Highlights
► KPNA2 has emerged as a potential cancer biomarker due to the elevated expression observed in a variety of cancer forms. ► The aberrant expression of KPNA2 is associated with poor survival and other adverse characteristics. ► Cellular studies have indicated that KPNA2 plays a role in the malignant transformation of cancer cells. ► KPNA2 shuttles a variety of proteins, including several which are intimately associated with carcinogenesis.
Keywords: Karyopherin α 2 (KPNA2), Importin, Biomarker, Carcinogenesis, Nuclear transport
Abstract
In recent years, Karyopherin α 2 (KPNA2) has emerged as a potential biomarker in multiple cancer forms. The aberrant high levels observed in cancer tissue have been associated with adverse patient characteristics, prompting the idea that KPNA2 plays a role in carcinogenesis. This notion is supported by studies in cancer cells, where KPNA2 deregulation has been demonstrated to affect malignant transformation. By virtue of its role in nucleocytoplasmic transport, KPNA2 is implicated in the translocation of several cancer-associated proteins. We provide an overview of the clinical studies that have established the biomarker potential of KPNA2 and describe its functional role with an emphasis on established associations with cancer.
1. Introduction
In cancer, dysfunction of the cellular transport machinery is commonly observed. The consequences for the localization and function of tumor-suppressors and oncogenes have lead to discussions of the therapeutic potential of targeting this machinery [1]. In this regard, one group of molecules of particular interest is the karyopherins. They act as carrier proteins in the selective bi-directional shuttling of proteins between the cytoplasm and the nucleus, termed nucleocytoplasmic transport.
Nucleocytoplasmic transport occurs through large Nuclear Pore Complexes (NPCs) in the nuclear membrane. Whereas some factors are able to diffuse passively through the pores, the shuttling of macromolecules larger than about 40 kDa is actively mediated by karyopherins [2]. The karyopherin family comprises both import factors (importins) and export factors (exportins) and more than 20 members of the family have been described [3]. They participate in several nuclear transport pathways into and out of the nucleus, with the classical nuclear protein import pathway as one of the best characterized pathways.
Nuclear import via the classical pathway is mediated by heterodimers consisting of importin β (or karyopherin β-1) and a member of the karyopherin α (importin α) family. The karyopherin α proteins recognize cargo proteins by virtue of their nuclear localization signal (NLS). Whereas the karyopherin α recognizes the cargo and acts as an adaptor, the importin β facilitates docking to and translocation through the NPC [3]. In the nucleus, importin β is bound by RanGTP which leads to the dissociation of the ternary complex and concurrent release of the cargo [2]. Importin β complexed with RanGTP is recycled to the cytoplasm on its own, whereas karyopherin α bind RanGTP as well as the export factor, CAS, another member of the Importin β family, before it is subsequently shuttled to the cytoplasm [2].
Karyopherin α 2 (KPNA2) is one of seven described members of the karyopherin α family [4]. The KPNA2 protein (also known as importin α-1 or RAG cohort 1) consists of 529 amino acids and weighs about 58 kDa. Its domain structure was initially delineated in the mid 90s [5], [6], [7], [8]. The protein comprise an N-terminal hydrophilic importin β-binding domain, a central hydrophobic region consisting of 10 armadillo (ARM) repeats, which binds the cargo’s NLS, and a short acidic C-terminus with no reported function. CAS, the factor responsible for the recycling of KPNA2 to the cytoplasm, binds the 10th ARM repeat [3]. The importin β-binding domain has been demonstrated to possess an auto-inhibitory function ensuring that KPNA2 is only translocated when bound to importin β as well as a cargo molecule [9], [10].
Several studies have linked KPNA2 to cancer. Here we present, for the first time, an overview of these reports. We describe the clinical studies which have demonstrated that KPNA2 is highly expressed in multiple cancer forms and that its aberrant expression is often tied to an adverse outcome for the patients. We also cover the studies of KPNA2 in cancer cells and present some of the cancer-associated proteins translocated by KPNA2.
2. Aberrant expression of KPNA2 in cancer – a marker of poor prognosis
In recent years elevated levels of KPNA2 have been observed in a variety of malignancies (summarized in Table 1 ). Following the discovery of the prognostic value of high KPNA2 expression in breast cancer by Dahl et al. [11], [12], [13], aberrant KPNA2 levels has been described in a variety of other cancer forms. These include melanoma [14], cervical cancer [15], esophageal cancer [16], lung cancer [17], ovarian cancer [18], prostate cancer [19], brain cancer [20], liver cancer [21] and bladder cancer [22].
Table 1.
Expression of KPNA2 in cancer tissue and its effect on patient outcome.
