Abstract
BackgroundDetection and characterization of viral RNA pathogens from fieldwork are challenging due to the instability of the RNA molecule. FTA cards® have proved useful for sample storage and latter identification of pathogens with importance for agricultural, animal and human health: however, for optimal handling, processing, and biosafety measures are not well-established.
ObjectiveThis systematic review aims to summarize the reported effectiveness of FTA cards® for storage and transport of viral RNA, as well as the conditions for their handling and use in downstream processes. Finally, the biosafety measures required to protect researchers and clinical lab workers are considered.
MethodsWe performed a systematic review following the PRISMA statement. We searched MEDLINE (PubMed), Scopus and Web of Science using the keywords “FTA cards” AND “RNA”. Articles were screened by title and abstract, and after examination of inclusion and exclusion criteria, relevant information was extracted. The quality of the studies was assessed, and the evidence was qualitatively summarized.
ResultsA total of 175 records were retrieved, and 11 additional documents were found by checking references of the eligible articles. A total of 47 articles were included. Samples from animals accounted for 38.3% of the publications, which identified viruses that cause disease in poultry, wild birds, suids, or bovids. Three different methods for RNA extraction were reported. Other factors that vary across reports include the size of RNA amplicon, storage temperature, and duration of storage. Only fourteen articles tested the inactivation of the virus on the FTA card®, and in one case, the virus remained infective.
ConclusionFTA cards® could be a suitable option for RNA virus storage and transport for fieldwork in areas where proper conditions for RNA preservation are difficult to achieve. Three different protocols have been used for RNA detection from this matrix. Biospecimens in the form of dried blood spots should be considered potentially infectious unless specifically treated to inactivate viral pathogens.
Keywords: FTA card, RNA, Virus, Isolation, Detection, Transport, RT-PCR
1. Introduction
1.1. Rationale
Flinders Technology Associates (FTA) cards® have been widely used for DNA preservation and analysis, particularly in forensic sciences and genetic studies (Hsiao et al., 1999; Smith and Burgoyne, 2004). This technology consists of filter paper impregnated with a patented chemical mixture that contains chemical denaturants and a free radical scavenger. The chemical mixture lyses cells and organelles, prevents overgrowth of bacteria, and denatures proteins. The DNA is captured in the matrix and remains tightly bound while proteins and inhibitors are washed. This keeps DNA stable during long-term storage at room temperature (GE Healthcare, 2019).
This method is attractive for several applications, including research of bacterial genetics, as DNA can be maintained at ambient temperature (Reeve et al., 2018). FTA cards® have also been used for storing samples and molecular detection of both DNA and RNA viruses. Recently, they have been proposed as an alternative to liquid media in the screening of cervical cancer for HPV detection (Dong et al., 2017), and improved biosafety has been cited as an advantage in the storage and transport of specimens containing avian influenza virus (Kraus et al., 2011). However, viral RNA is an unstable molecule, susceptible to ubiquitous RNases, complicating its transport and detection.
This challenge is particularly relevant in settings where proper technical support for RNA handling may not be available (hereafter referred to as fieldwork). Although some studies have evaluated the FTA® cards for viral RNA preservation (Ndunguru et al., 2005; Muthukrishnan et al., 2008; Flies et al., 2015), the optimal conditions for their handling have not been extensively studied. Notwithstanding, it is crucial to determine the appropriate temperature and maximum time for storing RNA samples on FTA cards, as well as their biosafety, in order to fieldwork with these pathogens.
1.2. Objectives
In this context, this systematic review aims to assess published studies that used FTA cards® as a mean for viral RNA preservation before molecular detection.
2. Methods
2.1. Literature search
For conducting this systematic review, we followed the recommendations of the PRISMA statement (Moher et al., 2009). In June 2018, MEDLINE (PubMed), Scopus and Science Citation Index (Web of Knowledge) were searched to identify potentially relevant articles using the search strategy (“FTA cards” AND “RNA”). No limit was set for the year, and only articles published in English or Spanish were assessed. The references of the included articles were also reviewed looking for eligible articles.
2.2. Study eligibility and selection
Two different authors conducted the review independently, and it was completed on July 1st, 2018. The resulting articles were initially selected by title and abstract to identify possibly eligible studies according to the selected criteria. Subsequently, duplicates were excluded, and two authors reviewed full-text of the eligible articles. When there was disagreement in the inclusion of a study, a third researcher made the final decision. We assessed studies that used FTA cards® for storage and later isolation of RNA of viral origin with subsequent amplification. Different conditions of storage, RNA extraction, or cDNA synthesis were separately summarized when the information was available. We excluded opinion articles, letters to the editor that did not present original data, reviews, as well as articles for which full-text was not accessible through our institutional subscriptions or their information was incomplete. The potentially eligible articles were entered in a database, screened based on title and abstract and reviewed independently by two research team members. Articles included in the review by either team member were submitted to full-text screening.
2.3. Data extraction
Data extraction was performed by a third author not involved in screening and selection. The data extracted were: author, title, year of the study, institution, source of sample (e.g. blood, serum, plasma, allantoic fluid, other tissues), origin of the sample (i.e. human, poultry, plants), method of extraction of RNA from the FTA card®, time and temperature of storage of FTA card, method of assessment of RNA integrity and purity, and amplification method of the viral genome fragment.
For studies that addressed different storage times, temperatures and amplicon sizes, the maximum time and maximum temperature at which the viral RNA was detectable as well as the largest amplicon size (bp) reported were recorded. Elution characteristics were compiled if RNA extraction was conducted through elution. If the study assessed the infectivity of the virus isolated from the FTA card®, it was included.
2.4. Quality assessment
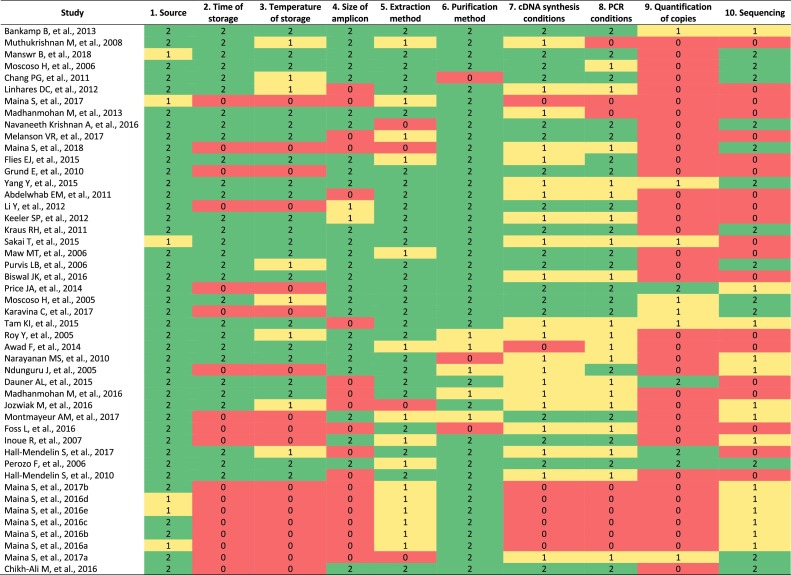
Because there is not a previously validated tool for assessment of the quality of studies assessing methods for sample preservation on FTA cards®, we built a checklist consisting of ten criteria that were scored using a discrete scoring. Each item was scored with 2 = if the information provided was complete or sufficient to replicate the methods, 1 = if the information was incomplete or unclear, or 0 = if no information was provided. For data visualization a heatmap was generated using the scores.
