Abstract
Recently found bat-derived influenza viruses (BatIVs) have hemagglutinin (HA) and neuraminidase (NA) gene segments distinct from those of previously known influenza A viruses. However, pathogenicities of these BatIVs remain unknown since infectious virus strains have not been isolated yet. To gain insight into the biological properties of BatIVs, we generated vesicular stomatitis viruses (VSVs) pseudotyped with the BatIV HA and NA. We found that VSVs pseudotyped with BatIV HAs and NAs efficiently infected particular bat cell lines but not those derived from primates, and that proteolytic cleavage with a trypsin-like protease was necessary for HA-mediated virus entry. Treatment of the susceptible bat cells with some enzymes and inhibitors revealed that BatIV HAs might recognize some cellular glycoproteins as receptors rather than the sialic acids used for the other known influenza viruses. These data provide fundamental information on the mechanisms underlying the cellular entry and host restriction of BatIVs.
Keywords: Bat influenza virus, Hemagglutinin, Pseudotyped vesicular stomatitis virus
Highlights
-
•
Particular bat cell lines may be susceptible to bat influenza viruses.
-
•
Bat influenza viruses do not use sialic acid receptors.
-
•
Some cell type-specific molecules may serve as bat influenza virus receptors.
-
•
Bat influenza viruses may have a limited host range.
Introduction
Influenza A viruses (IAVs), which belong to the family Orthomyxoviridae, have 8 segmented negative sense RNA genomes. IAV is one of the most important zoonotic pathogens, with high morbidity in humans, pigs, horses, and poultry. IAVs have two envelope glycoproteins, hemagglutinin (HA) and neuraminidase (NA), and are divided into subtypes based on antigenicity. IAVs of H1–16 HA and N1–9 NA subtypes have been isolated from water birds such as migratory ducks, the natural reservoir of IAVs (Fouchier et al., 2005, Kida and Yanagawa, 1979, Webster et al., 1992).
HAs are expressed as trimers on the virion surface (Wilson et al., 1981). HA is initially synthesized as an inactive precursor HA0 and subsequently cleaved into HA1 and HA2 subunits by trypsin-like proteases of host cells (Sakai et al., 2014). The proteolytic cleavage of the HA molecule is essential for IAVs to acquire infectivity (Lazarowitz et al., 1973, Wiley and Skehel, 1987). HA1 is responsible for virus binding to sialic acid receptors on the cell surface, and HA2 mediates membrane fusion under acidic conditions in endosomes, thereby delivering the viral genomic RNA into the cytoplasm of target cells (Matlin et al., 1981, Rust et al., 2004). NAs, expressed on the virion surface as tetramers, have sialidase activity that enables mature virus particles to be released from infected cells after budding (Colman, 1994, Webster et al., 1992).
Recently, IAV-like RNA genomes were detected in succession from 2 frugivorous bat species, little yellow-shouldered bats (Sturnira lilium) and flat-faced fruit bats (Artibeus planirostris) in Guatemala and Peru, respectively. The nucleotide sequences of the HA and NA of these bat-derived influenza viruses (BatIVs) were divergent from all previously known IAVs and new subtypes, H17N10 and H18N11, have been proposed (Tong et al., 2012, Tong et al., 2013). However, infectious viruses have not been isolated yet. Previous studies by others tried to rescue BatIVs using a reverse genetics approach, but failed to generate infectious BatIVs (Juozapaitis et al., 2014, Zhou et al., 2014). Thus, the information on the biological properties of BatIVs is mostly speculative and the possible functions of BatIV HAs and NAs are only hypothetical, based on structural analyses (Li et al., 2012, Tong et al., 2013, Zhu et al., 2012, Zhu et al., 2013).
In this study, we utilized a vesicular stomatitis virus (VSV) pseudotype system, enabling us to directly analyze the biological functions of the BatIV glycoproteins, which presumably play important roles in the replication cycle and pathogenicity. We found some bat cell lines susceptible to VSVs pseudotyped with BatIV HAs and NAs. Our data suggest that BatIVs do not use sialic acids as a viral receptor and may have a limited host range, at least considering receptor engagement.