| Cancer type | Sample size | Expression compared to normal | Main localization | Effect on survival for KPNA2-positive samples | Hazard ratioa | Main conclusion |
|---|---|---|---|---|---|---|
| Breast cancer [11] | 272 Paired samples | 56% of breast cancers and 1.8% of matched normals | Nucleus | OS in months: 101 [90–112] vs. 120 [110–129] | 2.42 [1.20–4.88] | KPNA2 overexpression is an independent risk factor |
| Melanoma [14] | 238 | NA | Cytoplasm | 4-year OS: 66% [57–75%] vs. 85% [74–95%] | NA | KPNA2 overexpression is a risk factor |
| Breast cancer [12] | 83 Matched normal, DCIS, invasive | 0% of adjacent benign tissues, 21.3% of DCIS and 31% of invasive carcinomas | Nucleus | RFS in months: 69 [47–92] vs. 118 [100–135] for invasive samples | NA | KPNA2 overexpression is a risk factor |
| Breast cancer [13] | 191 | NA | Nucleus | OS in months: 62.5 [32.3–89.7] vs. 103.6 [87.7–119.4] | OS: 1.86 [1.07–3.23] | KPNA2 overexpression is an independent risk factor irrespective of treatment intensity |
| Cervical cancer [15] | 26 | About 4.5× higher levels in cancer tissue by RT-PCR | Cytoplasm, nuclear envelope | NA | NA | KPNA2 is overexpressed in cancer tissue |
| Esophageal cancer [16] | 116 | No expression vs. expression in 51.7% (60/116) of cancers | Nucleus | 5-year OS: 41.6% vs. 62.3% | NA | KPNA2 overexpression is a risk factor |
| Lung cancer [17] | 66 Paired samples | 87.9% (58/66) of cancers vs. 4.5% (3/66) of matched normals | Nucleus | NA | NA | KPNA2 is overexpressed in tumors as well as serum |
| Ovarian cancer [18] | 102 | 8× normal expression by microarray. IHC: 0% (0/15) of normals and 49.0% (50/102) of cancers | Nucleus | 5-year OS: 60.5% vs. 73.1% | NA | KPNA2 overexpression is a risk factor |
| Prostate cancer [19] | 678 | Expression in 42.4% of the samples | Nucleus | 5 year RFS: 0.607 (SE = 0.055) vs. 0.794 (SE = 0.034) | 2.129 [1.332–3.403] | KPNA2 overexpression is an independent risk factor for recurrence |
| Liver cancer [21] | 124 | Expression in 36.3% of the samples | Nucleus | Early recurrence in 72.2% (13/18) vs. 31.6% (18/57) | RFS: 1.781 [1.188–2.730] | KPNA2 overexpression is an independent risk factor for survival and early recurrence |
| Bladder cancer [non-invasive] [22] | 234 | NA | Nucleus | Progression to muscle-invasive disease in 49% (59/120) vs. 25% (28/114) | PFS: 2.59 [1.49–4.49] | KPNA2 overexpression is an independent risk factor for progression |
| Bladder cancer [invasive] [22] | 377 | NA | Nucleus | Demonstrated as HRs | RFS: 1.66 [1.17–2.36] OS: 1.47 [1.07–2.03] | KPNA2 overexpression is an independent risk factor |
| Brain cancer [20] | 106 | Expression in all malign samples | Nucleus | Demonstrated as HRs | OS: 1.94 [1.01–3.7] PFS: 1.73 [0.92–3.22] | KPNA2 overexpression is an independent risk factor |
Paired samples designates that for each examined tumor sample a corresponding normal sample was also assayed. 95% confidence intervals have been indicated in brackets where applicable.
Abbreviations: DCIS: ductal carcinoma in situ, IHC: immunohistochemistry, NA: not applicable (e.g. not investigated), OS: overall survival, PFS: progression-free survival, RFS: recurrence-free survival, SE: standard error.
Portrayed hazard ratios are derived from multivariate analysis.
The majority of the studies have investigated the association between KPNA2 levels and patient outcome. Irrespective of cancer type, elevated expression of KPNA2 has been demonstrated to correlate with a poor prognosis. Importantly, several studies have established KPNA2 to be an independent prognostic factor compared to well-established clinical factors [11], [13], [19], [20], [21], [22]. Of potential interest is the observation in lung cancer where high levels of KPNA2 could also be detected in patient serum [17]. This raises the possibility that adverse KPNA2 levels can be detected in blood samples in other cancer forms as well.
The observed levels of KPNA2 in cancer tissues are markedly elevated compared to normal tissue. At the transcriptional level a 5–10-fold increase in KPNA2 expression has been reported [15], [18] and these findings are supported by immunohistochemical observations. In studies where normal samples have been included 0–5% of them stain for KPNA2 compared to 30–90% of the cancer samples [11], [12], [17], [18], [20]. As such, KPNA2 appears to be markedly upregulated in cancer tissue and the observed KPNA2 appears to be predominantly situated in the nucleus (see Table 1).
The reported association between KPNA2 expression and tumor stage and grade [11], [12], [13], [16], [18], [19], [20], [21], indicates that KPNA2 expression generally increases through tumor progression. However, studies do indicate that aberrant KPNA2 expression can be found in early lesions, such as the ductal carcinoma in situ (DCIS) in breast cancer [12] and non-invasive bladder cancer samples [22]. These findings are important, as they represent opportunities to gain prognostic information at an early stage. Furthermore, they suggest that KPNA2 could potentially participate in carcinogenesis.
The latter idea is augmented by findings which associates KPNA2 expression with an increased degree of malignancy. Besides the decrease in patient survival, high KPNA2 expression has also been correlated to an increased risk of recurrence in prostate cancer [19] and ovarian cancer [18] and an increased risk of progression in bladder cancer [22]. Furthermore, high KPNA2 expression has been associated with lymphatic spread and venous invasion in several cancers [12], [16], [17], [21], the latter of which is considered to be of particular importance in the process of developing distant metastases [23]. In accordance, high KPNA2 expression was coupled with a significantly increased risk of developing distant metastases in bladder cancer [22]. With further possible connections to cancer malignancy, high expression of KPNA2 has been correlated with high cellular proliferation in cancer tissue [13], [16], [17], [20], [22]. In summary, high levels of KPNA2 appear to be associated with increased malignancy across cancers, as witnessed by the correlation with adverse characteristics such as poor patient survival.
3. Investigations of KPNA2’s role in cancer cells
With the aim of elucidating the causes underlying the increased expression of KPNA2 in cancer, Leaner et al. recently examined the promoter activity in cervical cancer cells [24]. They located the promoter region responsible for the elevated promoter activity (situated at −180 to −24 bp), and identified highly conserved binding sites for E2F transcription factors. Due to the intimate relationship between E2Fs and the retinoblastoma protein, RB, the latter was suggested to ultimately cause the KPNA2 deregulation. The recent demonstration that E2F1 is translocated by KPNA2 [25] raises the possibility that a positive feedback loop exists, where elevated activity of E2Fs may lead to increased expression of KPNA2 which in turn increases the nuclear amounts of E2Fs. Previous studies of E2F and the cell cycle have identified KPNA2 as differentially expressed through the cell cycle with the highest levels occurring in the G2/M phases [26], [27]. Regulation of the KPNA2 promoter activity appears to be somewhat cell line specific as the high expression observed in embryonic stem cells was likely instigated by Klf2 and Klf4 at a separate promoter region [28].