The included items were: 1. Source of the sample: Report of the kind of sample obtained for storage in a FTA card®. 2. Time of storage: Report of the time the FTA card® was stored between sample collection and FTA card® processing for RNA isolation and RT-PCR. 3. Temperature of storage: Report of the temperature at which the FTA card® was stored. 4. Amplicon size: Report of the size (bp) of the targeted viral gene for amplification. If the work tried complete genome sequencing the answer to this question was also scored with two. 5. Extraction method: specification with precise detail to guarantee the reproducibility of the methods used for RNA extraction from the FTA card®. 6. Purification method: Specific information to assure the reproducibility of the methods used for RNA purification. 7. cDNA synthesis conditions: enough information about reagents and conditions to ensure the reproducibility of the methods for cDNA synthesis, including primers and cycling conditions. 8. PCR conditions: enough information concerning the methods and reagents used for PCR, including primers and cycling conditions. 9. Quantification of RNA copies: Report of quantification through RT-qPCR of the RNA isolated from the FTA card®. 10. Sequencing: Report about the sequencing of the desired amplicon and confirmed identity of the targeted virus, or confirmation through whole-genome sequencing with next-generation sequencing techniques (NGS).
2.5. Assessment of RNA isolation and definition of viral non-infectiousness
Experiments of viral RNA detection from FTA cards® were considered successful if the RNA fragment of interest was finally amplified from cDNA (Retro-transcription, RT) and PCR irrespective of the fragment size or if the RNA isolation allowed further genome sequencing through NGS. For studies assessing infectivity, we considered FTA card® specimens to be noninfectious when virus isolation was negative through cell culture or bio-assays using material obtained of the cards.
3. Results
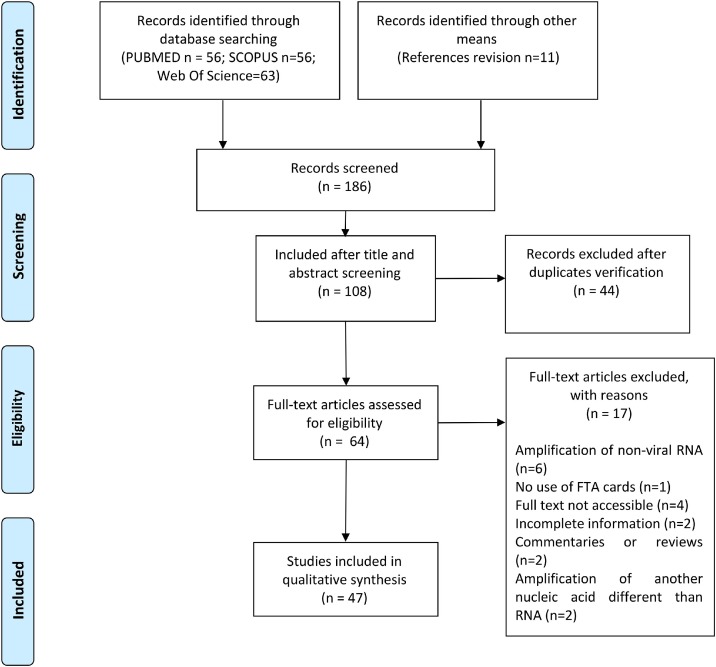
We found 175 records using the search strategy, and 11 additional records by checking references of the included articles. After title and abstract screening, 44 articles were excluded because they were duplicates, and 108 papers were selected for screening by title and abstract. After the examination of exclusion criteria, a total of 47 articles were finally included (Fig. 1 ). All the included articles were published in English, and all of them achieved viral RNA amplification. The assessment of the quality of the information in each article is shown in Fig. 2 .
Fig. 1.
PRISMA 2009 Flow Diagram of retrieved and selected articles during the systematic review.
Fig. 2.
Quality assessment chart. Each included study was assessed in order to determine the information provided for each variable. Each variable was scored with 2 = green, if the information provided was complete or sufficient to replicate the methods, 1 = yellow, if the information was incomplete or unclear, or 0 = red, if no information was provided. For data visualization a heatmap was generated using the scores (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
A total of 95.7% of the included studies extracted total RNA before targeted amplification of the viral RNA, while the 4.3% directly isolated viral RNA from the FTA card. The studies used a wide variety of sources and samples, which included samples from mammals, birds, insects, plants, cells, and humans (Table 1 ). The targeted viral genomes belonged to all classes of RNA virus of the Baltimore Classification except Class VI, with 8.5% of the studies reporting amplification of RNA fragments of Class III virus (dsRNA), 68.1% of Class IV virus ((+) ssRNA), 21.3% of Class V virus ((−) ssRNA), and 2.1% of the three classes mentioned above. An animal specimen source accounted for 38.3% of the reports (Moscoso et al., 2005; Maw et al., 2006; Moscoso et al., 2006; Perozo et al., 2006; Purvis et al., 2006; Inoue et al., 2007; Narayanan et al., 2010; Abdelwhab et al., 2011; Kraus et al., 2011; Keeler et al., 2012; Linhares et al., 2012; Madhanmohan et al., 2013; Awad et al., 2014; Biswal et al., 2016; Foss et al., 2016; Jozwiak et al., 2016; Madhanmohan et al., 2016; Manswr et al., 2018). Among them, twelve reports isolated RNA from viruses that can cause disease in poultry or wild birds, like Newcastle virus disease, avian influenza, or infectious bursal disease (Table 1). Six reports isolated RNA of viruses that can cause disease in suids or bovids, like respiratory syndrome virus, and foot-and-mouth disease virus (Table 1).
Table 1.
General characteristics of the included works that performed viral RNA amplification/sequencing from samples immobilized in FTA cards®. N/A = Not Apply, (−) = Information not provided.
