carboxyl-functionalized QDs and covalently bound them to influenza A virus antibodies. We selected subtypes H5 and H9 as the target pathogen and evaluated the detection time and sensitivity; we also measured the detection specificity and reproducibility of the QD-LFIAS.
2. Experimental
2.1. Materials
Cadmium oxide (CdO, 99.99%), zinc oxide (ZnO, 99.9%, powder), sulfur (S, 99.98%, powder), selenium (Se, 99.99%, powder), 1-octadecene (ODE, 90%), oleic acid (OA, 90%), and 2-(N-morpholino) ethanesulfonic acid (MES) were purchased from Sigma-Aldrich (USA). Sodium phosphate dibasic (Na2HPO4, 99%), sodium phosphate monobasic monohydrate (NaH2PO4·H2O, 98–102.0%), Tween-20, and bovine serum albumin (BSA) were purchased from Shanghai Sangon Ltd. (China). 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide (Sulfo-NHS) were purchased from Thermo Fisher Scientific (USA). Goat anti-mouse IgG antibody was purchased from Arista Biologicals (USA), and Influenza A virus subtype H5 and H9 antibodies (mAb IgG) were obtained from the Life Science Division of Tsinghua University. Influenza A virus subtype H9 antibody (coating, IgY) was donated by the Shenzhen Entry–Exit Inspection and Quarantine Bureau (China). Test antigens for influenza A subtype viruses and other HI respiratory tract viruses were purchased from Harbin Weike Biotechnology Development Company (China). All reagents were of analytical grade and were used as received without further purification.
2.2. Synthesis of water-soluble carboxyl-functionalized QDs
High-quality, hydrophobic, red (PL 620 nm)-emitting core–shell CdSe/ZnS QDs were prepared as described previously (Shen et al., 2009). Briefly, oleic acid (OA) was used as the hydrophobic capping agent. The as-synthesized QDs were precipitated in acetone followed by methanol and ultimately re-dissolved in methylbenzene prior to further treatment. Amphiphilic oligomer (polymaleic acid n-hexadecanolester, PMAH) was used to prepare water-soluble QDs. The PMAH have hydrophobic chains available for anchoring the hydrophobic CdSe/ZnS QDs and free carboxylic acid groups available for further surface modification. The hydrophobic QDs and PMAH bound to each other via the phase-transfer stem to form PMAH-stabilized PL QDs (Liu et al., 2013; Shen et al., 2011a , 2011c; Zhou et al., 2010). Briefly, 5.78 g of PMAH (0.32 mmol) was dissolved in 20 mL of chloroform. Subsequently, 2 mL of CdSe/Zns QDs (0.04 mmol, dispersed in chloroform) was added to the PMAH solution, which was then stirred for 24 h (room temperature) in a closed container. The chloroform was then slowly evaporated by rotary evaporation, and the remaining QD film was dispersed in ammonia water (pH 9.0) with sonication. The solutions were passed through a 0.22 μm Nylon syringe filter, washed, and then centrifuged four times at 20,000g to remove excess oligomers. The resulting PMAH-coated carboxyl-functionalized QDs (PMAH-QDs) readily redispersed into water.
The size of PMAH-QDs was characterized by transmission electron microscopy (TEM, Tecnai G2 Spirit, FEI) and dynamic light scattering (DLS, NanoZS 90, Malvern).
2.3. Antibodies functionalization of PMAH-QDs
The PMAH-QDs were conjugated to influenza A virus subtype H5 and H9 antibodies via an amide bond. To form an amine-reactive sulfo-NHS ester, 2 mg of the PMAH-QDs was mixed with 2 mM NHS and 5 mM EDC in 0.1 M MES-buffered saline at pH 4.7. After washing and centrifugation, the particles were dispersed in 50 mM borate buffer (pH=8.5). Subsequently, 0.08 mg of H5 or H9 antibody was added to the QDs. The solution was then incubated for 3 h at 37 °C, which resulted in the formation of a stable amide bond between the antibody and the PMAH-QDs. Residual active coupling sites were blocked by adding 5% BSA solution and incubating the mixture at 37 °C for 30 min. The antibody-functionalized QDs (QDs–Ab) were washed and centrifuged three times at 20,000g and stored at 4 °C before use.