As mentioned, the expression of KPNA2 in cancer tissue appears to be predominantly nuclear. This may be due to cellular stress, as previous reports have demonstrated how karyopherins, including KPNA2, accumulate in the nucleus following stressful conditions such as UV irradiation, heat shock and oxidative stress [29], [30], [31], [32]. As cells in advanced tumors frequently exhibit high levels of oxidative stress [33], this could explain the nuclear localization of KPNA2. The nuclear retention in response to cellular stress suppresses the nuclear transport and alter gene regulation [29], [30], [31], [32], [34], [35]. Further studies are required to determine how the rates of nuclear transport are altered in cancer tissue and whether such distortions are due to cellular stress.
With possible implications for the KPNA2 deregulation observed in cancer tissue, its importance has been examined in a variety of cancer cell lines. In a recent study, it was suggested that KPNA2 plays a role in the malignant transformation of cancer cells [36]. Increasing the expression of KPNA2 in a benign breast cancer cell line conferred several characteristics normally associated with malign cells such as an increase in colony formation and in cell migration activity. These findings were in line with previous studies of lung cancer cell lines, where the migratory capability had also been affected by altered KPNA2 levels [17].
In addition, knockdown of KPNA2 decreases the proliferation of cells derived from lung [17], liver [21] and prostate cancer [19]. Conversely, no effect on proliferation was observed in examined cervical cancer cells [15], [37]. Whereas studies in prostate cancer cells did not imply increased apoptosis as the cause [19], such a connection appeared to exist in breast cancer cells [36]. Importantly, increasing the level of KPNA2 could enhance proliferation of some breast cancer cells as well, giving further credit to the idea that KPNA2 levels do affect the viability of cancer cells [36].
4. KPNA2 in the translocation of cancer – associated proteins
A reasonable way, in which KPNA2 could affect carcinogenesis, is through the translocation of cancer-associated cargo proteins. In agreement with this concept, recent proteomic analysis demonstrated how knockdown of KPNA2 led to altered nuclear levels of several proteins [25]. In fact, KPNA2 has been demonstrated to interact with a variety of proteins that are associated with cancer, including proteins with tumor-suppressive as well as oncogenic properties.
One of the most prominent cargo proteins translocated by KPNA2 is the cell-cycle regulator Chk2 [38]. KPNA2 interacts with the NLS indispensable for Chk2 import and overexpression of KPNA2 correlates with an increased nuclear import [38]. KPNA2 has also been suggested to play a role in the nuclear translocation of BRCA1. BRCA1 is likely implicated in carcinogenesis due to its roles in DNA repair and cell cycle checkpoint control [39]. The importance of the NLS in BRCA1 translocation has been established along with direct interaction between KPNA2 and BRCA1 [40], [41]. The observation that a BRCA1 mutant lacking both NLSs could be observed in the nucleus [42], [43] led to discovery of an alternative way to import BRCA1 [44]. The importance of each of the two alternate pathways remains to be established. Furthermore, KPNA2 is responsible for the import of NBS1, another DNA repair protein which also partake in carcinogenesis [45], [46]. This interaction was supported by the consequences of KPNA2 knockdown, which impaired the cellular capability to form regular nuclear foci in response to DNA damage [45]. An interesting connection, especially with relations to prostate cancer, is the role of KPNA2 in the import of the androgen receptor (AR) [19], [47]. Binding of the active androgen to AR leads to its nuclear import where it activates the transcription of a range of target genes. As a consequence, it has been speculated that the high level of KPNA2 observed in prostate cancer tissue might be indicative of an increase in translocation of AR and therefore associated with hormone-refractory prostate cancer [19].
Earlier reports of potential associations between KPNA2, p53 [48] and c-Myc [49] were supported by a recent study, which confirmed the interaction with both p53 and c-Myc [25]. This study also found an overlap between p53 and c-Myc down-stream targets and proteins affected by KPNA2 knockdown. A separate study found alternate karyopherins to be responsible for the import of p53 [50] and further studies are needed to establish what role KPNA2 holds in the translocation and regulation of both of these crucial factors.
KPNA2 has been implicated in the translocation of additional transcription factors, including the all-ready mentioned E2F1 [25], two members of PLAG family of zinc-finger transcription factors, PLAG1 [51] and LOT1 [52], as well as the BTB/POZ transcription factor Kaiso [53]. As transcription factors, these proteins are involved in a variety of processes, however, deregulation in cancer plays a prominent role [54], [55], [56], [57], [58]. Furthermore, KPNA2 has been shown to abrogate the induction of the LOT1 downstream target p21WAF1/CIP1 which holds both tumor suppressive and oncogenic potential [52], [59]. Finally, the small Rho-GTPase RAC-1 is also translocated by KPNA2 [60]. RAC-1 could hold a role in carcinogenesis due to its functions in cell cycle progression [61] and cellular adhesion and migration [62], [63].
The cancer-associated CREB binding protein (CBP or p300) has been shown to acetylate KPNA2 on lysine 22 [64], [65]. Furthermore, the AMP-activated protein kinase (AMPK) has been demonstrated to mediate the phosphorylation and acetylation of KPNA2, with the latter probably being executed by CBP [66]. These post-translational modifications are presumably functional as they negatively affected the translocation of a downstream protein, HuR [66].
5. KPNA2 has been implicated in a multitude of cellular processes
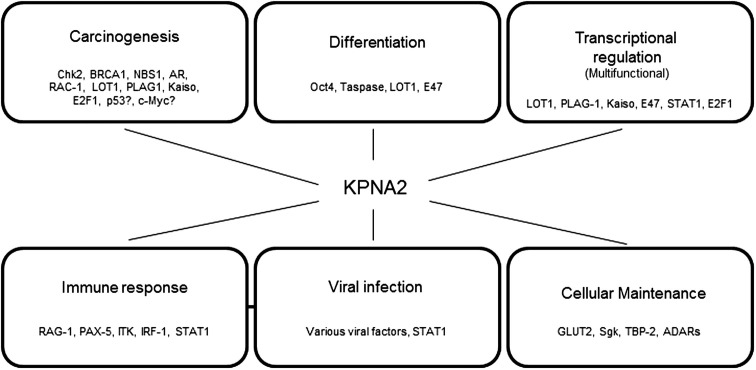
In addition to the interactions with cancer-associated proteins, KPNA2 has also been implicated in various other processes (an overview is presented in Fig. 1 ). It should be noted that albeit the described functions are not directly linked to cancer some of them could affect carcinogenesis e.g. through aberrant activation of differentiation pathways, viral induction of oncogenes or modulation of the immune system.