| Study | Year | Institution | RNA sample | Source | RNA isolated | Virus | Viral genome | Infectivity | Maximum time of storage | Temperature of storage | Extraction method | RT-PCR method | Primer strategy (RT) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bankamp B, et al.. | 2013 | Centers for Disease Control and Prevention | Vero/hSLAM cells | Cells | Total | Measles morbillivirus | ssRNA(−) | – | 6 months | 4 °C | Elution | one step RT-qPCR | Specific |
| Muthukrishnan M, et al. | 2008 | Indian Immunologicals Limited | BHK21 cell | Cells | Total | Foot-and-mouth disease virus | ssRNA(+) | No | 120 days | Room temperature | Disc cleaning | one step RT-qPCR | Specific |
| Elution | one step RT-qPCR | Specific | |||||||||||
| Manswr B, et al. | 2018 | University of Liverpool | Allantoic fluid | Animal | Total | Infectious bronchitis virus | ssRNA(+) | – | 21 days | Room temperature | Elution | two step RT-PCR | Specific |
| University of Liverpool | Allantoic fluid | Animal | Total | Infectious bronchitis virus | ssRNA(+) | – | 21 days | Room temperature | Elution | two step RT-PCR | Specific | ||
| Moscoso H, et al. | 2006 | The University of Georgia | Bursal tissue | Animal | Total | Infectious bursal disease virus | dsRNA | – | 8 months | −20 °C | Elution | one step RT-PCR | Specific |
| Chang PG, et al. | 2011 | Virginia Polytechnic Institute and State University | Leaf | Plant | Total | Tobacco etch virus, Soybean mosaic virus, Turnip mosaic virus, Cucumber mosaic virus, and Peanut stunt virus | ssRNA(+) | – | – | – | Disc cleaning | two-step RT-PCR | Specific |
| Linhares DC, et al | 2012 | College of Veterinary Medicine | Serum and lung | Animal | Total | Porcine reproductive and respiratory syndrome virus | ssRNA(+) | – | 14 days | Room temperature | Elution | one step RT-PCR | Specific |
| Maina S, et al. | 2017 | University of Western Australia | Leaf | Plant | Total | Cucurbit aphid-borne yellows virus | ssRNA(+) | – | – | N/S | Purification | RNA-seq | – |
| Madhanmohan M, et al. | 2013 | Indian Immunologicals Limited | Tongue epithelium | Animal | Total | Foot-and-mouth disease virus | ssRNA(+) | – | 1 month | 25 °C | Purification | RT-LAMP | Specific |
| Navaneeth Krishnan A, et al. | 2016 | Central Institute of Brackishwater Aquaculture | Larval homogenate, cell culture extract, milt extract, seawater spiked with larval homogenate | Animal | Total | Betanodavirus spp. | ssRNA(+) | – | 28 days | 4 °C | Purification | one step RT-PCR | Specific |
| Melanson VR, et al. | 2017 | United States Army Research Institute of Infectious Diseases | Ae. aegypti saliva | Insect | Total | Dengue virus | ssRNA(+) | – | – | – | – | one step RT-PCR | Specific |
| Maina S, et al. | 2018 | The University of Western Australia | Leaf | Plant | Total | Sweet potato feathery mottle virus and Sweet potato virus C | ssRNA(+) | – | – | – | Purification | one step RT-PCR | Specific |
| Flies EJ, et al. | 2015 | University of Tasmania | Mosquito saliva | Insect | Total | Arbovirus (Ross River virus, Barmah Forest virus, and Stratford virus) | ssRNA(+) | – | Seven days | 25 °C | Elution | two-step RT-PCR | Specific |
| Grund E, et al. | 2010 | University of Hamburg | Leaf | Plant | Total | Watermelon chlorotic stunt virus, Mycovirus China 9, Tomato spotted wilt virus, Tobacco mosaic virus, Cucumber mosaic virus, Little cherry virus 1, Little cherry virus 2, Potato spindle tuber viroid | dsRNA, ssRNA(+), ssRNA(−) | – | – | – | Elution | two-step RT-PCR | Specific |
| Yang Y, et al. | 2015 | Walter Reed Army Institute of Research | Blood | Insect | Total | Dengue virus type 2 | ssRNA(+) | – | 1 month | Room temperature | Elution | one step qRT-PCR, MiSeq | Specific and random |
| Abdelwhab EM, et al | 2011 | Free Berlin University | Amnio-allantoic fluid | Animal | Total | Avian influenza virus | ssRNA(−) | No | 5 months | Room temperature | Elution | one step RT-qPCR | Specific |
| Li Y, et al. | 2012 | Shandong Center for Disease Control and Prevention | HEp-2 Cells | Cells | Total | Enterovirus spp. | ssRNA(+) | – | – | – | Elution | one step qRT-PCR | Oligo dT and Random primers |
| Keeler SP, et al | 2012 | The University of Georgia | Cloacal swabs | Animal | Total | Avian influenza virus | ssRNA(−) | – | 90 days | Room temperature | Elution | one step RT-PCR | Specific |
| Kraus RH, et al | 2011 | Wageningen University | Cloacal swabs | Animal | Total | Avian influenza virus | ssRNA(−) | – | N/S | – | Elution | one step RT-PCR | Specific |
| Sakai T, et al | 2015 | Nihon University Veterinary Research Center | Vaccine | Vaccine | Viral | Rabies virus | ssRNA(−) | – | 4 months | −20 °C | Elution | one step RT-PCR | Specific |
| Maw MT, et al | 2006 | United Graduate School of Veterinary Sciences | Mucose | Animal | Total | Infectious bursal disease virus | dsRNA | Yes | 30 days | 37 °C | Elution | two-step RT-PCR | Specific |
| Purvis LB, et al | 2006 | University of Georgia | Bursal tissue | Animal | Total | Infectious bursal disease virus | dsRNA | – | 24 hours | Room temperature | Elution | one step RT-PCR | Specific |
| Biswal JK, et al | 2016 | ICAR- Directorate on Foot-and-mouth Disease | Clinical samples (cell culture isolates, tongue epithelial suspension, and impression smears) | Animal | Total | Foot-and-mouth disease virus | ssRNA(+) | – | 6 weeks | 4-37 °C | Elution | two-step RT-PCR | Oligo dT |
| Price JA, et al. | 2014 | Texas A&M AgriLife Research and Extension | Wheat curl mite | Animal | Total | Wheat streak mosaic virus | ssRNA(+) | – | 1 hour | Room temperature | Elution | one step RT-PCR | Specific |
| Moscoso H, et al. | 2005 | The University of Georgia | Allantoic fluid | Animal | Total | Infectious bronchitis virus | ssRNA(+) | – | 36 days | Room temperature | Elution | one step RT-PCR | Specific |
| Karavina C, et al | 2017 | University of KwaZulu-Natal | Leaf | Plant | Total | Tomato spotted wilt virus | ssRNA(−) | – | – | – | Elution | two step RT-mPCR | Specific |
| Tam KI, et al | 2015 | Centers for Disease Control and Prevention | Stool | Human | Total | Rotavirus A | dsRNA | – | 28 days | 37 °C | Elution | one step RT-PCR | Specific |
| Roy Y, et al. | 2005 | University of Guelph | Leaf and fruit | Plant | Total | Pepino mosaic virus and Rupestris stem pitting associated virus | ssRNA(+) | – | 8 months | Room temperature | Disc cleaning | one step RT-PCR | Specific |
| Awad F, et al. | 2014 | University of Liverpool | Tracheal organ cultures | Animal | Total | Avian metapneumovirus | ssRNA(−) | No | 2 months | 4 °C | Purification | RT-PCR | Specific |
| Narayanan MS, et al. | 2010 | Madras veterinary college | Allantoic fluid | Animal | Total | Avian avulavirus 1 | ssRNA(−) | No | 20 days | 4 °C | Disc cleaning | two-step RT-PCR | Specific |
| Ndunguru J, et al. | 2005 | International Laboratory of Tropical Agriculture Biotechnology | Leaf | Plant | Total | Geminivirus | ssRNA(+) | – | – | N/S | Elution | two step RT-PCR | Specific |
| Dauner AL, et al. | 2015 | Naval Medical Research Center | Blood | Human | Total | Dengue virus | ssRNA(+) | – | 21 days | 37 °C | Elution | one step RT-PCR | Specific |
| Madhanmohan M, et al. | 2016 | Indian Immunologicals | Smear | Animal | Total | Foot-and-mouth disease virus | ssRNA(+) | No | 56 days | 4 °C | Elution | one step RT-PCR | Specific |