2.4. Preparation of QD-LFIAS
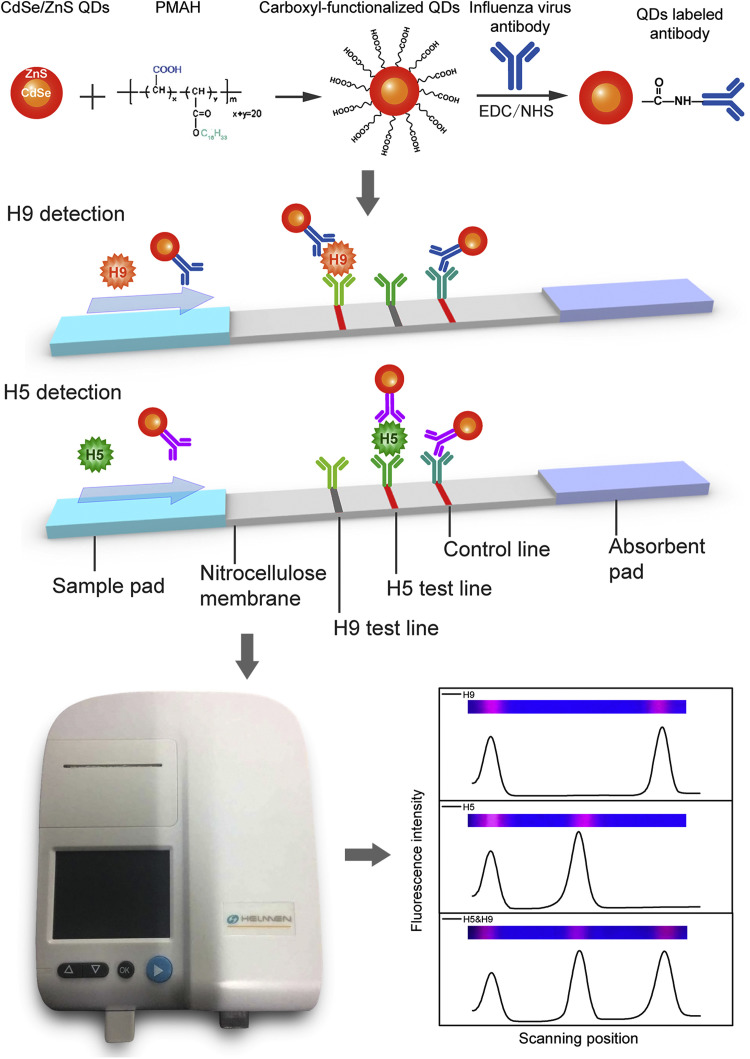
H5 and H9 coating antibodies were diluted in 20 mM sodium phosphate buffer (PBS) and striped at 2 mg/mL onto the nitrocellulose membrane (Hi Flow plus HF13504, Millipore Corporation) to generate test lines 1 and 2, respectively. Goat anti-mouse IgG antibodies were diluted in PBS and striped at 1 mg/mL onto the nitrocellulose membrane as the control line. These reagents were dispensed onto membrane using the XYZ Dispensing System (BioDot Inc., Irvine, CA). The striped nitrocellulose membranes were dried at 37 °C for 4 h in a vacuum oven. The sample pad was saturated with PBS containing BSA (1%, w/v) and Tween-20 and dried at 37 °C for 3 h in a vacuum oven. The standard configuration of the QD-LFIAS is shown in Fig. 1. The completed assay was cut into individual 3.5 mm strips using a CM4000 Guillotine Cutter (BioDot Inc., Irvine, CA). Each strip was incorporated into a plastic housing to facilitate the detection of the fluorescence intensity due to 365 nm ultra violet excitation using a fluorescence test strip scanner (Hangzhou He Mai Technology Co., Ltd.).
Fig. 1.
Schematic representation of the QD-LFIAS.