Results
Generation of VSVs pseudotyped with BatIV HAs and/or NAs
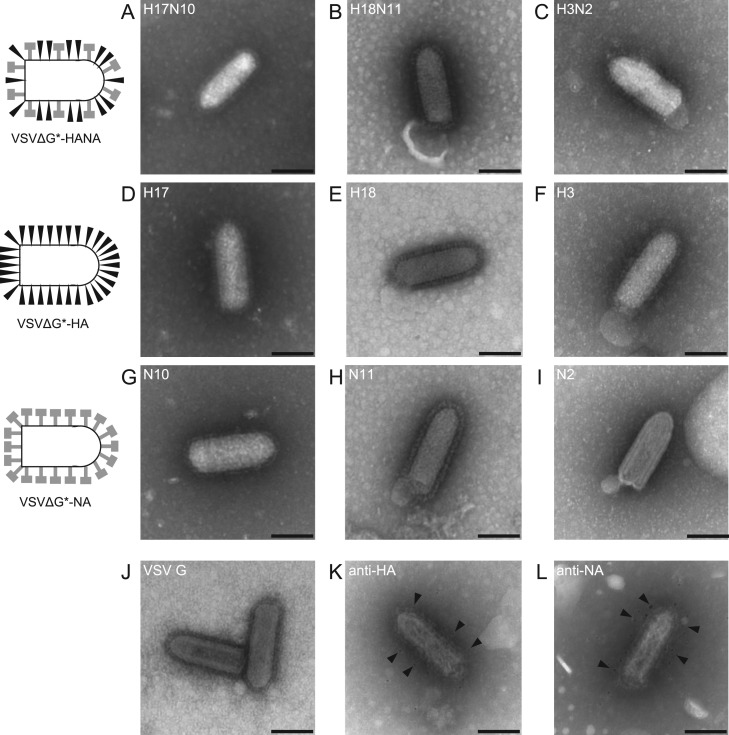
To investigate cellular entry mediated by BatIV glycoproteins, VSVs pseudotyped with BatIV HAs and/or NAs (VSVΔG*-H17N10, -H18N11, -H17, -H18, -N10, and -N11) were generated as described in Materials and Methods. We first observed the virions of these pseudotyped VSVs using transmission electron microscopy ( Fig. 1). We found that the virions of all of these pseudotyped VSVs showed characteristic morphology (i.e. a bullet-like shape) similar to parental VSVΔG*-G. It was noted that VSVs pseudotyped with BatIV HA and NA (Fig. 1A and B), HA alone (Fig. 1D and E), and NA alone (Fig. 1G and H) all had numerous spikes on their surfaces, as was the case with VSVs pseudotyped with IAV HA (H3) and NA (N2) (Fig. 1C), H3 HA alone (Fig. 1F), and N2 NA alone (Fig. 1I). Immune electron microscopy with anti-H17 HA and anti-N10 NA antibodies revealed that both BatIV HA and NA were efficiently incorporated into VSV particles (Fig. 1K and L). No difference was found in the overall morphology among these VSV virions. These data indicated that BatIV HAs and NAs were efficiently incorporated into the VSV particles.
Fig. 1.
Transmission electron microscopy of pseudotyped VSVs. VSVΔG*-H17N10 (A), -H18N11 (B), -H3N2 (C) -H17 (D), -H18 (E), -H3 (F), -N10 (G), -N11 (H), -N2 (I) and VSVΔG*-G (J) were fixed and stained as described in Materials and Methods. For immune transmission electron microscopy of VSVΔG*-H17N10, anti-HA2 monoclonal antibody (K) and anti-N10 NA mouse serum (L) were used. Scale bars represent 100 nm. Arrowheads indicate gold particles.
Cell lines susceptible to VSVs pseudotyped with BatIV glycoproteins
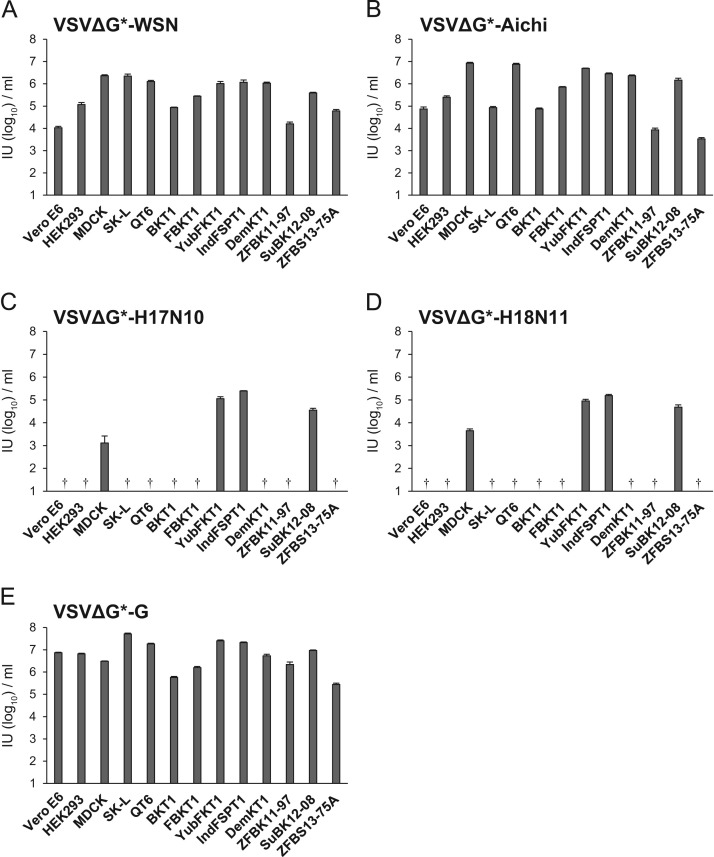
Since previous studies have suggested that cell lines commonly used for IAV propagation are nonpermissive for BatIVs, we screened various cell lines, including bat-derived cells, for susceptibility to pseudotyped VSVs ( Table 1) ( Fig. 2). VSVs pseudotyped with HAs and NAs of BatIVs and well-characterized IAV strains, A/WSN/1933 (H1N1) (WSN) and A/Aichi/2/1968 (H3N2) (Aichi), were generated and treated with trypsin before use, since BatIV HAs, like WSN and Aichi HAs, have a cleavage site potentially recognized by trypsin-like proteases (Tong et al., 2012, Tong et al., 2013). We found that VSVΔG*-WSN, -Aichi, and -VSV G infected all cell lines tested (Fig. 2A, B, and E). On the other hand, VSVΔG*-H17N10 and -H18N11 infected bat cell lines YubFKT1, IndFSPT1, and SuBK12-08, but not the other cell lines tested, except MDCK cells, which were much less susceptible than these bat cells. Since IndFSPT1 cells showed the highest susceptibility to VSVΔG*-H17N10 and -H18N11 (Fig. 2C and D), this cell line was used for the following experiments.
Table 1.