| Jozwiak M, et al. | 2016 | National Veterinary Research Institute | Allantoic fluid | Animal | Total | Avian influenza virus | ssRNA(-) | No | 90 days | Room temperature | – | one step RT-PCR | Specific |
| Montmayeur AM, et al. | 2017 | Centers for Disease Control and Prevention | Poliovirus culture | Virus | Viral | Poliovirus | ssRNA(+) | – | – | – | Purification | NGS sequencing | Random |
| Foss L, et al. | 2016 | California Department of Public Health | Oral swabs | Animal | Total | West Nile virus | ssRNA(+) | – | – | – | Elution | one step RT-PCR | Specific |
| Inoue R, et al. | 2007 | Kyoto Prefectural University | Blood | Animal | Total | Porcine reproductive and respiratory syndrome virus | ssRNA(+) | – | – | – | Disc cleaning | one step RT-PCR | Specific |
| Hall-Mendelin S, et al. | 2017 | Public Health Virology | Mosquito saliva | Animal | Total | Arbovirus (Zika virus, Chikungunya virus, Dengue virus, and Barmah Forest virus) | ssRNA(+) | – | 28 days | Room temperature | Elution | one step RT-PCR | Specific |
| Perozo F, et al | 2006 | University of Georgia | Allantoic fluid | Animal | Total | Newcastle disease virus | ssRNA(−) | No | 15 days | Room temperature | Purification | one step RT-PCR | Specific |
| Hall-Mendelin S, et al. | 2010 | University of Queensland | Mosquito saliva | Animal | Total | Arbovirus (West Nile river, Ross River virus, and chikungunya virus) | ssRNA(+) | – | 28 days | 23 °C | Elution | one step RT-PCR | Specific |
| Maina S, et al. | 2017 | University of Western Australia | Leaf | Plant | Total | Aphid lethal paralysis virus | ssRNA(+) | – | – | – | Purification | Sequencing (metagenomic) | N/A |
| Maina S, et al. | 2016 | University of Western Australia | – | Plant | Total | Sweet potato chlorotic fleck virus | ssRNA(+) | – | – | – | Purification | Genome sequencing | N/A |
| Maina S, et al. | 2016 | University of Western Australia | – | Plant | Total | Sweet potato virus 2 | ssRNA(+) | – | – | – | Purification | Genome sequencing | N/A |
| Maina S, et al. | 2016 | University of Western Australia | Leaf | Plant | Total | Suakwa aphid-borne yellows virus | ssRNA(+) | – | – | – | Purification | Genome sequencing | N/A |
| Maina S, et al. | 2016 | University of Western Australia | Leaf | Plant | Total | Bean common mosaic necrosis virus | ssRNA(+) | – | – | – | Purification | Genome sequencing | N/A |
| Maina S, et al. | 2016 | University of Western Australia | – | Plant | Total | Sweet potato virus G | ssRNA(+) | – | – | – | Purification | Genome sequencing | N/A |
| Maina S, et al. | 2017 | University of Western Australia | Leaf | Plant | Total | Papaya ringspot virus biotype W | ssRNA(+) | – | – | – | Purification | two step RT-PCR, RNA-seq | Specific |
| Chikh-Ali M, et al | 2016 | University of Idaho | Leaf | Plant | Total | Potato virus Y | ssRNA(+) | – | – | – | Elution | two step RT-PCR | Specific |
LAMP = Loop-mediated Isothermal Amplification.
Plant samples were assessed in 31.9% of the included articles, and samples obtained from insects corresponded to 14.9%. These works aimed to amplify viral RNA of arboviruses that can cause disease in humans, and also viruses capable of causing disease in fishes and plants (Hall-Mendelin et al., 2010; Price et al., 2014; Flies et al., 2015; Yang et al., 2015; Navaneeth Krishnan et al., 2016; Hall-Mendelin et al., 2017; Melanson et al., 2017; Chikh-Ali et al., 2016). Viral specimens were derived from cell culture or from a commercial inactivated vaccine in 10.6% of the reports (Muthukrishnan et al., 2008; Li et al., 2012; Bankamp et al., 2013; Sakai et al., 2015; Montmayeur et al., 2017). Finally, two reports isolated virus using human samples: stool and blood. (Dauner et al., 2015; Tam et al., 2015).
Infectious virus was isolated from one of only fourteen included studies that tested for infectivity of the virus from an FTA card® (Maw et al., 2006). In this study, ADVANTEC chromatography paper No. 526, (Toyo Roshi, Tokyo, Japan), ADVANTEC filter paper No. 2 (Toyo Roshi), and stationery copy paper were compared with FTA cards® for the isolation and inactivation of the dsRNA from the infectious bursal disease virus, a virus member of the Birnaviridae family, genus Avibirnavirus, with a segmented genome. In this study, every paper-based matrix allowed the isolation of viral RNA and infectious virus. However, virus inactivation was achieved after phenol fixation on the FTA card®. Thirteen other studies demonstrated a lack of infectious virus from the FTA card (Moscoso et al., 2005, Moscoso et al., 2006, Perozo et al., 2006, Muthukrishnan et al., 2008, Narayanan et al., 2010, Abdelwhab et al., 2011, Keeler et al., 2012, Linhares et al., 2012, Bankamp et al., 2013, Awad et al., 2014, Tam et al., 2015, Jozwiak et al., 2016, Madhanmohan et al., 2016) when examining (-) ssRNA viruses of the families Paramyxoviridae (Newcastle disease virus, genus Avulavirus, Measles morbillivirus, subfamily Orthoparamyxovirinae, genus Morbillivirus), Orthomyxoviridae (Avian influenza virus, genus Alphainfluenzavirus), and Pneumoviridae (Avian metapneumovirus, genus Metapneumovirus), (+) ssRNA virus members of the family Picornaviridae (Foot-and-mouth disease virus, genus Aphthovirus), Arteriviridae (Porcine reproductive and respiratory syndrome virus), and Coronaviridae (Avian coronavirus, genus Gammacoronavirus), and dsRNA virus of the family Birnaviridae (Infectious bursal disease virus, genus Avibirnavirus) and Reoviridae (Rotavirus A, genus Rotavirus). These studies attempted viral isolation from FTA cards impregnated with infected material using cell culture (Moscoso et al., 2006, Muthukrishnan et al., 2008, Keeler et al., 2012, Linhares et al., 2012, Bankamp et al., 2013, Awad et al., 2014, Tam et al., 2015, Madhanmohan et al., 2016), bioassays infecting specific pathogen-free embryonated chicken eggs (Moscoso et al., 2005, Perozo et al., 2006, Abdelwhab et al., 2011, Jozwiak et al., 2016) or both (Narayanan et al., 2010).
Regarding storage of the FTA cards® before further viral RNA purification, the cards were stored for periods that varied from one to eight months (median time: 30 days, IQR 21–90), at temperatures that ranged between −20 °C to 37 °C (median temperature: 25 °C, IQR 4 °C–25 °C). Three protocols for RNA amplification from the FTA card® were reported: 1. Elution of RNA from the FTA card®, 2. Direct viral RNA purification, and 3. Disk cleaning and soaking.
Elution of RNA from FTA cards® before RNA amplification was the most common method for RNA extraction. It was performed in 55.1% of the assessed studies. The elution conditions and eluents used in those studies are summarized in Table 2 . Briefly, viral RNA was obtained by soaking fragments of the FTA card® in an eluent solution, followed by incubation with or without agitation with temperatures that ranged between 4–95 °C. After elution, the RNA underwent a purification step using commercial kits (Moscoso et al., 2005; Ndunguru et al., 2005; Maw et al., 2006; Moscoso et al., 2006; Purvis et al., 2006; Abdelwhab et al., 2011; Keeler et al., 2012; Li et al., 2012; Bankamp et al., 2013; Sakai et al., 2015; Tam et al., 2015; Yang et al., 2015; Biswal et al., 2016; Foss et al., 2016; Madhanmohan et al., 2016; Karavina and Gubba, 2017; Manswr et al., 2018) or standard protocols like sodium acetate and isopropanol precipitation (Grund et al., 2010; Price et al., 2014). In one case, magnetic beads were used for purification and concentration of RNA (Linhares et al., 2012)
Table 2.