2.5. Analytical procedure
Sixty microliters of sample was mixed with 2 μL of H5 and H9 QD-Abs and then added onto the sample pad of the QD-LFIAS strip. Once the influenza A virus subtype H5 or H9 was added to the sample, the QD-labeled antibodies specifically bound the H5 or H9 subtype viruses and were then captured by the coating antibodies at test line 1 or 2 to form a sandwich complex; QD-labeled antibodies that were not bound to the H5 or H9 subtype virus were captured by the goat anti-mouse IgG antibodies at the control line. In the absence of influenza A virus subtype H5 or H9 in the sample, the QD-labeled antibodies were not captured by the coating antibodies at test line 1 or 2 but were only captured by the goat anti-mouse IgG antibodies at the control line. When only virus and antibodies were added onto the strip, the coating antibodies capture the complex well, but neither the test line nor the control line will show fluorescence signal. The captured QDs produced a bright fluorescent band in response to 365 nm ultraviolet excitation. The fluorescence signals from the captured QDs were scanned by a fluorescence test strip scanner. The fluorescence intensity is directly proportional to the amount of QD particles and virus complex on the test line of the strip, whereas the lowest fluorescence signal intensity closely correlates with the detection limit of the QD-LFIAS. The detection time was tested by adding H5 and H9 antigens at 16 hemagglutinating units (HAUs) onto the sample pad of the QD-LFIAS strip, which was scanned once per minute from 3 to 50 min by the fluorescence test strip scanner. Fifty negative samples were detected by QDs-LFIAS; the average signal value was calculated, and double this value was defined as the cutoff value. Specifically, the cutoff value for the H5 and H9 test lines were 205 a.u. and 170 a.u., respectively. Fluorescence intensities above and below these values were classified as positive and negative, respectively, for the respective tests. The limit of detection (LOD) of this system was identified by detecting samples containing various amounts of the H5 and H9 antigens. Briefly, high-concentration solutions of H5 and H9 antigens (128 HAU) were two-fold diluted with 20 mM PBS (64, 32, 16, 8, 4, 2, 1, 1/2, 1/4, 1/8, 1/16, 1/32, 1/64 and 1/128 HAU) and individually added onto the QD-LFIAS; 20 mM PBS was added as a negative control. The signals were detected by fluorescence test strip scanner. The LOD was then calculated accordingly. The LOD was defined as the minimal concentration that produced the lowest signal exceeding the cutoff value. The reproducibility of this QDs-LFIAS was tested by detecting 20 replicates of the H5 and H9 antigens at different concentrations (32, 4 and 1/2 HAU). The relative standard deviations were then calculated accordingly. The specificity of the QD-LFIAS was examined using influenza A subtype viruses (H1, H3, H5N1 re-4/6, H7N9, H9N2 re-2, and H9 SD696) and other respiratory tract viruses (IBV H52, IBDV Gt, and NDV La Sota) HI test antigens. Each type of the antigen was diluted to 64 HAU with PBS. Sixty microliters of antigen solution of each type was then added individually added onto the QD-LFIAS, followed by fluorescence test strip scanner detection.
2.6. Sample test
One hundred and forty-seven samples, including six H5N1-positive samples and eleven H9N2-positive samples collected and preserved by the Shenzhen Entry–Exit Inspection and Quarantine Bureau, were detected using QD-LFIAS. The avian cloacal swab samples were diluted in 500 μL of PBS solution with antibiotics, and sixty microliters of the dilution was then mixed with 2 μL of H5 and H9 QDs–Ab and added onto the QD-LFIAS. Serum samples were mixed with 2 μL of H5 and H9 QDs–Ab and directly added onto the QD-LFIAS. The QD-LFIAS were then detected by a fluorescence test strip scanner. In this study, a real-time PCR assay was utilized as a reference method to assess the accuracy of QDs- LFIAS.
3. Results and discussion
3.1. Formation of water-soluble carboxyl-functionalized QDs and labeled antibodies
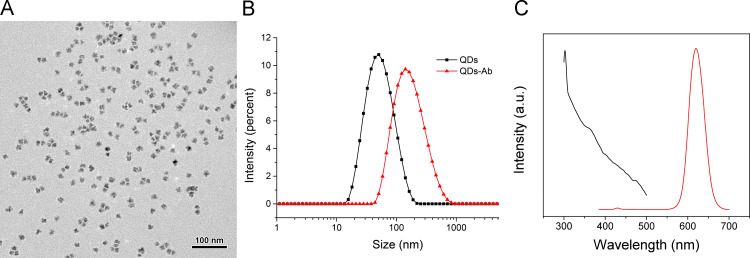
We have previously thoroughly described water-soluble CdSe/ZnS PMAH-QDs and their reaction mechanism (Shen et al., 2011a, Shen et al., 2011b, Shen et al., 2012). Based on the TEM characterization ( Fig. 2A), the PMAH-QDs presented a narrow size distribution and high monodispersity, with an average size at approximately 25 nm. The dynamic light scattering analysis in Fig. 2B shows that the hydrodynamic size of PMAH-QDs before conjugation is 46.7 nm, and this size increased to 145 nm after conjugation with antibodies, which indicates the successful conjugation of antibodies. We also inspected the fluorescence properties of the QDs, and the results showed that absorption was maximized at 365 nm, whereas emission was maximized at ~620 nm; these peaks were approximately symmetric (Fig. 2C). Furthermore, little overlap was observed between the absorption and emission spectrum, indicating excellent fluorescence properties.
Fig. 2.
Characterization of PMAH-QDs. (A) TEM result and (B) DLS data of PMAH-QDs and antibody-functionalized QDs. (C) Fluorescent absorption and emission of PMAH-QDs.
3.2. Performance analysis of QDs-LFIAS detection
The main advantage of a lateral flow immunoassay system is the integration of sample pre-treatment, separation, reaction and detection in one strip. However, quantifying the resultant signal is usually difficult, which restricts the utility of these assays. In this study, samples were mixed with QDs–Ab, and both H9 (or H5) subtype viruses in the sample and QDs–Ab moved forward to the absorbent pad when added to the sample pad of the strip and were captured by the immobilized capture antibodies in the test line to form a sandwich structure (Fig. 1); the fluorescence signals of QDs were measured by a handheld device (Fig. 1). Compared with the broad application of gold nanoparticle-based lateral flow immunoassay, our approach quantified the fluorescence intensity of bound QDs on test line (Fig. 1) and consequently was able to measure the related virus concentration.