Fig. 1.
Cellular processes and interaction partners associated with KPNA2. Details concerning the listed interaction partners can be found in the text. Note that, albeit interaction with KPNA2 has been demonstrated, alternate karyopherins may also partake in the translocation of the listed proteins.
First of all, expression of KPNA2 is linked to various differentiation processes. KPNA2 is highly expressed in embryonic stem cells from mice, possibly due to promoter activation by Krüppel-like factors Klf2 and Klf4 [28]. Furthermore, KPNA2 has been identified to translocate Taspase 1, a protease with developmentally important downstream targets [67], and Oct4 [68], [69], a major gatekeeper in development [70]. Other reports include the findings of varying levels of import factors through spermatogenesis [71], myogenesis [72] and early embryogenesis [73]. For brain development it has been demonstrated that an exquisite downregulation of KPNA2 is a prerequisite during neural differentiation of embryonic stem cells [69]. The already mentioned cargoes Oct4 and LOT1 have roles in brain differentiation [55], [69] as has yet another transcription factor, E47, which is also shuttled by KPNA2 [74]. Of potential interest is also the observation of aberrantly localized KPNA2 in hippocampal neurons in Alzheimer Disease [75].
KPNA2 plays a crucial role during viral infections mainly by importing parts of the viral machinery to the nucleus. It has so far been implicated in the shuttling of factors from a variety of viruses such as HIV [76], Epstein-Barr virus [77], polyomavirus [78] and HPV [79], [80], [81]. Furthermore, KPNA2 has been proposed to hold a role in regulating viral capsid assembly, preventing it from faultily occurring in the cytoplasm [82]. An alternate way of affecting viral infection has been demonstrated for the SARS coronavirus which sequesters KPNA2 to the endoplasmic reticulum and Golgi membrane, thereby preventing nuclear translocation of STAT1, an important messenger in host-defense against viruses [83]. Interestingly, karyopherins have also been proposed to define host specificity of viruses as demonstrated recently for the influenza virus [84].
The KPNA2-mediated translocation of PAX-5, ITK, IRF-1 and RAG-1 provide a series of associations with the immune system [85], [86], [87], [88], [89]. Whereas PAX-5 is an important regulator of B-cell differentiation [90], ITK and IRF-1 hold more general roles in the immune response [91], [92], [93] and RAG-1 plays a key role in V(D)J recombination [94]. Possibly related is the report that the activation of lymphocytes leads to highly increased expression of KPNA2 and redistribution from the cytoplasm to the nucleus and the plasma membrane [95].
As for general metabolic roles it has been established that KPNA2 participates in the response to glucose addition via an interaction with GLUT2 [96], [97] and that it is responsible for the serum-induced translocation of Sgk [98], a kinase which functions in the regulation of epithelial ion transport [99]. KPNA2 activity might also be regulated by Ca2+ levels via interactions with the S100A6 protein [100]. Furthermore, KPNA2 is involved in controlling the intracellular redox environment through translocation of the thioredoxin-binding protein-2 (TBP-2) [101]. Regarding RNA metabolism, KPNA2 has been shown to import RNA-editing adenosine deaminases (ADARs) [102], partake in the formation of RNA stress granules [103] and interact with the Poly(A) Binding Protein (PABPC) involved in the regulation of mRNA levels [104]. The increasing number of interaction partners for KPNA2 indicates that it is truly a jack-of-all-trades.
6. Concluding remarks
Due to its function in nucleocytoplasmic transport, where it mediates the translocation of a multitude of proteins, KPNA2 is involved in many cellular processes. Nonetheless, the association with cancer appears to be very prominent, considering the aberrantly high levels of KPNA2 observed across cancers of diverse origin. Importantly, the altered expression pattern is associated with adverse patient characteristics. This spurs the idea that KPNA2 may hold a role in carcinogenesis. This idea is supported by the cellular studies of KPNA2 which have indicated that the protein may play a role in the malignant transformation of cells. The association with adverse cancer characteristics establishes KPNA2 as a potentially relevant therapeutic target. However, targeting of the protein will be complicated by the great number of conventional cellular processes which are also associated with KPNA2. Instead, utilization as a cancer biomarker may well be the first clinical implementation of KPNA2. Considering the results described here, obtained in a variety of cancer forms, this vision appears promising. Further prospective clinical studies are required to fully demonstrate the biomarker potential of KPNA2, and, in fact, KPNA2 is part of a translational effort in bladder cancer and is currently being investigated in ongoing multi-center prospective studies [105].
The high levels of KPNA2 in cancer cells would presumably perturb a variety of processes through the altered shuttling of nuclear proteins. Elucidating the interactions that connect KPNA2 to carcinogenesis is complicated by the fact that karyopherins can have overlapping preferences for cargo proteins [3]. We have presented some of the currently identified interaction partners. However, it should be kept in mind that, at present, many interaction partners might remain unknown, and these cargo proteins could act individually as well as in concert with known as well as unknown interaction partners. Nonetheless, the pronounced deregulation of KPNA2 in cancer, and the suggested role in malignant transformation, warrants further studies of this protein and its function in carcinogenesis.