Characteristics of studies that performed RNA elution from FTA cards® before RT-PCR reaction. (-) Not performed.
| Study | Eluent | Elution volume (μL) | Elution time (min) | Agitation | Elution temperature | Purification method |
|---|---|---|---|---|---|---|
| Bankamp B, et al. | PBS + AVL buffer | 750 | 10 | – | Room temperature | QIAamp Viral RNA Mini Kit |
| Manswr B, et al. | TE buffer | 1000 | 15 | – | Room temperature | QIAamp viral RNA Mini Kit |
| TE buffer | 1000 | 15 | – | Room temperature | QIAamp viral RNA Mini Kit | |
| Moscoso H, et al. | Tris-HCl + EDTA | 200 | 10 | – | Room temperature | High Pure Viral RNA Kit |
| Linhares DC, et al. | RNA rapid extraction solution | 100 | 5 | – | Room temperature | Magnetic bead-based technology |
| Flies EJ, et al. | GM + 3%FBS | 1000 | 20 | Vortex every 5min | On ice | EZ1 Virus Mini 109 Kit v2.0 (Qiagen, Victoria, AU) |
| Grund E, et al. | Elution buffer | 400 | 15 | Agitation every 5min | 4 °C | Sodium acetate and isopropanol precipitation |
| Yang Y, et al. | AVL buffer + RNA carrier | 566 | 5 | Continuous vortex +5 min centrifugation | Room temperature | Qiagen QIAamp Viral RNA kit |
| Abdelwhab EM, et al | TE buffer | 200 | 10 | – | Room temperature | Qiagen Viral RNA Mini Kit |
| Li Y, et al. | Deionized sterile water + TE-1 + Tris–HCl + Buffer EB (Tris–Cl, elution buffer from the QIAamp Viral RNA Mini Kit) | 30 | 40 | Vortex at 100x during 30 min, 10 min vortex at 200xg in intervals of 1 min | 65 °C or 95 °C | Roche Pure Viral RNA kit |
| Keeler SP, et al. | RNA rapid extraction solution | 75 | 60 | – | Room temperature | MagMAX™-96 AI/ND Viral RNA Isolation Kit |
| RNA rapid extraction solution | 75 | 60 | – | Room temperature | MagMAX™-96 AI/ND Viral RNA Isolation Kit | |
| Kraus RH, et al. | RNA rapid extraction solution | 70 | 5 | Agitation | Room temperature | MagMAX-96 viral RNA isolation kit |
| Sakai T, et al. | Nuclease-free water | 150 | 1440 | – | Room temperature | QIAamp® Viral RNA Mini Kit |
| TE buffer | 150 | 1440 | – | Room temperature | QIAamp® Viral RNA Mini Kit | |
| RNA rapid extraction solution | 150 | 30 | – | Room temperature | QIAamp® Viral RNA Mini Kit | |
| TRIzol Reagent | 150 | 30 | – | Room temperature | QIAamp® Viral RNA Mini Kit | |
| AVL buffer | 150 | 1440 | – | Room temperature | QIAamp® Viral RNA Mini Kit | |
| Maw MT, et al. | Nuclease-free water | 75 | 5 | Agitation | Room temperature | TRIzol Reagent |
| Purvis LB, et al. | Whatman purification reagent | 200 | 20 | Vortex every 5min | Room temperature | High Pure RNA isolation kit |
| Biswal JK, et al. | AVL buffer | 560 | 10 | – | Room temperature | QIAmp Viral RNA mini kit |
| Price JA, et al. | TE buffer | 400 | 15 | Vortex every 5min | Ice incubation | Sodium acetate and isopropanol precipitation |
| Moscoso H, et al. | Tris-HCl and EDTA | 200 | 10 | – | Room temperature | High Pure Viral RNA Kit |
| Karavina C, et al. | Nuclease-free water | 50 | 30 | Vortex 15 seg | 95 °C | – |
| Tam KI, et al. | TE buffer | 400 | 240 | – | Room temperature | MagMAX™ Viral RNA isolation kit |
| Ndunguru J, et al. | RNA processing buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 800 U/mL RNase Out™ 200–250 μg/mL glycogen and 2 mM DTT) | 500 | 30 | Vortex every 5min | Room temperature | Sodium acetate and isopropanol precipitation |
| Dauner AL, et al. | AVL buffer | 300 | 60 | Shaking | Room temperature | QIAamp viral RNA Mini Kit |
| Madhanmohan M, et al. | VMM | 460 | 900 | – | 4 °C | RNeasy® Mini Kit |
| Foss L, et al. | ABI Magmax lysis binding solaution | 1000 | 900 | – | Room temperature | – |
| Hall-Mendelin S, et al. | GM + 3%FBS | 1000 | 20 | Vortex every 5min | On ice | QIAamp® Virus BioRobot® MDx Kit |
| Hall-Mendelin S, et al. | GM + 3%FBS | 1000 | 20 | Vortex every 5min | On ice | QIAamp® Virus BioRobot® MDx Kit |
| Chikh-Ali M, et al | RNA processing buffer | 500 | 30 | Vortex every 5min | Room temperature | Sodium acetate and isopropanol precipitation |
Direct virus RNA purification was performed in 30.6% of the studies. It consisted of a protocol in which FTA card® punches were used for RNA purification without a previous step of elution from the FTA card®. The punches were directly soaked in the lysis and extraction solutions of different commercial kits, TRIzol reagent® or Guanidinium thiocyanate-phenol-chloroform method (Table 1). Most of the studies used methods for total RNA extraction and only one study used a commercial kit for viral RNA isolation (Montmayeur et al., 2017). In this particular case, the genome of polioviruses, (Enterovirus C, genus Enterovirus, family Picornaviridae), a virus with a (+) ssRNA non-segmented genome of 6.7–10.1 kb with a poly(A) tail, was isolated from FTA cards®. This method allowed the use of NGS techniques for complete genome sequencing with a total of 206,156 reads (90% of reads mapping to poliovirus, giving 99% genome coverage) (Montmayeur et al., 2017).
On the other hand, 8.2% of the included studies used disk cleaning and soaking for RNA purification, which consisted of punching a disk from the card and placing in a solution to remove contaminants before proceeding one or two-step RT-PCR by placing the cleaned disk into a second solution (the reaction mix). Note that this method did not include an RNA purification step (Roy and Nassuth, 2005; Inoue et al., 2007; Narayanan et al., 2010; Chang et al., 2011).
The third strategy was to perform one or two-step RT-PCR after RNA isolation or preparation from the FTA cards®. In 57.1% of these studies, the targeted viral genes were amplified by one-step RT-PCR. Most of them (96.4%) used specific primers for the RT step and the PCR step. Only one study used random primers for RT and then specific primers for PCR (Yang et al., 2015). The reaction mix composition, as well as the cycling conditions in each work, varied depending on the PCR kit used and the desired amplicon. Eleven studies quantified the product of the RT-PCR, 26 used the product of these reactions for sequencing, and in one study, RNA-seq was implemented for viral identification. (Maina et al., 2016a,c,b,d,e; Maina et al., 2017a, b; Maina et al., 2017c; Montmayeur et al., 2017).
4. Discussion
The findings of this systematic review support that viral RNA from different types of RNA viruses spotted in a FTA card® can be purified and then amplified for viral identification. The RNA can be isolated from a large number of sources and samples, and stored over a wide range of periods of time and temperatures. This highlights the fact that FTA cards® could be used as means for transport and storage of RNA pathogens. However, storage time and temperature are critical variables affecting RNA preservation. Studies reviewed here suggest that viral RNA could be stable in a wide range of temperatures for several months. Cards stored at 4 °C allowed the amplification of fragments up to ˜1200 bp, and cards stored for six months allowed amplification of fragments up to ˜800 bp.