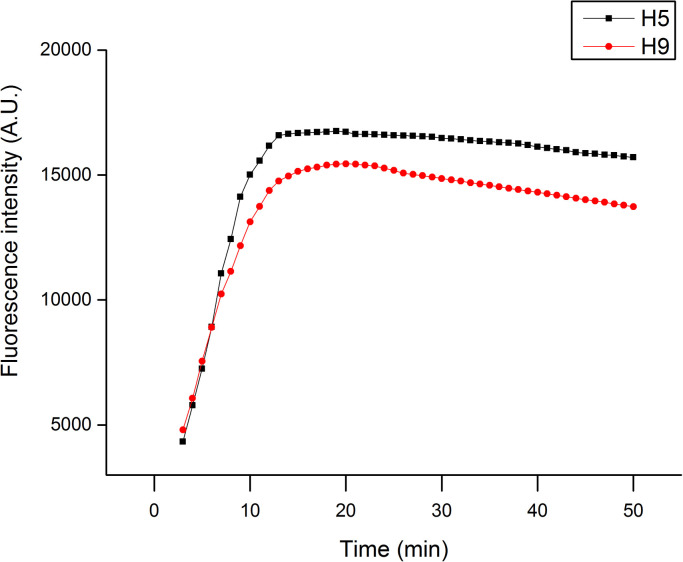
We first evaluated the duration of QD-LFIAS detection. H5 and H9 subtype viruses with concentrations of 16 HAU were added to the sample pad, and the fluorescence of QDs was measured every minute from 3 to 50 min. Fig. 3 shows that the fluorescence intensity of QDs from both the H9 and H5 samples dramatically increased from 3 min to 15 min, but this decrease was attenuated as the detection time was extended further and the fluorescence signal ultimately reached a plateau. Therefore, the optimum detection time is 15 min after the addition of the sample. General PCR-based influenza virus detection requires several hours to complete the procedure, whereas the QD-LFIAS can be completed in less than 20 min.
Fig. 3.
Time performance of QD-LFIAS at a HAU concentration of 16 for H5 and H9.
3.3. Limit of detection of QDs-LFIAS
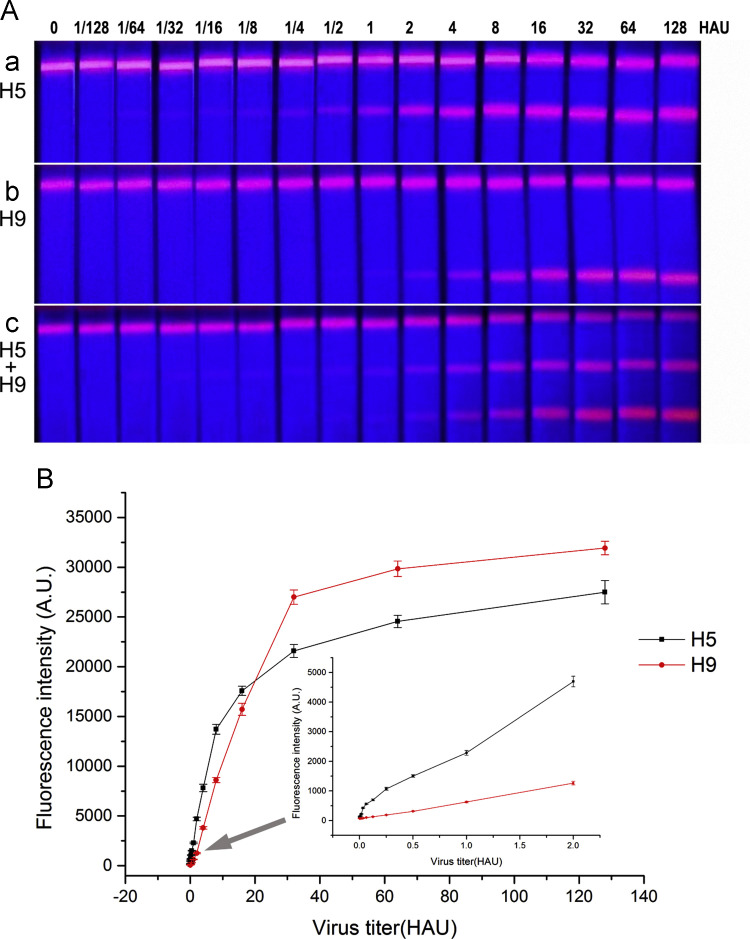
To evaluate the LOD of the QD-LFIAS for the detection of the influenza A virus H5 and H9 subtypes, the QD-LFIAS was used to detect three replicate samples with virus titers ranging from 1/128 to 128 HAU. Fifteen minutes after the addition of the samples, the signals were scanned using a fluorescence test strip scanner. As demonstrated in Fig. 4A, the virus concentration directly correlated with the fluorescence intensity on the test lines. The fluorescence from test line of H5 at 0.125 (1/8) HAU remained visible with the naked eye, whereas the lowest concentration of H9 detected with the naked eye was 0.5 (1/2) HAU; however, the handheld device was able to detect low titers of both the H5 and H9 virus subtypes. As show in Fig. 4B, the QD-LFIAS was able to detect influenza A virus subtype H5 at a concentration of 1/64 HAU or higher because the resultant fluorescence intensities were higher than the cutoff value (205), implying that the LOD of this method for H5 was 1/64 HAU. For the detection of influenza A virus subtype H9, the QD-LFIAS was able to detect a minimum concentration of 1/4 HAU because the resultant fluorescence intensity was higher than the cutoff value (170), implying that the LOD of this method for H9 was 1/4 HAU. Most importantly, the signals did not interfere with each other when simultaneously analyzing H5 and H9 on a same strip. Thus, this assay was able to simultaneously detect two subtypes of type A influenza virus on one strip with high sensitivity. In order to measure non-specific adsorption, 