Origins of cell lines used in this study.
| Cell line | Species | Zoological name | Organ |
|---|---|---|---|
| Vero E6 | African green monkey | Chlorocebus sp. | Kidney |
| HEK293 | Human | Homo sapiens | Kidney |
| MDCK | Dog | Canis lupus familiaris | Kidney |
| SK-L | Pig | Sus scrofa domesticus | Kidney |
| QT6 | Japanese quail | Coturnix japonica | Muscle |
| BKT1 | Greater horseshoe bata | Rhinolophus ferrumequinum | Kidney |
| FBKT1 | Yaeyama flying foxa | Pteropus dasymallus yayeyamae | Kidney |
| YubFKT1 | Eastern bent-winged bata | Miniopterus fuliginosus | Kidney |
| IndFSPT1 | Indian flying foxa | Pteropus giganteus | Spleen |
| DemKT1 | Leschenault׳s rousettea | Rousettus leschenaultii | Kidney |
| ZFBK11-97 | Gambian epauletted fruit bata | Epomophorus gambianus | Kidney |
| SuBK12-08 | Schreiber׳s bata | Miniopterus schreibersii | Kidney |
| ZFBS13-75A | Straw-colored fruit batb | Eidolon helvum | Spleen |
Previously described (Maruyama et al., 2014).
Determined by habitat and morphology.
Fig. 2.
Infectivities of pseudotyped VSVs in several cell lines. VSVΔG*-WSN, -Aichi, -H17N10, -H18N11, and VSVΔG*-G were inoculated into several cell lines (Table 1). Infectious units (IUs) of each virus in different cell lines were determined by counting the number of GFP-expressing cells. Each experiment was performed three times, and averages and standard deviations are shown. Infectivities of VSVΔG*-H17N10 and -H18N11 in some cell lines were under the limit of detection (†). Significant differences (student׳s t-test) were found between MDCK and any of the bat cell lines (P<0.01).
Trypsin requirement for the HA function and the dispensability of NA in virus entry
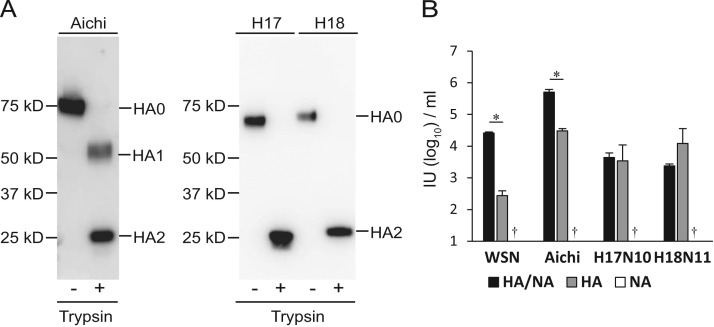
IAV HAs are known to be cleaved into HA1 and HA2 subunits by trypsin-like proteases to acquire the ability to mediate membrane fusion (Klenk and Rott, 1988). Western blotting revealed that both H17 and H18 HAs were cleaved into HA1 and HA2 by trypsin treatment ( Fig. 3A). Thus, we investigated the requirement of HA cleavage for infectivity of pseudotyped VSVs. As expected, VSVs pseudotyped with BatIV glycoproteins did not infect IndFSPT1 cells without trypsin treatment, in a manner consistent with other IAVs (data not shown), whereas trypsin-treated viruses efficiently infected this cell line (Fig. 2). These data indicated that the HA cleavage was a prerequisite for BatIV infectivity. Next, to clarify whether BatIV HAs was responsible for virus entry, VSVΔG*-H17N10, -H18N11,-H17, -H18, -N10, and -N11 were inoculated to IndFSPT1 cells and their infectivities were compared (Fig. 3B). We found that VSVΔG*-H17 and -H18 infected IndFSPT1 cells as efficiently as VSVΔG*-H17N10 and -H18N11, whereas the infectivity of VSV pseudotyped with WSN or Aichi HA alone was much lower than that of VSVs pseudotyped with both HA and NA of the respective viruses. VSVΔG*-N10 and -N11 showed no infectivity, similarly to VSVs pseudotyped with NAs of WSN and Aichi. These results suggest that BatIV HA is the only glycoprotein mediating both virus attachment and membrane fusion and that BatIV NA is dispensable during the entry into cells.
Fig. 3.
Infectivities of pseudotyped VSVs with BatIV HAs and/or NAs in IndFSPT1 cells. (A) VSVΔG*-Aichi, -H17N10, and -H18N11 were treated with or without trypsin (final concentration 0.0005%) for 30 min at 37 °C and then mixed with SDS-PAGE sample buffer with 5% 2-mercaptoethanol. After SDS-PAGE, separated proteins were detected by western blotting with anti-H3 chicken antiserum and anti-HA2 monoclonal antibody 3N12-6-4. (B) VSVs pseudotyped with HAs and/or NAs of WSN and Aichi and VSVΔG*-H17N10, -H17, -N10, -H18N11, -H18, and -N11 were inoculated to IndFSPT1 cells. Infectious units (IUs) were determined by counting the number of GFP-expressing cells. Each experiment was performed three times, and averages and standard deviations are shown. Infectivities of VSV pseudotyped with NAs alone were under the limit of detection (†). Statistical significance was calculated using student׳s t-test (*P<0.01).