Use of FTA cards to work with biospecimens appears to be relatively safe since isolation of infectious virus was rarely reported. Nevertheless, the possibility remains that some infectious virus could be present in this type of sample, as shown by the study of bursal disease virus. In this case, additional chemical fixation was used to render the sample completely noninfectious (Maw et al., 2006).
More data is required to explain why the bursal disease virus remained infectious when such a wide variety of viruses were inactivated after spotting on FTA cards® (Perozo et al., 2006; Narayanan et al., 2010; Abdelwhab et al., 2011; Awad et al., 2014; Jozwiak et al., 2016; Madhanmohan et al., 2016). Possible explanations include that infectious bursal disease virus is a non-enveloped virus. While enveloped viruses can persist viable in dried surfaces for five days, non-enveloped viruses can persist for weeks (Firquet et al., 2015). However, the inability to isolate viable foot-and-mouth disease virus from FTA cards®, another non-enveloped virus, is inconsistent with this argument (Madhanmohan et al., 2016). Storage time and temperature and card manipulation and processing during viral isolation are other possible factors that could affect viral infectiousness. It is remarkable that Maw et al., 2006 isolated infective virus after seven days of storage at room temperature, but Jozwiak et al., 2016 were unable of performing viral isolation even as earlier as 1 h after sample spotting on FTA card, and Linhares et al., 2012 after storage at 4 °C for 24 h. In this context, viral viability testing after immobilization on FTA cards® seems like an advisable practice before considering FTA cards® isolates innocuous. Then, FTA cards® should be managed and stored following the required biosafety precautions.
Different methods for extraction and purification were represented in the literature. The elution and subsequent purification of the RNA are the most widely used, and different elution solutions in a wide range of volumes, time, agitation and temperatures have been tested. Based on these protocols, we advise to keep the elution volume below 500 μL and the elution time below 30 min, with vortex or agitation every 5 min at room temperature. Then perform RNA purification using commercial kits or standard protocols before subsequent RT-PCR. This method was effective for amplification of fragments up to ˜1700 bp.
For disc cleaning protocols it is advisable to add 200 μL of FTA purification reagent in a tube containing the discs, mix thoroughly for 3 min at room temperature, and repeat this step twice. Then, rinse the discs with 200 μL of TE Buffer and, finally, add the disc to the proper PCR reaction mix. When the source material likely contains PCR inhibitors, an additional step of cleaning with 200 μL of Ethanol 70% can be performed (Roy and Nassuth, 2005; Inoue et al., 2007; Narayanan et al., 2010; Chang et al., 2011). These protocols and direct purification protocols allowed amplification of fragments of ˜900 bp, even when the storage times of the FTA card® were up to eight months at room temperature. This suggests that probably the three reported methods for RNA extraction from the FTA card® have similar effectiveness and that their selection could be related to reagent availability or personal preferences.
4.1. Limitations
Since the stability and integrity of viral RNA for molecular detection is a significant concern during fieldwork, this systematic review focused on the evaluation of FTA cards® as a strategy for detection of these pathogens. However, successful applications of these cards have been also reported for DNA viruses and bacteria (Dong et al., 2017; Reeve et al., 2018). For instance, the use of FTA cards® have been proved effective as a means of sample storage and transport for genotyping isolates of Campylobacter jejuni obtained from poultry and stored for three months at room temperature (Sierra-Arguello et al., 2016).
It is possible that publication bias, the lack of publishing reports with negative results, could lead to failure in identifying studies that were unable to amplify RNA extracted from the FTA cards®. The search strategy (“FTA cards” AND “RNA”) was used instead of (“FTA card” AND “RNA”) since the first retrieved more records in two of the assessed databases, and the same number of records in Scopus. Additionally, though we conducted an extensive search in different databases, and the screened works were searched through different institutional subscriptions, four papers were not accessible for full-text assessment. Nevertheless, the search identified more than forty reports evaluating FTA cards® as RNA immobilization matrix with sufficient information to summarize their protocols for potential replication.
Although the goal of this review was to consider fieldwork conditions, some of the identified studies used the FTA card® for RNA spotting in controlled laboratory conditions. Therefore, the effectiveness of this matrix could vary when used in fieldwork and associated handling and shipping conditions. In these particular cases, fieldwork testing should be performed. However, some of the studies identified tested their effectiveness submitting these matrices to transport and shipping, similar to fieldwork conditions (Madhanmohan et al., 2016).
5. Conclusions
The effectiveness of FTA cards® for sample collection and recovery of viral RNA for downstream analysis was demonstrated by several studies. Use of FTA cards® may be an attractive approach for fieldwork involving molecular detection and/or characterization of viral pathogens, particularly when simple and cheap sample collection and transport systems are desirable. The stability of RNA genomic material of many different viruses under a wide range of conditions further supports the advantage of FTA cards® use, even for metagenomic analysis (Yang et al., 2015; Maina et al., 2017b). Though many viruses will become inactivated in the collection and storage process, we suggest that all specimens should be considered potentially infectious unless proven otherwise. Thus, the available evidence suggests that FTA cards® are a good option for virus storage and transport for molecular detection and identification of RNA viral pathogens, although additional field testing would be advisable to further validate and standardize these methods for use in the fieldwork.
Funding
This work was supported by Sistema General de Regalías de Colombia, Departamento Nacional de Planeación (Código BPIN 2012000100050), and Vicerrectoría de Investigaciones, Innovación y Extensión, Universidad Tecnológica de Pereira [Grant name: Desarrollo de Capacidades Científicas y Tecnológicas en Biotecnología Aplicadas a los Sectores de la Salud y la Agroindustria en el Departamento de Risaralda]. The sponsoring institution did not have a role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Acknowledgments
We acknowledge Augusto Zuluaga Ph.D. (c) at Universidad Tecnológica de Pereira for his help in data analysis and Dr. Matthew H. Collins at Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University for English revision.