20 mM PBS was striped onto the nitrocellulose membrane as the test line 1 and line 2, 1 mg/mL goat anti-mouse IgG was striped as the control line, then assemble them into strips. H5 and H9 antigens at different concentrations were tested with those strips, The results showed very low fluorescence intensity was detected at the test line 1 and line 2 (data not show here). This proved that the fluorescence intensity variation was due to the interaction between the immobilized antibodies and the virus.
Fig. 4.
(A) Images of tested QD-LFIAS in response to excitation with 365 nm ultraviolet light. (a) Strips containing samples of influenza A virus subtype H5. (b) Strips containing samples of influenza A virus subtype H9. (c) Strips containing mixed samples of influenza A virus subtype H5 and H9. (B) Fluorescence intensity scans at different concentrations of H5 and H9 measured by the fluorescence strip scanning device.
3.4. Reproducibility of QDs-LFIAS
The reproducibility of this QD-LFIAS was test by detecting the signals produced by 20 replicates of different concentrations of the H5 and H9 antigens (32, 4 and 0.5 HAU). The relative standard deviations (RSD) were then calculated. Table 1 shows that the RSD values are less than 10%, which demonstrates that the QD-LFIAS is reproducible.
Table 1.
Reproducibility QD-LFIAS.
| Samples | Concentrations (HAU) | Fluorescence intensity average (a.u.) | Std. dev | RSD/% n=20 |
|---|---|---|---|---|
| H5 | 0.5 | 1573.5 | 91.4 | 5.8 |
| 4 | 7603.5 | 411.2 | 5.4 | |
| 32 | 22,131 | 976.5 | 4.4 | |
| H9 | 0.5 | 336.2 | 23.3 | 6.9 |
| 4 | 3715.5 | 214.3 | 5.8 | |
| 32 | 26,473 | 1139.6 | 4.3 |
3.5. Specificity of QDs-LFIAS
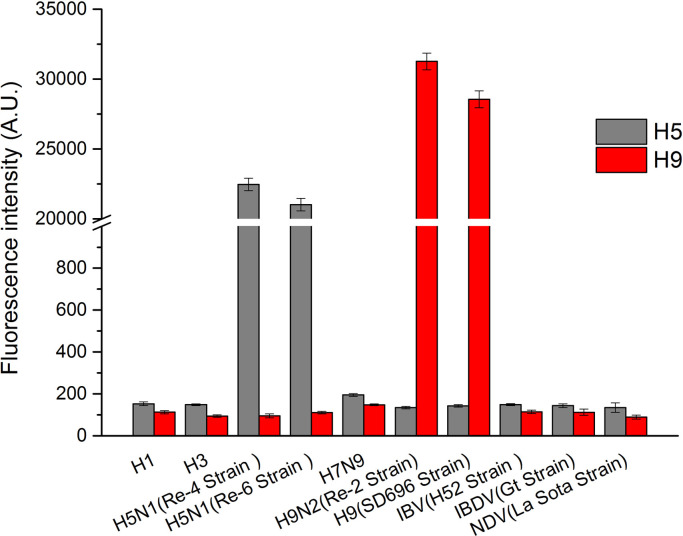
The specificity of the QD-LFIAS was evaluated using other subtypes of type A influenza viruses (H1, H3, H5N1 re-4/6, H7N9, H9N2 re-2, and H9 SD696) and HI respiratory tract viruses test antigens (IBV H52, IBDV Gt, and NDV La Sota). The concentrations of all of these HI test antigens were maintained at a relatively high level (64 HAU). Fig. 5 shows that at the same virus titer, the fluorescent signal from the H5 and H9 QD-LFIAS tests reached ultrahigh levels (20,000–25,000 for H5 subtypes and 28,000–32,000 for H9 subtypes), whereas those of the other viruses were below the cutoff value. This result demonstrated that the QD-LFIAS does not cross-react with other type A influenza viruses and respiratory tract viruses.