Effects of chemical and enzymatic treatments of cells on susceptibility to VSVs pseudotyped with BatIVs
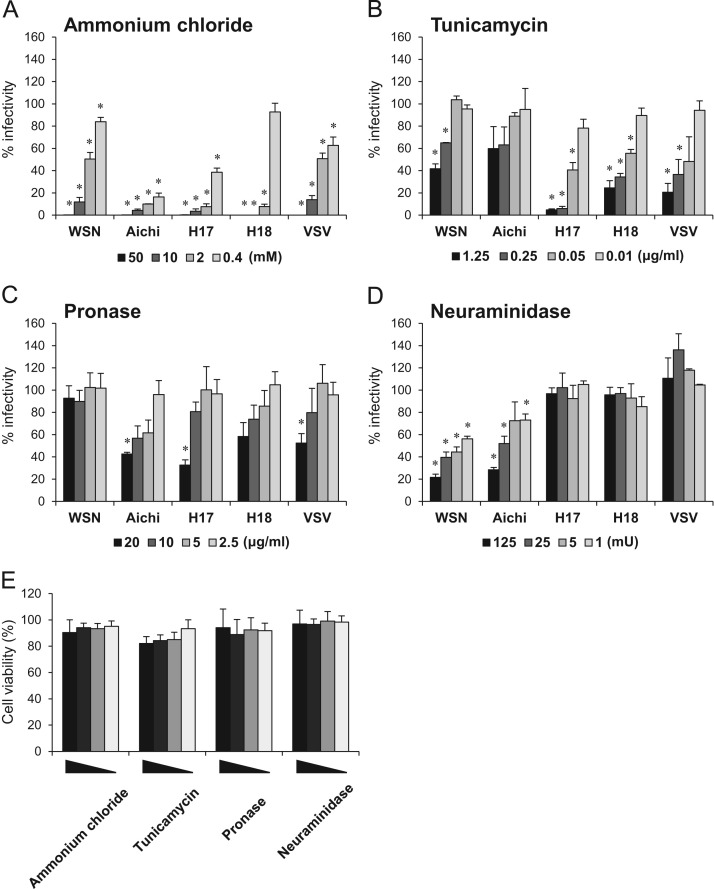
It is generally known that IAV HAs mediate membrane fusion in a low pH-dependent manner (Kida et al., 1983, Rust et al., 2004). To investigate the requirement of endosomal acidification for BatIV HA-mediated membrane fusion, IndFSPT1 cells were treated with ammonium chloride, which is known to neutralize the pH of acidic intracellular compartments, and then infected with VSVΔG*-WSN, -Aichi, -H17N10, -H18N11, and VSVΔG*-G. Treatment of the cells with ammonium chloride markedly reduced the infectivity of VSVΔG*-H17N10 and -H18N11, as was the case with VSVΔG*-G, -WSN, and -Aichi, in a dose-dependent manner, suggesting that BatIV HAs require a low pH for membrane fusion, consistent with the other IAV HAs ( Fig. 4A). To obtain information on the biological characteristics of cellular receptors for BatIVs, IndFSPT1 cells were pretreated with tunicamycin, pronase, or neuraminidase (i.e., an N-linked glycosylation inhibitor, mixture of proteases, and sialidase, respectively), and then infected with pseudotyped VSVs (Fig. 4B–D). Tunicamycin treatment markedly reduced the infectivities of VSVΔG*-G, -H17N10, and -H18N11, but less significantly those of VSVΔG*-WSN and -Aichi (Fig. 4B). Preincubation of cells with pronase reduced the infectivities of the pseudotyped VSVs, except for VSVΔG*-WSN (Fig. 4C). Neuraminidase treatment reduced VSVΔG*-WSN and -Aichi infectivities, but interestingly did not affect the infectivities of VSVΔG*-H17N10 and -H18N11 (Fig. 4D). We confirmed that no remarkable cytotoxicity was observed during these treatments (Fig. 4E). These results suggest that BatIV HAs do not recognize sialic acids which are critical components of the IAV receptor and some other molecules such as glycoproteins may serve as BatIV receptors.
Fig. 4.
Effects of chemical and enzymatic modification on infectivities of pseudotyped VSVs. IndFSPT1 cells were treated with ammonium chloride (A), tunicamycin (B), pronase (C), or neuraminidase (D) as described in Materials and Methods. Treated cells were then infected with VSVΔG*-WSN, -Aichi, -H17N10, -H18N11, and VSVΔG*-G appropriately diluted to yield 200–1000 IUs. The percentages of infectivity were determined by setting the number of the untreated cells to 100%. Each experiment was performed three times, and averages and standard deviations are shown. Cell viabilities were measured by the alamar blue assay (E). The percentages of fluorescence were determined by setting the number of the untreated cells to 100%. Each experiment was performed three times, and averages and standard deviations are shown. Statistical significances compared to untreated cells were calculated using student׳s t-test (*P<0.01).
Discussion
In recent years, particular attention has been paid to bat-derived viruses since some species of bats have been reported to be reservoirs of several viral zoonotic pathogens (e.g., lyssavirus, henipavirus, SARS coronavirus, and Marburgvirus) (Calisher et al., 2006, Smith and Wang, 2013, Wang et al., 2011, Wong et al., 2007). Although the zoonotic potential of BatIVs has not been fully evaluated yet, recent studies generated reassortant viruses that had HA and NA gene segments of well-characterized IAVs (i.e., H1, H3, and H7 HAs and N1, N2, and N7 NAs) and the other gene segments derived from BatIVs, and demonstrated that the reassortant viruses replicated in cultured cells and caused severe disease in mice (Juozapaitis et al., 2014, Zhou et al., 2014). However, characterization of BatIV HAs and NAs remains an open research problem, since reassortant viruses carrying the BatIV HA and NA gene segments have not been rescued due to the lack of information on cells susceptible to this novel virus. In this study, we first determined the potentially permissive bat cell lines using VSVs pseudotyped with BatIV HAs and NAs.