References
- 1.Kau T.R., Way J.C., Silver P.A. Nuclear transport and cancer: from mechanism to intervention. Nat. Rev. Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 2.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 3.Goldfarb D.S., Corbett A.H., Mason D.A., Harreman M.T., Adam S.A. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 4.Kelley J.B., Talley A.M., Spencer A., Gioeli D., Paschal B.M. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010;11:63. doi: 10.1186/1471-2121-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radu A., Blobel G., Moore M.S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weis K., Mattaj I.W., Lamond A.I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 7.Gorlich D., Henklein P., Laskey R.A., Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 8.Weis K., Ryder U., Lamond A.I. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 9.Chook Y.M., Blobel G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 10.Harreman M.T., Cohen P.E., Hodel M.R., Truscott G.J., Corbett A.H., Hodel A.E. Characterization of the auto-inhibitory sequence within the N-terminal domain of importin alpha. J. Biol. Chem. 2003;278:21361–21369. doi: 10.1074/jbc.M301114200. [DOI] [PubMed] [Google Scholar]
- 11.Dahl E., Kristiansen G., Gottlob K. Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin. Cancer Res. 2006;12:3950–3960. doi: 10.1158/1078-0432.CCR-05-2090. [DOI] [PubMed] [Google Scholar]
- 12.Dankof A., Fritzsche F.R., Dahl E., Pahl S., Wild P., Dietel M., Hartmann A., Kristiansen G. KPNA2 protein expression in invasive breast carcinoma and matched peritumoral ductal carcinoma in situ. Virchows Arch. 2007;451:877–881. doi: 10.1007/s00428-007-0513-5. [DOI] [PubMed] [Google Scholar]
- 13.Gluz O., Wild P., Meiler R. Nuclear karyopherin alpha2 expression predicts poor survival in patients with advanced breast cancer irrespective of treatment intensity. Int. J. Cancer. 2008;123:1433–1438. doi: 10.1002/ijc.23628. [DOI] [PubMed] [Google Scholar]
- 14.Winnepenninckx V., Lazar V., Michiels S. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J. Natl. Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 15.van der Watt P.J., Maske C.P., Hendricks D.T., Parker M.I., Denny L., Govender D., Birrer M.J., Leaner V.D. The karyopherin proteins, Crm1 and karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int. J. Cancer. 2009;124:1829–1840. doi: 10.1002/ijc.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai M., Sohda M., Miyazaki T., Suzuki S., Sano A., Tanaka N., Inose T., Nakajima M., Kato H., Kuwano H. Significance of karyopherin-{alpha} 2 (KPNA2) expression in esophageal squamous cell carcinoma. Anticancer Res. 2010;30:851–856. [PubMed] [Google Scholar]
- 17.Wang C.I., Wang C.L., Wang C.W., Chen C.D., Wu C.C., Liang Y., Tsai Y.H., Chang Y.S., Yu J.S., Yu C.J. Importin subunit alpha-2 is identified as a potential biomarker for non-small cell lung cancer by integration of the cancer cell secretome and tissue transcriptome. Int. J. Cancer. 2011;128:2364–2372. doi: 10.1002/ijc.25568. [DOI] [PubMed] [Google Scholar]
- 18.Zheng M., Tang L., Huang L., Ding H., Liao W.T., Zeng M.S., Wang H.Y. Overexpression of karyopherin-2 in epithelial ovarian cancer and correlation with poor prognosis. Obstet. Gynecol. 2010;116:884–891. doi: 10.1097/AOG.0b013e3181f104ce. [DOI] [PubMed] [Google Scholar]
- 19.Mortezavi A., Hermanns T., Seifert H.H. KPNA2 expression is an independent adverse predictor of biochemical recurrence after radical prostatectomy. Clin. Cancer Res. 2011;17:1111–1121. doi: 10.1158/1078-0432.CCR-10-0081. [DOI] [PubMed] [Google Scholar]
- 20.Gousias K., Becker A.J., Simon M., Niehusmann P. Nuclear karyopherin a2: a novel biomarker for infiltrative astrocytomas. J. Neurooncol. 2012;109:545–553. doi: 10.1007/s11060-012-0924-2. [DOI] [PubMed] [Google Scholar]
- 21.Yoshitake K., Tanaka S., Mogushi K. Importin-alpha1 as a novel prognostic target for hepatocellular carcinoma. Ann. Surg. Oncol. 2011 doi: 10.1245/s10434-011-1569-7. [DOI] [PubMed] [Google Scholar]
- 22.Jensen J.B., Munksgaard P.P., Sorensen C.M., Fristrup N., Birkenkamp-Demtroder K., Ulhoi B.P., Jensen K.M., Orntoft T.F., Dyrskjot L. High expression of karyopherin-alpha2 defines poor prognosis in non-muscle-invasive bladder cancer and in patients with invasive bladder cancer undergoing radical cystectomy. Eur. Urol. 2011 doi: 10.1016/j.eururo.2011.01.048. [DOI] [PubMed] [Google Scholar]
- 23.Valastyan S., Weinberg R.A. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van D.W., Ngarande E., Leaner V.D. Overexpression of kpn beta 1 and kpn alpha 2 importin proteins in cancer derives from deregulated E2F activity. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C.I., Chien K.Y., Wang C.L., Liu H.P., Cheng C.C., Chang Y.S., Yu J.S., Yu C.J. Quantitative proteomics reveals regulation of karyopherin subunit alpha-2 (KPNA2) and its potential novel cargo proteins in nonsmall cell lung cancer. Mol. Cell. Proteomics. 2012;11:1105–1122. doi: 10.1074/mcp.M111.016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishida S., Huang E., Zuzan H., Spang R., Leone G., West M., Nevins J.R. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu W., Giangrande P.H., Nevins J.R. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 2004;23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamikawa Y., Yasuhara N., Yoneda Y. Cell type-specific transcriptional regulation of the gene encoding importin-alpha 1. Exp. Cell Res. 2011;317:1970–1978. doi: 10.1016/j.yexcr.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 29.Stochaj U., Rassadi R., Chiu J. Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J. 2000;14:2130–2132. doi: 10.1096/fj.99-0751fje. [DOI] [PubMed] [Google Scholar]