Fig. 5.
Cross-reactivity tests of influenza subtype viruses and other respiratory tract viruses.
3.6. Sample tests
One hundred and forty-seven samples, including six H5N1-positive samples and eleven H9N2-positive samples collected and preserved by the Shenzhen Entry–Exit Inspection and Quarantine Bureau, were detected using QD-LFIAS. A real-time PCR assay was utilized as a reference method to assess the accuracy of the QD-based LFIAS. Table 2 shows that the results obtained via either QD-LFIAS or real-time PCR matched 100%, suggesting that QD-LFIAS accurately detects the influenza A virus H5 and H9 subtypes. As a preliminary screening method, QD-LFIAS was able to rapidly analyze samples with a high accuracy. Thus, it is a less time-consuming method for influenza A virus H5 and H9 subtype monitoring than PCR and does not require the purchase of expensive equipment.
Table 2.
Sample tests using QD-LFIAS or real-time PCR.
| Samples | Number of samples | QDs-LFIAS result (P/N) | Real-time PCR result (P/N) |
|---|---|---|---|
| Avian cloacal swab | 100 | 0/100 | 0/100 |
| Human serum | 30 | 0/30 | 0/30 |
| H5 positive sample | 6 | 6/0 | 6/0 |
| H9 positive sample | 11 | 11/0 | 11/0 |
3.7. Advantages of QDs-LFIAS
The above results demonstrate the high sensitivity and specificity of the QD-LFIAS. Furthermore, this assay is easy to conduct and rapidly produces results, making it a promising candidate for preliminary screening. The assay consists of only one step: adding the sample and the QDs–Ab mixtures onto the QDs-LFIAS strip and checking the result presented by the equipment in 15 min, which eliminates the need for professional laboratory technicians and expensive equipment and permits high-throughput screening. Alternatively, nucleic acid-based detection methods were proved to be highly efficient, specific and accurate detection methods for viruses (Deng et al., 2011; Grabowska et al., 2014; Lai et al., 2012; Payungporn et al., 2006; Zhao et al., 2010). However, nucleic acid isolation requires a clean environment and professional technicians and usually requires multiple steps. Virus isolation and identification, the golden standard of virus detection, requires special serum and complex procedures. Traditional POCT methods, such as colloidal gold immune chromatography strips, relies on color depth and is consequently associated with subjective judgment error. Furthermore, the complexity of the enzyme-linked immunosorbent assay (ELISA) may affect the detection accuracy. Conversely, the objectivity of the QD-LFIAS avoids the subjectivity associated with detecting color depth with the human eye and can detect an unobservable signal in just one simple step. Because the fluorescence immunoassay was more sensitive than ELISA, its use is promising in POCT diagnosis (Li et al., 2012).
According to the above results, the QD-LFIAS can be used to detect both influenza A virus subtypes H5 and H9; the prudent selection of the labeling and capturing antibody-pair prevent conflict when using this system to simultaneously detect the two virus subtypes. Therefore, the QD-LFIAS can be used to diagnose influenza caused by H5 and H9 subtype viruses with high sensitivity.
Given the recent boom in social networking, an epidemic infectious disease, such Middle East respiratory syndrome (MERS), may induce a global panic (Kupferschmidt, 2015). Therefore, convenient but reliable diagnostic technologies and devices need to be developed. Our approach, which harnesses a hand-held device to measure a fluorescent signal from strips exposed to different pathogens, meets this need and may be utilized for in-home healthcare. Thus, QD-LFIAS can be used for pathogen surveillance and relieving public concern.
4. Conclusions
In the current study, a sensitive QD-LFIAS for the simultaneous detection of influenza A virus subtypes H5 and H9 was developed. This assay was able to rapidly analyze the sample in one simple step and yielded objective results via analysis on an inexpensive, low-maintenance instrument within 15 min. The LODs of QD-LFIAS for the detection of influenza A virus subtypes H5 and H9 were 0.016 HAU and 0.25 HAU, respectively. Moreover, these two virus subtypes can be simultaneously detected on only one LFIAS strip without interferences. The specificity and reproducibility were both good, and the accuracy was as high as that of real-time PCR. By changing the conjugation and capturing antibody pairs, this system can be conveniently extended to detect other types of pathogens. Overall, the QD-LFIAS is a sensitive method to rapidly detect influenza A virus subtypes H5 and H9 and may be used for diagnosis in field tests.