We demonstrated that VSVΔG*-H17N10 and -H18N11 efficiently infected the bat-derived cell lines IndFSPT1, YubFKT1, and SuBK12-08. While IndFSPT1 was derived from Pteropus giganteus (family Pterodidae), YubFKT1 and SuBK12-08 were prepared from bats belonging to the same species (Miniopterus sp., family Miniopteridae). Based on a phylogenetic study of bats (Agnarsson et al., 2011), Miniopteridae belongs to the same cluster as Phyllostomidae, from which H17N10 and H18N11 BatIVs were detected, little yellow-shouldered bats (S. lilium) and flat-faced fruit bats (A. planirostris), respectively (Tong et al., 2012, Tong et al., 2013). Thus, BatIV HAs appear to recognize cell surface molecules shared among the bats at least in Miniopteridae and Phyllostomidae families. IndFSPT1 should also have such molecules since it showed the highest susceptibility to BatIV HA-pseudotyped VSVs. It was noted that VSVΔG*-H17N10 and -H18N11 also infected MDCK cells, although less efficiently than these bat cell lines. This result might contradict a previous report that H17 HA did not bind to the surface of MDCK cells (Sun et al., 2013). However, it is conceivable that the binding affinity of BatIV HA to MDCK cell surface molecules is quite low and thus below the level of detection in the assay used in the previous study. In the present study, MDCK cells indeed showed much lower susceptibility to BatIV HA-pseudotyped VSVs than YubFKT1, IndFSPT1, and SuBK12-08. Nonetheless, it would be interesting to clarify whether MDCK cells express some BatIV receptor molecules shared with the bat cell lines.
It is also noteworthy that VSVΔG*-H17N10 and -H18N11 did not infect Vero E6, HEK293, SK-L, and QT6 cells. Previous studies show that quails can act as an intermediate host in the interspecies spread of avian IAVs (Makarova et al., 2003, Perez et al., 2003, Uchida et al., 2011). Furthermore, pigs are thought to serve as “mixing vessels” for the production of reassortant viruses between avian and human IAVs (Chang et al., 2009, Hinshaw et al., 1981, Ito et al., 1998, Kida et al., 1994, Scholtissek et al., 1985). Our results suggest that BatIVs do not readily infect humans, pigs, or birds and support that notion that these viruses have limited zoonotic potential (Juozapaitis et al., 2014, Zhou et al., 2014).
It is known that VSV G protein and IAV HA recognize ubiquitous cell surface molecules for virus entry. VSV G recognizes various cell surface molecules and thus VSV exhibits remarkably robust and pantropic infectivity (Finkelshtein et al., 2013, Johannsdottir et al., 2009, Lichty et al., 2004, Roche et al., 2008). IAV HAs recognize sialic acids typically occupying the terminal positions of glycoproteins or glycolipids (Gambaryan et al., 2005, Suzuki et al., 2000). Accordingly, VSVΔG*-G, -WSN, and -Aichi infected all cell lines used in this study, whereas we found that VSVΔG*-H17N10 and -H18N11 infected only particular bat cell lines and that neuraminidase treatment did not affect the infectivities of VSVΔG*-H17N10 and -H18N11. This result was in agreement with previous results based on the crystal structure analysis and surface plasmon resonance of sialylated glycans with α2,3-linkage or α2,6-linkage (Sun et al., 2013, Zhu et al., 2013). Glycan microarray analyses also showed that H17 HA did not display obvious avidity to any glycans (Sun et al., 2013). Interestingly, we found that the infectivities of VSVΔG*-H17N10 and -H18N11 were markedly reduced by the treatment of IndFSPT1 cells with tunicamycin, which inhibits N-linked glycosylation, leading to unfolding or misfolding of proteins and inhibition of glycoprotein expression. Pretreatment of the cells with pronase also reduced the infectivities of VSVΔG*-H17N10 and -H18N11. Taken together, our data suggest that some particular glycoprotein(s) serve as receptors for BatIVs.
Previous studies indicated that N10 NA did not have sialidase activity (Li et al., 2012, Zhu et al., 2012). It was also shown that most of the amino acid residues responsible for NA activity were substituted, and proposed that N10 NA protein should be termed an NA-like protein (Zhu et al., 2012). In this study, we found that VSVs pseudotyped with BatIV NAs alone were not infectious, confirming that NA did not play a central role in IAV entry into cells. However, it should be noted that the production efficiency of pseudotyped VSVs bearing WSN and Aichi HAs alone was much lower than that of VSVs pseudotyped with both HAs and NAs, suggesting that NA activity facilitated virus release from infected cells and/or increased the HA function (Su et al., 2009). By contrast, no remarkable difference was found in the infectivity between VSVs pseudotyped with BatIV glycoproteins (i.e., HA alone vs. HA and NA). These data suggest that, unlike the other IAVs, the target molecules of BatIV HAs and NAs are different and that the “HA–NA balance” concept proposed for IAVs does not be applied to BatIVs.
Because H17N10 and H18N11 BatIVs have never been isolated, their ability to infect humans and other mammals and the pathogenic potential for these hosts can only be hypothesized. In this study, the replication-incompetent VSV pseudotype system enabled us to investigate the cellular tropism controlled by the interaction between BatIV HA and its cellular ligand, which might be some glycoproteins. Although a reverse genetics approach and in vivo experiments for the infectious BatIV are needed to provide direct evidence of its pathogenicity and host specificity, our data suggest that BatIV may preferentially infect particular bat species.