- 30.Furuta M., Kose S., Koike M., Shimi T., Hiraoka Y., Yoneda Y., Haraguchi T., Imamoto N. Heat-shock induced nuclear retention and recycling inhibition of importin alpha. Genes Cells. 2004;9:429–441. doi: 10.1111/j.1356-9597.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- 31.Kodiha M., Chu A., Matusiewicz N., Stochaj U. Multiple mechanisms promote the inhibition of classical nuclear import upon exposure to severe oxidative stress. Cell Death Differ. 2004;11:862–874. doi: 10.1038/sj.cdd.4401432. [DOI] [PubMed] [Google Scholar]
- 32.Miyamoto Y., Saiwaki T., Yamashita J., Yasuda Y., Kotera I., Shibata S., Shigeta M., Hiraoka Y., Haraguchi T., Yoneda Y. Cellular stresses induce the nuclear accumulation of importin alpha and cause a conventional nuclear import block. J. Cell Biol. 2004;165:617–623. doi: 10.1083/jcb.200312008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 34.Yasuda Y., Miyamoto Y., Yamashiro T., Asally M., Masui A., Wong C., Loveland K.L., Yoneda Y. Nuclear retention of importin alpha coordinates cell fate through changes in gene expression. EMBO J. 2012;31:83–94. doi: 10.1038/emboj.2011.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto Y., Loveland K.L., Yoneda Y. Nuclear importin alpha and its physiological importance. Commun. Integr. Biol. 2012;5:220–222. doi: 10.4161/cib.19194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noetzel E., Rose M., Bornemann J., Gajewski M., Knuchel R., Dahl E. Nuclear transport receptor karyopherin-alpha2 promotes malignant breast cancer phenotypes in vitro. Oncogene. 2011:2101–2114. doi: 10.1038/onc.2011.403. [DOI] [PubMed] [Google Scholar]
- 37.Quensel C., Friedrich B., Sommer T., Hartmann E., Kohler M. In vivo analysis of importin alpha proteins reveals cellular proliferation inhibition and substrate specificity. Mol. Cell. Biol. 2004;24:10246–10255. doi: 10.1128/MCB.24.23.10246-10255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zannini L., Lecis D., Lisanti S., Benetti R., Buscemi G., Schneider C., Delia D. Karyopherin-alpha2 protein interacts with Chk2 and contributes to its nuclear import. J. Biol. Chem. 2003;278:42346–42351. doi: 10.1074/jbc.M303304200. [DOI] [PubMed] [Google Scholar]
- 39.Narod S.A., Foulkes W.D. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 40.Thakur S., Zhang H.B., Peng Y. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol. Cell. Biol. 1997;17:444–452. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C.F., Li S., Chen Y., Chen P.L., Sharp Z.D., Lee W.H. The nuclear localization sequences of the BRCA1 protein interact with the importin-alpha subunit of the nuclear transport signal receptor. J. Biol. Chem. 1996;271:32863–32868. doi: 10.1074/jbc.271.51.32863. [DOI] [PubMed] [Google Scholar]
- 42.Wilson C.A., Payton M.N., Elliott G.S., Buaas F.W., Cajulis E.E., Grosshans D., Ramos L., Reese D.M., Slamon D.J., Calzone F.J. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene. 1997;14:1–16. doi: 10.1038/sj.onc.1200924. [DOI] [PubMed] [Google Scholar]
- 43.Huber L.J., Yang T.W., Sarkisian C.J., Master S.R., Deng C.X., Chodosh L.A. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol. Cell. Biol. 2001;21:4005–4015. doi: 10.1128/MCB.21.12.4005-4015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabbro M., Rodriguez J.A., Baer R., Henderson B.R. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J. Biol. Chem. 2002;277:21315–21324. doi: 10.1074/jbc.M200769200. [DOI] [PubMed] [Google Scholar]
- 45.Tseng S.F., Chang C.Y., Wu K.J., Teng S.C. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J. Biol. Chem. 2005;280:39594–39600. doi: 10.1074/jbc.M508425200. [DOI] [PubMed] [Google Scholar]
- 46.Teng S.C., Wu K.J., Tseng S.F., Wong C.W., Kao L. Importin KPNA2, NBS1, DNA repair and tumorigenesis. J. Mol. Histol. 2006;37:293–299. doi: 10.1007/s10735-006-9032-y. [DOI] [PubMed] [Google Scholar]
- 47.Cutress M.L., Whitaker H.C., Mills I.G., Stewart M., Neal D.E. Structural basis for the nuclear import of the human androgen receptor. J. Cell. Sci. 2008;121:957–968. doi: 10.1242/jcs.022103. [DOI] [PubMed] [Google Scholar]
- 48.Kim I.S., Kim D.H., Han S.M. Truncated form of importin alpha identified in breast cancer cell inhibits nuclear import of p53. J. Biol. Chem. 2000;275:23139–23145. doi: 10.1074/jbc.M909256199. [DOI] [PubMed] [Google Scholar]
- 49.Nadler S.G., Tritschler D., Haffar O.K., Blake J., Bruce A.G., Cleaveland J.S. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J. Biol. Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- 50.Marchenko N.D., Hanel W., Li D., Becker K., Reich N., Moll U.M. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha 3 binding. Cell Death Differ. 2010;17:255–267. doi: 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braem C.V., Kas K., Meyen E., Debiec-Rychter M., Van De Ven W.J., Voz M.L. Identification of a karyopherin alpha 2 recognition site in PLAG1, which functions as a nuclear localization signal. J. Biol. Chem. 2002;277:19673–19678. doi: 10.1074/jbc.M112112200. [DOI] [PubMed] [Google Scholar]
- 52.Huang S.M., Huang S.P., Wang S.L., Liu P.Y. Importin alpha1 is involved in the nuclear localization of Zac1 and the induction of p21WAF1/CIP1 by Zac1. Biochem. J. 2007;402:359–366. doi: 10.1042/BJ20061295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly K.F., Otchere A.A., Graham M., Daniel J.M. Nuclear import of the BTB/POZ transcriptional regulator kaiso. J. Cell Sci. 2004;117:6143–6152. doi: 10.1242/jcs.01541. [DOI] [PubMed] [Google Scholar]
- 54.Chen H., Tsai S., Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdollahi A. LOT1 (ZAC1/PLAGL1) and its family members: Mechanisms and functions. J. Cell. Physiol. 2007;210:16–25. doi: 10.1002/jcp.20835. [DOI] [PubMed] [Google Scholar]