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 Program, No. 2013AA032204).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bios.2015.10.002.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- Alberini I., Del Tordello E., Fasolo A., Temperton N.J., Galli G., Gentile C., Montomoli E., Hilbert A.K., Banzhoff A., Del Giudice G., Donnelly J.J., Rappuoli R., Capecchi B. Vaccine. 2009;27(43):5998–6003. doi: 10.1016/j.vaccine.2009.07.079. [DOI] [PubMed] [Google Scholar]
- Antarasena C., Sirimujalin R., Prommuang P., Promkuntod N., Prommuang P., Blacksell S.D. Res. Vet. Sci. 2007;83(2):279–281. doi: 10.1016/j.rvsc.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Chan P.K.S. Clin. Infect. Dis. 2002;34:S58–S64. doi: 10.1086/338820. [DOI] [PubMed] [Google Scholar]
- Chen H.T., Zhang J., Sun D.H., Ma L.N., Liu X.T., Cai X.P., Liu Y.S. J. Virol. Methods. 2008;151(2):200–203. doi: 10.1016/j.jviromet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Deng M.J., Xiao X.Z., Zhang Y.M., Wu X.H., Zhu L.H., Xin X.Q., Wu D.L. Mol. Biol. Rep. 2011;38(3):1941–1948. doi: 10.1007/s11033-010-0315-8. [DOI] [PubMed] [Google Scholar]
- Grabowska I., Stachyra A., Gora-Sochacka A., Sirko A., Olejniczak A.B., Lesnikowski Z.J., Radecki J., Radecka H. Biosens. Bioelectron. 2014;51:170–176. doi: 10.1016/j.bios.2013.07.026. [DOI] [PubMed] [Google Scholar]
- Guzelian A.A., Katari J.E.B., Kadavanich A.V., Banin U., Hamad K., Juban E., Alivisatos A.P., Wolters R.H., Arnold C.C., Heath J.R. J. Phys. Chem. 1996;100(17):7212–7219. [Google Scholar]
- He Q., Velumani S., Du Q., Lim C.W., Ng F.K., Donis R., Kwang J. Clin. Vaccine Immunol. 2007;14(5):617–623. doi: 10.1128/CVI.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw V.S., Air G.M., Gibbs A.J., Graves L., Prescott B., Karunakaran D. J. Virol. 1982;42(3):865–872. doi: 10.1128/jvi.42.3.865-872.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H.T., Qian H.L., He F., Meng T., Szyporta M., Prabhu N., Prabakaran M., Chan K.P., Kwang J. Clin. Vaccine Immunol. 2009;16(5):726–732. doi: 10.1128/CVI.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X.P., Jiang T., Li Y.Q., Lin F., Liu H., Chang G.H., Zhu Q.Y., Qin E.D., Qin C.F., Yang Y.H. Virol. J. 2010;7:113. doi: 10.1186/1743-422X-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Sasono P., Fox A., Nguyen V.K., Le Q.M., Pham Q.T., Nguyen T.H., Nguyen T.L., Horby P., Golding H. J. Virol. 2011;85(23):12455–12463. doi: 10.1128/JVI.06023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M., Wilbrink B., Conyn M., Natrop G., van der Nat H., Vennema H., Meijer A., van Steenbergen J., Fouchier R., Osterhaus A., Bosman A. Lancet. 2004;363(9409):587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. Science. 2015;348(6240):1183–1184. doi: 10.1126/science.348.6240.1183. [DOI] [PubMed] [Google Scholar]
- Lai W.A., Lin C.H., Yang Y.S., Lu M.S. Biosens. Bioelectron. 2012;35(1):456–460. doi: 10.1016/j.bios.2012.02.049. [DOI] [PubMed] [Google Scholar]
- Li X., Lu D., Sheng Z., Chen K., Guo X., Jin M., Han H. Talanta. 2012;100:1–6. doi: 10.1016/j.talanta.2012.08.041. [DOI] [PubMed] [Google Scholar]
- Lin Y.P., Shaw M., Gregory V., Cameron K., Lim W., Klimov A., Subbarao K., Guan Y., Krauss S., Shortridge K., Webster R., Cox N., Hay A. Proc. Natl. Acad. Sci. USA. 2000;97(17):9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Wu F., Zhou C., Shen H., Yuan H., Du Z., Ma L., Li L.S. Sens. Actuators B: Chem. 2013;186:235–243. [Google Scholar]
- Lu Z., Zhu Z., Zheng X., Qiao Y., Guo J., Li C.M. Nanotechnology. 2011;22(15):155604. doi: 10.1088/0957-4484/22/15/155604. [DOI] [PubMed] [Google Scholar]