Materials and methods
Cells
HEK293, HEK293T, and Vero E6 cells were grown in Dulbecco׳s modified Eagle׳s medium (DMEM) with 10% fetal calf serum (FCS) and penicillin–streptomycin. MDCK cells were grown in Dulbecco׳s modified Eagle׳s medium (DMEM) with 10% calf serum, L-glutamine, and penicillin–streptomycin. Bat cell lines BKT1, FBKT1, YubFKT1, IndFSPT1, DemKT1, ZFBK11-97, SuBK12-08, and ZFBS13-76A were established as described previously (Maeda et al., 2008, Maruyama et al., 2014). All bat cell lines were grown in RPMI-1640 medium with 10% FCS, L-glutamine, and penicillin–streptomycin.
Construction of plasmids expressing HAs and NAs
Coding regions of the HAs and NAs of BatIVs were synthesized in vector pUC19 or pUCFa, based on the nucleotide sequences of GenBank (accession numbers for H17 HA, N10 NA, H18 HA, and N11 NA: CY103892, CY103894, CY125945, and CY125947, respectively) (FASMAC). Each coding region of the viral proteins was amplified by PCR with primers including restriction sites, the kozak sequence, and the stop codon. After digestion by restriction enzymes, each gene was cloned into the mammalian expression vector pCAGGS (Niwa et al., 1991). H1 HA and N1 NA of A/WSN/1933 (H1N1) (WSN), and H3 HA and N2 NA of A/Aichi/2/1968 (H3N2) (Aichi) were cloned into pCAGGS as described previously (Muramatsu et al., 2013).
Vesicular stomatitis viruses (VSVs) pseudotyped with HAs and/or NAs
Using VSV containing the green fluorescent protein (GFP) gene instead of the receptor-binding VSV G protein gene (VSVΔG*-G), pseudotyped viruses with HAs and/or NAs of BatIVs, WSN, and Aichi were generated as described previously (Takada et al., 1997). VSVs pseudotyped with IAV glycoproteins were pretreated with trypsin (final concentration 0.0005%) for 30 min at 37 °C, followed by incubation with an anti-VSV G monoclonal antibody, VSV-G(N)1-9, to abolish the background infectivity of parental VSVΔG*-G (Nakayama et al., 2011). For virus titration, 10-fold diluted pseudotyped VSVs were inoculated into confluent monolayers of each cell line on 96-well plates, and the infectious unit (IU) in each cell line was determined 20 hours later by counting the number of GFP-expressing cells under a fluorescent microscope.
Electron microscopy
Transmission electron microscopy was carried out as described previously (Maruyama et al., 2014). Pseudotyped VSVs fixed with 0.25% glutaraldehyde were adsorbed to collodion-carbon-coated copper grids and negatively stained with 2% phosphotungstic acid solution (pH=5.8). For immune transmission electron microscopy, we used an anti-HA2 monoclonal antibody (3N12-6-4) broadly cross-reactive to group 1 HA subtypes, anti-N10 NA mouse serum (FM0137) produced by immunization with a synthetic peptide corresponding to amino acid residues 328–343 (AQEKGEGGIQGFILDE) of N10 NA, and an immunogold-conjugated goat anti-mouse IgG (H+L) polyclonal antibody (BB International). Samples were examined with an H-7650 electron microscope (Hitachi) at 80 kV.
SDS-PAGE and western blotting
Pseudotyped VSVs were treated with or without trypsin (final concentration 0.0005%) for 30 min at 37 °C and then mixed with SDS-PAGE sample buffer with 5% 2-mercaptoethanol and boiled for 5 minutes. After electrophoresis on 5–20% SuperSep (Wako), separated proteins were blotted on a polyvinylidene difluoride membrane (Millipore). The membrane was incubated with an anti-H3N2 chicken polyclonal antiserum or anti-HA2 monoclonal antibody 3N12-6-4, which reacts to H1, H2, H5, H6, H17, and H18 HAs, followed by incubation with peroxidase-conjugated rabbit anti-chicken IgY (H+L) or goat anti-mouse IgG (H+L) (Jackson ImmunoResearch). The bound antibodies were visualized with Immobilon Western (Millipore).
Cell treatment with enzymes and inhibitors
IndFSPT1 cells were preincubated with the medium containing an endosomal acidification inhibitor, ammonium chloride (Wako), at 37 °C for 2 h in a CO2 incubator, and then infected with pseudotyped VSVs appropriately diluted to yield 200–1000 IUs, followed by incubation in the presence of ammonium chloride. IndFSPT1 cells were also pretreated with pronase (a mixture of endo- and exoproteases from Streptomyces griseus) (Calbiochem) (Narahashi et al., 1968), for 20 min, an N-glycosylation inhibitor (tunicamycin from Streptomyces sp.) (Sigma) for 8 h, which blocks the reaction of UDP-GlcNAc and dolichol phosphate in the first step of glycoprotein synthesis, thus inhibiting the synthesis of N-linked glycoproteins, or neuraminidase from Vibrio cholerae (Roche) (Uchida et al., 1977) for 1 h at 37 °C in a CO2 incubator. Treated cells were washed with serum free RPMI-1640 medium 3 times, and then incubated with pseudotyped VSVs appropriately diluted to yield 200–1000 IUs for 1 h. After adsorption of the virus, the inoculum was aspirated and the growth medium (10% FCS RPMI-1640 medium) was added. Cells were incubated for 20 h, and infected cells were counted under a fluorescent microscope. Cell viabilities were assessed by the alamar blue assay. After treatments of each enzyme and inhibitor, cells were incubated with FCS-free RPMI-1640 medium containing 10% Alamar blue (Biosource) for 2 h, and fluorescence with excitation wavelength at 530–560 nm was measured using EnVision (PerkinElmer).