- 56.van Roy F.M., McCrea P.D. A role for kaiso-p120ctn complexes in cancer? Nat. Rev. Cancer. 2005;5:956–964. doi: 10.1038/nrc1752. [DOI] [PubMed] [Google Scholar]
- 57.Prokhortchouk A., Sansom O., Selfridge J. Kaiso-deficient mice show resistance to intestinal cancer. Mol. Cell Biol. 2006;26:199–208. doi: 10.1128/MCB.26.1.199-208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spring C.M., Kelly K.F., O’Kelly I., Graham M., Crawford H.C., Daniel J.M. The catenin p120ctn inhibits kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp. Cell Res. 2005;305:253–265. doi: 10.1016/j.yexcr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 59.Gartel A.L. P21(WAF1/CIP1) and cancer: a shifting paradigm? Biofactors. 2009;35:161–164. doi: 10.1002/biof.26. [DOI] [PubMed] [Google Scholar]
- 60.Sandrock K., Bielek H., Schradi K., Schmidt G., Klugbauer N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic. 2010;11:198–209. doi: 10.1111/j.1600-0854.2009.01015.x. [DOI] [PubMed] [Google Scholar]
- 61.Olson M.F., Ashworth A., Hall A. An essential role for rho, rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 62.Pankov R., Endo Y., Even-Ram S., Araki M., Clark K., Cukierman E., Matsumoto K., Yamada K.M. A rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li A., Ma Y., Yu X. Rac1 drives melanoblast organization during mouse development by orchestrating pseudopod driven motility and cell-cycle progression. Dev. Cell. 2011;21:722–734. doi: 10.1016/j.devcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bannister A.J., Miska E.A., Gorlich D., Kouzarides T. Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr. Biol. 2000;10:467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- 65.Ryan C.M., Harries J.C., Kindle K.B., Collins H.M., Heery D.M. Functional interaction of CREB binding protein (CBP) with nuclear transport proteins and modulation by HDAC inhibitors. Cell Cycle. 2006;5:2146–2152. doi: 10.4161/cc.5.18.3207. [DOI] [PubMed] [Google Scholar]
- 66.Wang W., Yang X., Kawai T., Lopez de Silanes I., Mazan-Mamczarz K., Chen P., Chook Y.M., Quensel C., Kohler M., Gorospe M. AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1: involvement in the nuclear import of RNA-binding protein HuR. J. Biol. Chem. 2004;279:48376–48388. doi: 10.1074/jbc.M409014200. [DOI] [PubMed] [Google Scholar]
- 67.Bier C., Knauer S.K., Docter D., Schneider G., Kramer O.H., Stauber R.H. The importin-alpha/nucleophosmin switch controls taspase1 protease function. Traffic. 2011;12:703–714. doi: 10.1111/j.1600-0854.2011.01191.x. [DOI] [PubMed] [Google Scholar]
- 68.Li X., Sun L., Jin Y. Identification of karyopherin-alpha 2 as an Oct4 associated protein. J. Genet. Genomics. 2008;35:723–728. doi: 10.1016/S1673-8527(08)60227-1. [DOI] [PubMed] [Google Scholar]
- 69.Yasuhara N., Shibazaki N., Tanaka S., Nagai M., Kamikawa Y., Oe S., Asally M., Kamachi Y., Kondoh H., Yoneda Y. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat. Cell. Biol. 2007;9:72–79. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- 70.Pesce M., Scholer H.R. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 71.Hogarth C.A., Calanni S., Jans D.A., Loveland K.L. Importin alpha mRNAs have distinct expression profiles during spermatogenesis. Dev. Dynam. 2006;235:253–262. doi: 10.1002/dvdy.20569. [DOI] [PubMed] [Google Scholar]
- 72.Hall M.N., Griffin C.A., Simionescu A., Corbett A.H., Pavlath G.K. Distinct roles for classical nuclear import receptors in the growth of multinucleated muscle cells. Dev. Biol. 2011;357 doi: 10.1016/j.ydbio.2011.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cabot R.A., Prather R.S. Cleavage stage porcine embryos may have differing developmental requirements for karyopherins alpha2 and alpha3. Mol. Reprod. Dev. 2003;64:292–301. doi: 10.1002/mrd.10238. [DOI] [PubMed] [Google Scholar]
- 74.Mehmood R., Yasuhara N., Fukumoto M., Oe S., Tachibana T., Yoneda Y. Cross-talk between distinct nuclear import pathways enables efficient nuclear import of E47 in conjunction with its partner transcription factors. Mol. Biol. Cell. 2011;22 doi: 10.1091/mbc.E10-10-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee H.G., Ueda M., Miyamoto Y., Yoneda Y., Perry G., Smith M.A., Zhu X. Aberrant localization of importin alpha1 in hippocampal neurons in alzheimer disease. Brain Res. 2006;1124:1–4. doi: 10.1016/j.brainres.2006.09.084. [DOI] [PubMed] [Google Scholar]
- 76.Gallay P., Stitt V., Mundy C., Oettinger M., Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fischer N., Kremmer E., Lautscham G., Mueller-Lantzsch N., Grasser F.A. Epstein-barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin alpha2. J. Biol. Chem. 1997;272:3999–4005. doi: 10.1074/jbc.272.7.3999. [DOI] [PubMed] [Google Scholar]
- 78.Qu Q., Sawa H., Suzuki T., Semba S., Henmi C., Okada Y., Tsuda M., Tanaka S., Atwood W.J., Nagashima K. Nuclear entry mechanism of the human polyomavirus JC virus-like particle: role of importins and the nuclear pore complex. J. Biol. Chem. 2004;279:27735–27742. doi: 10.1074/jbc.M310827200. [DOI] [PubMed] [Google Scholar]
- 79.Merle E., Rose R.C., LeRoux L., Moroianu J. Nuclear import of HPV11 L1 capsid protein is mediated by karyopherin alpha2beta1 heterodimers. J. Cell Biochem. 1999;74:628–637. [PubMed] [Google Scholar]
- 80.Nelson L.M., Rose R.C., LeRoux L., Lane C., Bruya K., Moroianu J. Nuclear import and DNA binding of human papillomavirus type 45 L1 capsid protein. J. Cell Biochem. 2000;79:225–238. [PubMed] [Google Scholar]