- Medintz I.L., Uyeda H.T., Goldman E.R., Mattoussi H. Nat. Mater. 2005;4(6):435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Moore C., Telles J.N., Corden S., Gao R.B., Vernet G., Van Aarle P., Shu Y.L. J. Virol. Methods. 2010;170(1–2):173–176. doi: 10.1016/j.jviromet.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Payungporn S., Chutinimitkul S., Chaisingh A., Damrongwantanapokin S., Buranathai C., Amonsin A., Theamboonlers A., Poovorawan Y. J. Virol. Methods. 2006;131(2):143–147. doi: 10.1016/j.jviromet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Pepin K.M., Wang J., Webb C.T., Hoeting J.A., Poss M., Hudson P.J., Hong W., Zhu H., Guan Y., Riley S. PLoS One. 2013;8(2):e56157. doi: 10.1371/journal.pone.0056157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapountzi E.A., Tragoulias S.S., Kalogianni D.P., Ioannou P.C., Christopoulos T.K. Anal. Chim. Acta. 2015;864:48–54. doi: 10.1016/j.aca.2015.01.020. [DOI] [PubMed] [Google Scholar]
- Shahsavandi S., Salmanian A.H., Ghorashi S.A., Masoudi S., Fotouhi F., Ebrahimi M.M. J. Virol. Methods. 2011;171(1):260–263. doi: 10.1016/j.jviromet.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Shen H., Wang H., Tang Z., Niu J.Z., Lou S., Du Z., Li L.S. CrystEngComm. 2009;11(8):1733. [Google Scholar]
- Shen H., Wang H., Zhou C., Niu J.Z., Yuan H., Ma L., Li L.S. Dalton Trans. 2011;40(36):9180–9188. doi: 10.1039/c1dt10865d. [DOI] [PubMed] [Google Scholar]
- Shen H., Yuan H., Niu J.Z., Xu S., Zhou C., Ma L., Li L.S. Nanotechnology. 2011;22(37):375602. doi: 10.1088/0957-4484/22/37/375602. [DOI] [PubMed] [Google Scholar]
- Shen H., Yuan H., Wu F., Bai X., Zhou C., Wang H., Lu T., Qin Z., Ma L., Li L.S. J. Mater. Chem. 2012;22(35):18623. [Google Scholar]
- Shen H., Zhou C., Xu S., Yu C., Wang H., Chen X., Li L.S. J. Mater. Chem. 2011;21(16):6046–6053. [Google Scholar]
- Xie Z., Pang Y.S., Liu J., Deng X., Tang X., Sun J., Khan M.I. Mol. Cell. Probes. 2006;20(3–4):245–249. doi: 10.1016/j.mcp.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Xiong W.W., Yang G.H., Wu X.C., Zhu J.J. ACS Appl. Mater. Interfaces. 2013;5(16):8210–8216. doi: 10.1021/am402328t. [DOI] [PubMed] [Google Scholar]
- Yager P., Domingo G.J., Gerdes J. Annu. Rev. Biomed. Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- Yang S.Y., Chieh J.J., Wang W.C., Yu C.Y., Lan C.B., Chen J.H., Horng H.E., Hong C.Y., Yang H.C., Huang W. J. Virol. Methods. 2008;153(2):250–252. doi: 10.1016/j.jviromet.2008.07.025. [DOI] [PubMed] [Google Scholar]
- Yea C., Petric M., Pasick J., Tellier R. Mol. Cell. Probes. 2010;24(6):364–369. doi: 10.1016/j.mcp.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Yuan Z., Zhu W., Chen Y., Zhou P., Cao Z., Xie J., Zhang C., Ke C., Qi W., Su S., Zhang G. Microb. Pathog. 2013;64:39–42. doi: 10.1016/j.micpath.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Zhao J., Tang S., Storhoff J., Marla S., Bao Y.P., Wang X., Wong E.Y., Ragupathy V., Ye Z., Hewlett I.K. BMC Biotechnol. 2010;10:74. doi: 10.1186/1472-6750-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Shen H., Guo Y., Xu L., Niu J., Zhang Z., Du Z., Chen J., Li L.S. J. Colloid Interface Sci. 2010;344(2):279–285. doi: 10.1016/j.jcis.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Zhou C., Yuan H., Shen H., Guo Y., Li X., Liu D., Xu L., Ma L., Li L.S. J. Mater. Chem. 2011;21(20):7393. [Google Scholar]
- Zhou C., Yuan H., Shen H., Guo Y., Li X., Liu D., Xu L., Ma L., Li L.S. J. Mater. Chem. 2011;21(20):7393–7400. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material