Acknowledgments
This work was supported by KAKENHI (15H01249), a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Society for the Promotion of Science (JSPS) (14J00026), Japan, and partly by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) (15FM0108008H0001) and the Japan Science and Technology Agency (JST) and Japan International Cooperation Agency (JICA) within the framework of the Science and Technology Research Partnership for Sustainable Development (SATREPS) (15JM0110005H0004). Funding was also provided by the Program for Leading Graduate Schools from MEXT, Japan.
References
- Agnarsson I., Zambrana-Torrelio C.M., Flores-Saldana N.P., May-Collado L.J. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia) PLoS Curr. 2011;3 doi: 10.1371/currents.RRN1212. RRN1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.Y., Shih S.R., Shao P.L., Huang D.T., Huang L.M. Novel swine-origin influenza virus A (H1N1): the first pandemic of the 21st century. J. Formos. Med. Assoc. 2009;108:526–532. doi: 10.1016/S0929-6646(09)60369-7. [DOI] [PubMed] [Google Scholar]
- Colman P.M. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci. 1994;3:1687–1696. doi: 10.1002/pro.5560031007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Munster V., Wallensten A., Bestebroer T.M., Herfst S., Smith D., Rimmelzwaan G.F., Olsen B., Osterhaus A.D. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaryan A., Yamnikova S., Lvov D., Tuzikov A., Chinarev A., Pazynina G., Webster R., Matrosovich M., Bovin N. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology. 2005;334:276–283. doi: 10.1016/j.virol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Hinshaw V.S., Webster R.G., Easterday B.C., Bean W.J., Jr. Replication of avian influenza A viruses in mammals. Infect. Immun. 1981;34:354–361. doi: 10.1128/iai.34.2.354-361.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Couceiro J.N., Kelm S., Baum L.G., Krauss S., Castrucci M.R., Donatelli I., Kida H., Paulson J.C., Webster R.G., Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsdottir H.K., Mancini R., Kartenbeck J., Amato L., Helenius A. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 2009;83:440–453. doi: 10.1128/JVI.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juozapaitis M., Aguiar Moreira E., Mena I., Giese S., Riegger D., Pohlmann A., Hoper D., Zimmer G., Beer M., Garcia-Sastre A., Schwemmle M. An infectious bat-derived chimeric influenza virus harbouring the entry machinery of an influenza A virus. Nat. Commun. 2014;5:4448. doi: 10.1038/ncomms5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H., Ito T., Yasuda J., Shimizu Y., Itakura C., Shortridge K.F., Kawaoka Y., Webster R.G. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 1994;75(Pt 9):2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- Kida H., Webster R.G., Yanagawa R. Inhibition of virus-induced hemolysis with monoclonal antibodies to different antigenic areas on the hemagglutinin molecule of A/seal/Massachusetts/1/80 (H7N7) influenza virus. Arch. Virol. 1983;76:91–99. doi: 10.1007/BF01311693. [DOI] [PubMed] [Google Scholar]
- Kida H., Yanagawa R. Isolation and characterization of influenza a viruses from wild free-flying ducks in Hokkaido, Japan. Zentralbl. Bakteriol. Orig. A. 1979;244:135–143. [PubMed] [Google Scholar]
- Klenk H.D., Rott R. The molecular biology of influenza virus pathogenicity. Adv. Virus Res. 1988;34:247–281. doi: 10.1016/S0065-3527(08)60520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz S.G., Compans R.W., Choppin P.W. Proteolytic cleavage of the hemagglutinin polypeptide of influenza virus. Function of the uncleaved polypeptide HA. Virology. 1973;52:199–212. doi: 10.1016/0042-6822(73)90409-1. [DOI] [PubMed] [Google Scholar]
- Li Q., Sun X., Li Z., Liu Y., Vavricka C.J., Qi J., Gao G.F. Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus. Proc. Natl. Acad. Sci. USA. 2012;109:18897–18902. doi: 10.1073/pnas.1211037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty B.D., Power A.T., Stojdl D.F., Bell J.C. Vesicular stomatitis virus: re-inventing the bullet. Trends Mol. Med. 2004;10:210–216. doi: 10.1016/j.molmed.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Maeda K., Hondo E., Terakawa J., Kiso Y., Nakaichi N., Endoh D., Sakai K., Morikawa S., Mizutani T. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae) Emerg. Infect. Dis. 2008;14:347–349. doi: 10.3201/eid1402.070932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova N.V., Ozaki H., Kida H., Webster R.G., Perez D.R. Replication and transmission of influenza viruses in Japanese quail. Virology. 2003;310:8–15. doi: 10.1016/s0042-6822(03)00094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama J., Miyamoto H., Kajihara M., Ogawa H., Maeda K., Sakoda Y., Yoshida R., Takada A. Characterization of the envelope glycoprotein of a novel filovirus, lloviu virus. J. Virol. 2014;88:99–109. doi: 10.1128/JVI.02265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin K.S., Reggio H., Helenius A., Simons K. Infectious entry pathway of influenza virus in a canine kidney cell line. J. Cell Biol. 1981;91:601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Yoshida R., Miyamoto H., Tomabechi D., Kajihara M., Maruyama J., Kimura T., Manzoor R., Ito K., Takada A. Heterosubtypic antiviral activity of hemagglutinin-specific antibodies induced by intranasal immunization with inactivated influenza viruses in mice. PLoS One. 2013;8:e71534. doi: 10.1371/journal.pone.0071534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama E., Tomabechi D., Matsuno K., Kishida N., Yoshida R., Feldmann H., Takada A. Antibody-dependent enhancement of Marburg virus infection. J. Infect. Dis. 2011;204(Suppl 3):S978–S985. doi: 10.1093/infdis/jir334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narahashi Y., Shibuya K., Yanagita M. Studies on proteolytic enzymes (pronase) of Streptomyces griseus K-1. II. Separation of exo- and endopeptidases of pronase. J. Biochem. 