Abstract
It is likely that gene-based vaccines will enter the human vaccine area soon. A few veterinary vaccines employing this concept have already been licensed, and a multitude of clinical trials against infectious diseases or different forms of cancer are ongoing. Highly important when developing novel vaccines are the safety aspects and also new adjuvants and delivery techniques needs to be carefully investigated so that they meet all short- and long-term safety requirements. One novel in vivo delivery method for plasmid vaccines is electroporation, which is the application of short pulses of electric current immediately after, and at the site of, an injection of a genetic vaccine. This method has been shown to significantly augment the transfection efficacy and the subsequent vaccine-specific immune responses. However, the dramatic increase in delivery efficacy offered by electroporation has raised concerns of potential increase in the risk of integration of plasmid DNA into the host genome. Here, we demonstrate the safety and lack of integration after immunization with a high dose of a multigene HIV-1 vaccine delivered intradermally using the needle free device Biojector 2000 together with electroporation using Derma Vax™ DNA Vaccine Skin Delivery System. We demonstrate that plasmids persist in the skin at the site of injection for at least four months after immunization. However, no association between plasmid DNA and genomic DNA could be detected as analyzed by qPCR following field inversion gel electrophoresis separating heavy and light DNA fractions. We will shortly initiate a phase I clinical trial in which healthy volunteers will be immunized with this multiplasmid HIV-1 vaccine using a combination of the delivery methods jet-injection and intradermal electroporation.
Keywords: Electroporation, Skin, Dermal, HIV, Plasmid DNA, Bioject
1. Introduction
Clinical investigations of plasmid DNA have primarily been focused on two in vivo applications: gene therapy and immunization. In order to verify the safety and efficacy, the two applications require different approaches. For plasmid immunization, the aim is to rapidly induce an immune response, while for safety reasons, the plasmid should preferably be cleared rapidly from the body/tissue. In contrast, gene therapy applications aim at introducing the genetic material in the target tissue without evoking an immune response, which could serve to eliminate cells expressing the therapeutic agent.
During the efforts to prove the safety of plasmid DNA, the biodistribution of plasmids has been studied in several animal species, and the results from published experiments all indicate that the plasmid is rapidly cleared from the body (Table 1 [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11]). Directly after injection into skin or muscle, low levels of plasmids are transported via the blood stream and can therefore often be detected in various organs at early time points (Table 1). However, the plasmids are eventually, and often rapidly, cleared from the organs and are normally found exclusively at the site of injection at later time points [12]. For safety reasons, the presence of plasmid in gonads has been given special attention, but in no study has plasmid been detected in testis or ovaries for more than one week after immunization (Table 1). Moreover, there is no indication that either the inserted gene or the plasmid backbone itself plays any major role for the distribution or persistence of the plasmid [9]. The introduction of in vivo electroporation for increasing the uptake and immunogenicity of plasmid vaccines has re-energized the field of genetic immunization and currently several electroporation devices are being evaluated in clinical trials against cancer and infectious diseases (http://clinicaltrials.gov/ct2/results?term=electroporation). However, since electroporation dramatically increases the transfection efficacy, concerns have been raised that the method might affect the persistence of plasmid DNA in the target tissue. This may result in increased risk of integration of foreign DNA into the host chromosomal DNA as reported in a single publication [13].
Table 1.
Previous experiments analyzing plasmid persistence after immunization.
| Antigen | Dose/route | Species/sex | Max local conc. at the site of injection | Liver | Spleen | Gonads |
|---|---|---|---|---|---|---|
| HIV GagPol (Winegar et al. [10]) | 400 μg i.m. | Rabbit ♀, ♂ | 6 × 106 c/μg @4 h | 0/5 > 10 c/μg @4 h | 0/5 > 10 c/μg @4 h | 0/5 > 10 c/μg @4 h |