References
- Abdelwhab E.M., Luschow D., Harder T.C., Hafez H.M. The use of FTA(R) filter papers for diagnosis of avian influenza virus. J. Virol. Methods. 2011;174:120–122. doi: 10.1016/j.jviromet.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Awad F., Baylis M., Jones R.C., Ganapathy K. Evaluation of flinders technology associates cards for storage and molecular detection of avian metapneumoviruses. Avian Pathol. 2014;43:125–129. doi: 10.1080/03079457.2014.885114. [DOI] [PubMed] [Google Scholar]
- Bankamp B., Byrd-Leotis L.A., Lopareva E.N., Woo G.K., Liu C., Jee Y., Ahmed H., Lim W.W., Ramamurty N., Mulders M.N., Featherstone D., Bellini W.J., Rota P.A. Improving molecular tools for global surveillance of measles virus. J. Clin. Virol. 2013;58:176–182. doi: 10.1016/j.jcv.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal J.K., Subramaniam S., Ranjan R., Pattnaik B. Evaluation of FTA((R)) card for the rescue of infectious foot-and-mouth disease virus by chemical transfection of extracted RNA in cultured cells. Mol. Cell. Probes. 2016;30:225–230. doi: 10.1016/j.mcp.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Chang P.G., McLaughlin W.A., Tolin S.A. Tissue blot immunoassay and direct RT-PCR of cucumoviruses and potyviruses from the same NitroPure nitrocellulose membrane. J. Virol. Methods. 2011;171:345–351. doi: 10.1016/j.jviromet.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Chikh-Ali M., Bosque-Perez N.A., Vander Pol D., Sembel D., Karasev A.V. Occurrence and molecular characterization of recombinant potato virus Y(NTN) isolates from Sulawesi, Indonesia. Plant. Dis. 2016;(100):269–275. doi: 10.1094/pdis-07-15-0817-re. [DOI] [PubMed] [Google Scholar]
- Dauner A.L., Gilliland T.C., Jr, Mitra I., Pal S., Morrison A.C., Hontz R.D., Wu S.J. Evaluation of nucleic acid stabilization products for ambient temperature shipping and storage of viral RNA and antibody in a dried whole blood format. Am. J. Trop. Med. Hyg. 2015;93:46–53. doi: 10.4269/ajtmh.15-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Lin C., Li L., Wang M., Cui J., Feng R., Liu B., Wu Z., Lian J., Liao G., Chen W., Qiao Y. An evaluation of clinical performance of FTA cards for HPV 16/18 detection using cobas 4800 HPV Test compared to dry swab and liquid medium. J. Clin. Virol. 2017;94:67–71. doi: 10.1016/j.jcv.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Firquet S., Beaujard S., Lobert P.E., Sane F., Caloone D., Izard D., Hober D. Survival of enveloped and non-enveloped viruses on inanimate surfaces. Microbes Environ. 2015;30:140–144. doi: 10.1264/jsme2.ME14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flies E.J., Toi C., Weinstein P., Doggett S.L., Williams C.R. Converting mosquito surveillance to arbovirus surveillance with honey-baited nucleic acid preservation cards. Vector Borne Zoonotic Dis. 2015;15:397–403. doi: 10.1089/vbz.2014.1759. [DOI] [PubMed] [Google Scholar]
- Foss L., Reisen W.K., Fang Y., Kramer V., Padgett K. Evaluation of nucleic acid preservation cards for west nile virus testing in dead birds. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GE Healthcare . GE Healthcare; Japan: 2019. FTA Cards: High-quality Media for Storage and Transport of DNA.https://www.gelifesciences.co.jp/catalog/1454.html [Google Scholar]
- Grund E., Darissa O., Adam G. Application of FTA (R) cards to sample microbial plant pathogens for PCR and RT-PCR. J Phytopathol. 2010;158:750–757. doi: 10.1111/j.1439-0434.2010.01695.x. [DOI] [Google Scholar]
- Hall-Mendelin S., Hewitson G.R., Genge D., Burtonclay P.J., De Jong A.J., Pyke A.T., van den Hurk A.F. FTA cards facilitate storage, shipment, and detection of arboviruses in infected Aedes aegypti collected in adult mosquito traps. Am. J. Trop. Med. Hyg. 2017;96:1241–1243. doi: 10.4269/ajtmh.16-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Mendelin S., Ritchie S.A., Johansen C.A., Zborowski P., Cortis G., Dandridge S., Hall R.A., Van den Hurk A.F. Exploiting mosquito sugar feeding to detect mosquito-borne pathogens. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11255–11259. doi: 10.1073/pnas.1002040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K.M., Lin H.M., Pan H., Li T.C., Chen S.S., Jou S.B., Chiu Y.L., Wu M.F., Lin C.C., Li S.Y. Application of FTA sample collection and DNA purification system on the determination of CTG trinucleotide repeat size by PCR-based Southern blotting. J. Clin. Lab. Anal. 1999;13:188–193. doi: 10.3958/059.039.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue R., Tsukahara T., Sunaba C., Itoh M., Ushida K. Simple and rapid detection of the porcine reproductive and respiratory syndrome virus from pig whole blood using filter paper. J. Virol. Methods. 2007;141:102–106. doi: 10.1016/j.jviromet.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Jozwiak M., Wyrostek K., Domanska-Blicharz K., Olszewska-Tomczyk M., Smietanka K., Minta Z. application of FTA (R) cards for detection and storage of avian influenza virus. J. Vet. Res. 2016;60:1–6. doi: 10.1515/jvetres-2016-0001. [DOI] [Google Scholar]
- Karavina C., Gubba A. Detection and characterization of tomato spotted wilt virus infecting field and greenhouse-grown crops in Zimbabwe. Eur. J. Plant Pathol. 2017;149:933–944. doi: 10.1007/s10658-017-1243-4. [DOI] [Google Scholar]
- Keeler S.P., Ferro P.J., Brown J.D., Fang X., El-Attrache J., Poulson R., Jackwood M.W., Stallknecht D.E. Use of FTA sampling cards for molecular detection of avian influenza virus in wild birds. Avian Dis. 2012;56:200–207. doi: 10.1637/9862-072611-Reg.1. [DOI] [PubMed] [Google Scholar]
- Kraus R.H., van Hooft P., Waldenstrom J., Latorre-Margalef N., Ydenberg R.C., Prins H.H. Avian influenza surveillance with FTA cards: field methods, biosafety, and transportation issues solved. J. Vis. Exp. 2011;(54):1–6. doi: 10.3791/2832. UNSP e2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yoshida H., Wang L., Tao Z., Wang H., Lin X., Xu A. An optimized method for elution of enteroviral RNA from a cellulose-based substrate. J. Virol. Methods. 2012;186:62–67. doi: 10.1016/j.jviromet.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Linhares D.C., Rovira A., Torremorell M. Evaluation of flinders Technology associates cards for collection and transport of samples for detection of Porcine reproductive and respiratory syndrome virus by reverse transcription polymerase chain reaction. J. Vet. Diagn. Invest. 2012;24:328–332. doi: 10.1177/1040638711429492. [DOI] [PubMed] [Google Scholar]
- Madhanmohan M., Nagendrakumar S.B., Manikumar K., Yuvaraj S., Parida S., Srinivasan V.A. Development and evaluation of a real-time reverse transcription-loop-mediated isothermal amplification assay for rapid serotyping of foot-and-mouth disease virus. J. Virol. Methods. 2013;187:195–202. doi: 10.1016/j.jviromet.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Madhanmohan M., Yuvaraj S., Manikumar K., Kumar R., Nagendrakumar S.B., Rana S.K., Srinivasan V.A. Evaluation of the flinders technology associates cards for storage and temperature challenges in field conditions for foot-and-mouth disease virus surveillance. Transbound. Emerg. Dis. 2016;63:675–680. doi: 10.1111/tbed.12316. [DOI] [PubMed] [Google Scholar]
- Maina S., Coutts B.A., Edwards O.R., de Almeida L., Ximenes A., Jones R.A.C. Papaya ringspot virus populations from east timorese and northern australian cucurbit crops: biological and molecular properties, and absence of genetic connectivity. Plant Dis. 2017;101:985–993. doi: 10.1094/Pdis-10-16-1499-Re. [DOI] [PubMed] [Google Scholar]