- 81.Le Roux L.G., Moroianu J. Nuclear entry of high-risk human papillomavirus type 16 E6 oncoprotein occurs via several pathways. J. Virol. 2003;77:2330–2337. doi: 10.1128/JVI.77.4.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bird G., O’Donnell M., Moroianu J., Garcea R.L. Possible role for cellular karyopherins in regulating polyomavirus and papillomavirus capsid assembly. J. Virol. 2008;82:9848–9857. doi: 10.1128/JVI.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gabriel G., Klingel K., Otte A. Differential use of importin-alpha isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2011;2:156. doi: 10.1038/ncomms1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Umegaki N., Tamai K., Nakano H., Moritsugu R., Yamazaki T., Hanada K., Katayama I., Kaneda Y. Differential regulation of karyopherin alpha 2 expression by TGF-beta1 and IFN-gamma in normal human epidermal keratinocytes: Evident contribution of KPNA2 for nuclear translocation of IRF-1. J. Invest. Dermatol. 2007;127:1456–1464. doi: 10.1038/sj.jid.5700716. [DOI] [PubMed] [Google Scholar]
- 86.Kovac C.R., Emelyanov A., Singh M., Ashouian N., Birshtein B.K. BSAP (Pax5)-importin alpha 1 (Rch1) interaction identifies a nuclear localization sequence. J. Biol. Chem. 2000;275:16752–16757. doi: 10.1074/jbc.M001551200. [DOI] [PubMed] [Google Scholar]
- 87.Perez-Villar J.J., O’Day K., Hewgill D.H., Nadler S.G., Kanner S.B. Nuclear localization of the tyrosine kinase itk and interaction of its SH3 domain with karyopherin alpha (Rch1alpha) Int. Immunol. 2001;13:1265–1274. doi: 10.1093/intimm/13.10.1265. [DOI] [PubMed] [Google Scholar]
- 88.Cuomo C.A., Kirch S.A., Gyuris J., Brent R., Oettinger M.A. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl. Acad. Sci. USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spanopoulou E., Cortes P., Shih C., Huang C.M., Silver D.P., Svec P., Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 90.O’Brien P., Morin P., Jr., Ouellette R.J., Robichaud G.A. The pax-5 gene: a pluripotent regulator of B-cell differentiation and cancer disease. Cancer Res. 2011;71:7345–7350. doi: 10.1158/0008-5472.CAN-11-1874. [DOI] [PubMed] [Google Scholar]
- 91.Bouker K.B., Skaar T.C., Riggins R.B., Harburger D.S., Fernandez D.R., Zwart A., Wang A., Clarke R. Interferon regulatory factor-1 (IRF-1) exhibits tumor suppressor activities in breast cancer associated with caspase activation and induction of apoptosis. Carcinogenesis. 2005;26:1527–1535. doi: 10.1093/carcin/bgi113. [DOI] [PubMed] [Google Scholar]
- 92.Kroger A., Koster M., Schroeder K., Hauser H., Mueller P.P. Activities of IRF-1. J. Interferon Cytokine Res. 2002;22:5–14. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- 93.Sahu N., August A. ITK inhibitors in inflammation and immune-mediated disorders. Curr. Top. Med. Chem. 2009;9:690–703. doi: 10.2174/156802609789044443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matthews A.G., Oettinger M.A. RAG: A recombinase diversified. Nat. Immunol. 2009;10:817–821. doi: 10.1038/ni.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Andrade R., Alonso R., Pena R., Arlucea J., Arechaga J. Localization of importin alpha (Rch1) at the plasma membrane and subcellular redistribution during lymphocyte activation. Chromosoma. 2003;112:87–95. doi: 10.1007/s00412-003-0247-3. [DOI] [PubMed] [Google Scholar]
- 96.Guillemain G., Munoz-Alonso M.J., Cassany A., Loizeau M., Faussat A.M., Burnol A.F., Leturque A. Karyopherin alpha2: a control step of glucose-sensitive gene expression in hepatic cells. Biochem. J. 2002;364:201–209. doi: 10.1042/bj3640201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cassany A., Guillemain G., Klein C., Dalet V., Brot-Laroche E., Leturque A. A karyopherin alpha2 nuclear transport pathway is regulated by glucose in hepatic and pancreatic cells. Traffic. 2004;5:10–19. doi: 10.1046/j.1398-9219.2003.0143.x. [DOI] [PubMed] [Google Scholar]
- 98.Maiyar A.C., Leong M.L., Firestone G.L. Importin-alpha mediates the regulated nuclear targeting of serum- and glucocorticoid-inducible protein kinase (sgk) by recognition of a nuclear localization signal in the kinase central domain. Mol. Biol. Cell. 2003;14:1221–1239. doi: 10.1091/mbc.E02-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Loffing J., Flores S.Y., Staub O. Sgk kinases and their role in epithelial transport. Ann. Rev. Physiol. 2006;68:461–490. doi: 10.1146/annurev.physiol.68.040104.131654. [DOI] [PubMed] [Google Scholar]
- 100.Takata M., Shimamoto S., Yamaguchi F., Tokuda M., Tokumitsu H., Kobayashi R. Regulation of nuclear localization signal-importin alpha interaction by Ca2+/S100A6. FEBS Lett. 2010;584:4517–4523. doi: 10.1016/j.febslet.2010.09.052. [DOI] [PubMed] [Google Scholar]
- 101.Nishinaka Y., Masutani H., Oka S., Matsuo Y., Yamaguchi Y., Nishio K., Ishii Y., Yodoi J. Importin alpha1 (Rch1) mediates nuclear translocation of thioredoxin-binding protein-2/vitamin D(3)-up-regulated protein 1. J. Biol. Chem. 2004;279:37559–37565. doi: 10.1074/jbc.M405473200. [DOI] [PubMed] [Google Scholar]
- 102.Maas S., Gommans W.M. Identification of a selective nuclear import signal in adenosine deaminases acting on RNA. Nucl. Acids Res. 2009;37:5822–5829. doi: 10.1093/nar/gkp599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fujimura K., Suzuki T., Yasuda Y., Murata M., Katahira J., Yoneda Y. Identification of importin alpha1 as a novel constituent of RNA stress granules. Biochim. Biophys. Acta. 1803;2010:865–871. doi: 10.1016/j.bbamcr.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 104.Kumar G.R., Shum L., Glaunsinger B.A. Importin alpha-mediated nuclear import of cytoplasmic Poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol. Cell. Biol. 2011;31:3113–3125. doi: 10.1128/MCB.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dyrskjot L., Reinert T., Novoradovsky A. Analysis of molecular intra-patient variation and delineation of a prognostic 12-gene signature in non-muscle invasive bladder cancer; technology transfer from microarrays to PCR. Br. J. Cancer. 2012;107:1392–1398. doi: 10.1038/bjc.2012.412. [DOI] [PMC free article] [PubMed] [Google Scholar]