1968;64:427–437. doi: 10.1093/oxfordjournals.jbchem.a128914. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Perez D.R., Lim W., Seiler J.P., Yi G., Peiris M., Shortridge K.F., Webster R.G. Role of quail in the interspecies transmission of H9 influenza A viruses: molecular changes on HA that correspond to adaptation from ducks to chickens. J. Virol. 2003;77:3148–3156. doi: 10.1128/JVI.77.5.3148-3156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S., Albertini A.A., Lepault J., Bressanelli S., Gaudin Y. Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited. Cell. Mol. Life Sci. 2008;65:1716–1728. doi: 10.1007/s00018-008-7534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Lakadamyali M., Zhang F., Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 2004;11:567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Ami Y., Tahara M., Kubota T., Anraku M., Abe M., Nakajima N., Sekizuka T., Shirato K., Suzaki Y., Ainai A., Nakatsu Y., Kanou K., Nakamura K., Suzuki T., Komase K., Nobusawa E., Maenaka K., Kuroda M., Hasegawa H., Kawaoka Y., Tashiro M., Takeda M. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J. Virol. 2014;88:5608–5616. doi: 10.1128/JVI.03677-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C., Burger H., Kistner O., Shortridge K.F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- Smith I., Wang L.F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013;3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B., Wurtzer S., Rameix-Welti M.A., Dwyer D., van der Werf S., Naffakh N., Clavel F., Labrosse B. Enhancement of the influenza A hemagglutinin (HA)-mediated cell–cell fusion and virus entry by the viral neuraminidase (NA) PLoS One. 2009;4:e8495. doi: 10.1371/journal.pone.0008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Shi Y., Lu X., He J., Gao F., Yan J., Qi J., Gao G.F. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Rep. 2013;3:769–778. doi: 10.1016/j.celrep.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Ito T., Suzuki T., Holland R.E., Jr., Chambers T.M., Kiso M., Ishida H., Kawaoka Y. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 2000;74:11825–11831. doi: 10.1128/jvi.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Robison C., Goto H., Sanchez A., Murti K.G., Whitt M.A., Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Li Y., Rivailler P., Conrardy C., Castillo D.A., Chen L.M., Recuenco S., Ellison J.A., Davis C.T., York I.A., Turmelle A.S., Moran D., Rogers S., Shi M., Tao Y., Weil M.R., Tang K., Rowe L.A., Sammons S., Xu X., Frace M., Lindblade K.A., Cox N.J., Anderson L.J., Rupprecht C.E., Donis R.O. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Zhu X., Li Y., Shi M., Zhang J., Bourgeois M., Yang H., Chen X., Recuenco S., Gomez J., Chen L.M., Johnson A., Tao Y., Dreyfus C., Yu W., McBride R., Carney P.J., Gilbert A.T., Chang J., Guo Z., Davis C.T., Paulson J.C., Stevens J., Rupprecht C.E., Holmes E.C., Wilson I.A., Donis R.O. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y., Tsukada Y., Sugimori T. Distribution of neuraminidase in Arthrobacter and its purification by affinity chromatography. J. Biochem. 1977;82:1425–1433. doi: 10.1093/oxfordjournals.jbchem.a131830. [DOI] [PubMed] [Google Scholar]
- Uchida Y., Kanehira K., Mase M., Takemae N., Watanabe C., Usui T., Fujimoto Y., Ito T., Igarashi M., Ito K., Takada A., Sakoda Y., Okamatsu M., Yamamoto Y., Nakamura K., Kida H., Hiromoto Y., Tsuda T., Saito T. Genetic characterization and susceptibility on poultry and mammal of H7N6 subtype avian influenza virus isolated in Japan in 2009. Vet. Microbiol. 2011;147:1–10. doi: 10.1016/j.vetmic.2010.05.037. [DOI] [PubMed] [Google Scholar]
- Wang L.F., Walker P.J., Poon L.L. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr. Opin. Virol. 2011;1:649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D.C., Skehel J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Wong S., Lau S., Woo P., Yuen K.Y. Bats as a continuing source of emerging infections in humans. Rev. Med. Virol. 2007;17:67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Ma J., Liu Q., Bawa B., Wang W., Shabman R.S., Duff M., Lee J., Lang Y., Cao N., Nagy A., Lin X., Stockwell T.B., Richt J.A., Wentworth D.E., Ma W. Characterization of uncultivable bat influenza virus using a replicative synthetic virus. PLoS Pathog. 2014;10:e1004420. doi: 10.1371/journal.ppat.1004420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Yang H., Guo Z., Yu W., Carney P.J., Li Y., Chen L.M., Paulson J.C., Donis R.O., Tong S., Stevens J., Wilson I.A. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc. Natl. Acad. Sci. USA. 2012;109:18903–18908. doi: 10.1073/pnas.1212579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Yu W., McBride R., Li Y., Chen L.M., Donis R.O., Tong S., Paulson J.C., Wilson I.A. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc. Natl. Acad. Sci. USA. 2013;110:1458–1463. doi: 10.1073/pnas.1218509110. [DOI] [PMC free article] [PubMed] [Google Scholar]