- Maina S., Edwards O.R., Barbetti M.J., de Almeida L., Ximenes A., Jones R.A. Deep sequencing reveals the complete genome sequence of sweet potato virus G from East Timor. Genome Announc. 2016;4(5):e00957–16. doi: 10.1128/genomeA.00957-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina S., Edwards O.R., de Almeida L., Ximenes A., Jones R.A. Complete genome sequences of the carlavirus sweet potato chlorotic fleck virus from East Timor and Australia. Genome Announc. 2016;4(3):e00414–16. doi: 10.1128/genomeA.00414-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina S., Edwards O.R., de Almeida L., Ximenes A., Jones R.A. Complete genome sequences of the potyvirus sweet potato virus 2 from East Timor and Australia. Genome Announc. 2016;4(3):e00504–16. doi: 10.1128/genomeA.00504-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina S., Edwards O.R., de Almeida L., Ximenes A., Jones R.A. First complete genome sequence of bean common mosaic necrosis virus from East Timor. Genome Announc. 2016;4(5):e01049–16. doi: 10.1128/genomeA.01049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina S., Edwards O.R., de Almeida L., Ximenes A., Jones R.A. First complete genome sequence of suakwa aphid-borne yellows virus from East Timor. Genome Announc. 2016;4(4):e00718–16. doi: 10.1128/genomeA.00718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina S., Edwards O.R., de Almeida L., Ximenes A., Jones R.A. Metagenomic Analysis of Cucumber RNA from East Timor Reveals an Aphid Lethal Paralysis Virus. Genome Announc. 2017;5(2):e01445–16. doi: 10.1128/genomeA.01445-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina S., Edwards O.R., de Almeida L., Ximenes A., Jones R.A.C. Analysis of an RNA-seq strand-specific library from an east timorese cucumber sample reveals a complete cucurbit aphid-borne yellows virus genome. Genome Announc. 2017;5(19):e00320–17. doi: 10.1128/genomeA.00320-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manswr B., Ball C., Forrester A., Chantrey J., Ganapathy K. Evaluation of full S1 gene sequencing of classical and variant infectious bronchitis viruses extracted from allantoic fluid and FTA cards. Avian Pathol. 2018:1–9. doi: 10.1080/03079457.2018.1471196. [DOI] [PubMed] [Google Scholar]
- Maw M.T., Yamaguchi T., Kasanga C.J., Terasaki K., Fukushi H. A practical tissue sampling method using ordinary paper for molecular detection of infectious bursal disease virus RNA by RT-PCR. Avian Dis. 2006;50:556–560. doi: 10.1637/7537-032806R.1. [DOI] [PubMed] [Google Scholar]
- Melanson V.R., Jochim R., Yarnell M., Ferlez K.B., Shashikumar S., Richardson J.H. Improving vector-borne pathogen surveillance: a laboratory-based study exploring the potential to detect dengue virus and malaria parasites in mosquito saliva. J. Vector Borne Dis. 2017;54:301–310. doi: 10.4103/0972-9062.225834. [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J. Clin. Epidemiol. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Montmayeur A.M., Ng T.F., Schmidt A., Zhao K., Magana L., Iber J., Castro C.J., Chen Q., Henderson E., Ramos E., Shaw J., Tatusov R.L., Dybdahl-Sissoko N., Endegue-Zanga M.C., Adeniji J.A., Oberste M.S., Burns C.C. High-throughput next-generation sequencing of polioviruses. J. Clin. Microbiol. 2017;55:606–615. doi: 10.1128/JCM.02121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso H., Alvarado I., Hofacre C.L. Molecular analysis of infectious bursal disease virus from bursal tissues collected on FTA filter paper. Avian Dis. 2006;50:391–396. doi: 10.1637/7505-011306R.1. [DOI] [PubMed] [Google Scholar]
- Moscoso H., Raybon E.O., Thayer S.G., Hofacre C.L. Molecular detection and serotyping of infectious bronchitis virus from FTA filter paper. Avian Dis. 2005;49:24–29. doi: 10.1637/7220. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan M., Singanallur N.B., Ralla K., Villuppanoor S.A. Evaluation of FTA cards as a laboratory and field sampling device for the detection of foot-and-mouth disease virus and serotyping by RT-PCR and real-time RT-PCR. J. Virol. Methods. 2008;151:311–316. doi: 10.1016/j.jviromet.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Narayanan M.S., Parthiban M., Sathiya P., Kumanan K. Molecular detection of Newcastle disease virus using flinders tehnology associates-PCR. Vet. Arh. 2010;80:51–60. [Google Scholar]
- Navaneeth Krishnan A., Bhuvaneswari T., Ezhil Praveena P., Jithendran K.P. Paper-based archiving of biological samples from fish for detecting betanodavirus. Arch. Virol. 2016;161:2019–2024. doi: 10.1007/s00705-016-2875-y. [DOI] [PubMed] [Google Scholar]
- Ndunguru J., Taylor N.J., Yadav J., Aly H., Legg J.P., Aveling T., Thompson G., Fauquet C.M. Application of FTA technology for sampling, recovery and molecular characterization of viral pathogens and virus-derived transgenes from plant tissues. Virol. J. 2005;2:45. doi: 10.1186/1743-422X-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo F., Villegas P., Estevez C., Alvarado I., Purvis L.B. Use of FTA filter paper for the molecular detection of Newcastle disease virus. Avian Pathol. 2006;35:93–98. doi: 10.1080/03079450600597410. [DOI] [PubMed] [Google Scholar]
- Price J.A., Simmons A., Bass J., Rush C.M. Use of FTA Technology to extract wheat streak mosaic virus and candidatus liberibacter solanacearum from single vectors. Southwest. Entomol. 2014;39:223–236. doi: 10.3958/059.039.0203. [DOI] [Google Scholar]
- Purvis L.B., Villegas P., Perozo F. Evaluation of FTA paper and phenol for storage, extraction and molecular characterization of infectious bursal disease virus. J. Virol. Methods. 2006;138:66–69. doi: 10.1016/j.jviromet.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Reeve B.W.P., McFall S.M., Song R., Warren R., Steingart K.R., Theron G. Commercial products to preserve specimens for tuberculosis diagnosis: a systematic review. Int. J. Tuberc. Lung Dis. 2018;22:741–753. doi: 10.5588/ijtld.17.0816. [DOI] [PubMed] [Google Scholar]
- Roy Y., Nassuth A. Detection of plant genes, gene expression and viral RNA from tissue prints on FTA cards. Plant Mol. Biol. Report. 2005;23:383–395. doi: 10.1007/bf02788886. [DOI] [Google Scholar]
- Sakai T., Ishii A., Segawa T., Takagi Y., Kobayashi Y., Itou T. Establishing conditions for the storage and elution of rabies virus RNA using FTA((R)) cards. J. Vet. Med. Sci. 2015;77:461–465. doi: 10.1292/jvms.14-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra-Arguello Y.M., Faulkner O., Tellez G., Hargis B.M., Pinheiro do Nascimento V. The use of FTA cards for transport and detection of gyrA mutation of Campylobacter jejuni from poultry. Poult. Sci. 2016;95:798–801. doi: 10.3382/ps/pev384. [DOI] [PubMed] [Google Scholar]
- Smith L.M., Burgoyne L.A. Collecting, archiving and processing DNA from wildlife samples using FTA databasing paper. BMC Ecol. 2004;4:4. doi: 10.1186/1472-6785-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam K.I., Esona M.D., Williams A., Ndze V.N., Boula A., Bowen M.D. Evaluation of BBL Sensi-Discs and FTA(R) cards as sampling devices for detection of rotavirus in stool samples. J. Virol. Methods. 2015;222:41–46. doi: 10.1016/j.jviromet.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Garver L.S., Bingham K.M., Hang J., Jochim R.C., Davidson S.A., Richardson J.H., Jarman R.G. Feasibility of using the mosquito blood meal for rapid and efficient human and animal virus surveillance and discovery. Am. J. Trop. Med. Hyg. 2015;93:1377–1382. doi: 10.4269/ajtmh.15-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]