| Malaria PfCSP (Parker et al. [6]) | 50 μg i.m. | Mouse ♀, ♂ | >100 c/μg @1 h | 5/6 < 100 c/μg@1 h 0/6 < 100 c/μg @2 d | 5/6<100 c/μg@1 h 0/6 < 100 c/μg @2 d | 3/6 < 100 c/μg @1 h 0/6 < 100 c/μg @2 d |
| HIV Gag (Manam et al. [5]) | 320 μg i.m. | Mouse♀, ♂ | ND | >16 c/μg @2 d 0 c/μg @6 w | 1–2 c/μg @2 d 0 c/μg @6 w | 0–40 c/μg @2 d 0 c/μg @6 w |
| HIV Gag/Env/Pol/Vif/Nef/Tat/Vpu (Kim et al. [4]) | 100 μg i.m., i.v. | Mouse♂ | 150 ng/mg tissue @5 min | 5/5 > 0.5 pg @1,5 h 0/5 > 0.5 pg @8 h | 3/5 > 0.5 pg @1.5 h, 0/5 > 0.5 pg @ 8 h | 4/5 > 0.5pg @1.5 h, 0/5 > 0.5pg @8 h |
| hDel-1 (Quezada et al. [7]) | 16 μg i.v. | Mouse♀, ♂ | ND | 2/6 > 1000 c/μg DNA@24 h 0/6 > 1000 c/μg DNA@1 w | 2/6 > 1000 c/μg DNA@24 h 0/6 > 1000 c/μg DNA@1 w | 1/6 > 1000 c/μg DNA@24 h 0/6 > 1000 c/μg DNA@1 w |
| Foot and mouth disease (Zhang et al. [11]) | 200 μg i.m. | Mouse ♀, ♂ | 53,000 c/μg@10 min | 35 ± 27 c/μg @10 min 0/6 > 10 c/μg @1 w | 6/6 > 10 c/μg @10 min 4/6 > 10 c/μg @1 w | 4/6 > 10 c/μg @10 min 0/6 > 10 c/μg @24 h |
| MVA-HIVA (multigene) (Hanke et al. [3]) | 50 μg i.m. | Mouse♀, ♂ | ND | 0/3 > 10 c/μg @46 d | 0/3 > 10 c/μg @46 d | 0/3 > 10 c/μg @46 d |
| 10 plasmids HIV, Ebola v, SARS v, WNV, Malaria (Sheets et al. [9]) | 100 μg 2000 μg i.m., i.d. | Mouse, rabbit ♀, ♂ | 104 to 106 μg/DNA@9 d | All < 50 c/μg @30 d | All < 50 c/μg @30 d | All < 50 c/μg @30 d |
| Porcine herpes virus (Gravier et al. [2]) | 481 μg i.m. | Pig ♀ | ND | ND | ND | 1/3 > 500 c/μg @1 d 0/3 > 500 c/μg @17 d |
| Measles virus (Ramirez et al. [8]) | 1840 μg i.d. | Rabbit ♀, ♂ | 106 c/μg DNA@9,31,60 d | 0/15 > 10 c/μg @9,31,60 d | 0/15 > 10 c/μg @9,31,60 d | 0/15 > 10 c/μg @9,31,60 d |
| Hepatitis C virus (Bacardi et al. [1]) | 50 μg i.m. | Mouse ♀ | 5/5 > 100 c/μg @1 h; 5/5 > 100 c/μg @17 h: 5/5 > 100 c/μg @30 d | 5/5 > 100 c/μg @1 h; 5/5 > 100 c/μg @17 h: 0/5 > 100 c/μg @30 d | ND | 5/5 > 100 c/μg @1 h; 0/5 > 100 c/μg @17 h |
c/μg: plasmid copy number/μg total DNA, X/Y: X indicates X positive animals out of Y analyzed.
ND: not determined; i.m.: intramuscular; i.d.: intradermal; i.v. intravenous; w: week; d: day; MVA: Modified Vaccinia virus Ankara; WNV: West Nile Virus; SARSv: Severe Acute Respiratory Syndrome virus; hDel-1: human developmentally regulated endothelial locus-1 protein.
In the present study, a multigene vaccine against HIV [14], [15] was delivered intradermally by needle free jet-injection followed by electroporation. This vaccine has previously been evaluated (without the addition of electroporation) in several clinical trials and has been shown safe and immunogenic [15], [16]. Here, toxicology, biodistribution, persistence and potential integration of the 7 plasmid HIV vaccine, were analyzed.
2. Materials and methods
2.1. Vaccine constructs and immunizations
The vaccine includes seven plasmids (Fig. 1 ) vectoring the following HIV-1 genes: envelope genes env A, env B and env C (pKCMVgp160 A, B and C, respectively), rev (pKCMVrev), RT (reverse transcriptase pKCMVRTmut), gag A and gag B (pKCMVp37BA and B, respectively) and have been previously described [14], [17], [18]. The pKCMVgp160B encodes gp160 of subtype B, a fusion protein of gp120 and gp 41, while the pKCMVgp160B/A and pKCMVgp160B/C encode chimeric gp160B proteins with the hypervariable loops (V1-V5) exchanged for subtype A or C sequences, respectively. pKCMVp37B encodes p17 and p24 of HIV subtype B, while in pKCMVp37BA the p24 subtype B gene is exchanged for a gene of a subtype A. Regulatory protein rev is native while the enzyme RT has been mutated in its enzyme-active site; both proteins derive from HIV subtype B. The Good Manufacturing Practice (GMP) grade DNA vaccine was produced by Vecura at the Karolinska Hospital, Sweden.
Fig. 1.
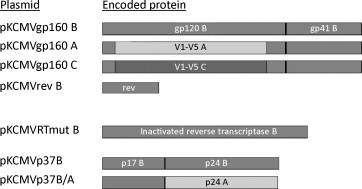
Schematic overview of the antigen encoded by the seven different plasmids. V: variable loop; A, B, C – indicates that the protein, or indicated part of the protein, derives from HIV-1 of subtype A, B, or C, respectively.
2.2. Immunizations
In all experiments described, the immunization procedure was the same and as described below. The seven plasmids, were delivered as two entities; one containing the gag p37 (BA and B) and RT genes and the other containing env gp160 (A, B and C) and rev genes, in order to avoid interference between antigens [19]. Mice were sedated with isoflurane, shaved at the lower back and immunized intradermally at the shaved area with two separate jet injections using the Biojector 2000 (Bioject Medical Technologies, Inc. OR, USA) with syringe no. 2 and intradermal spacer. Animals were injected with 100 μg DNA of each vaccine entity at a concentration of 1 mg DNA/ml saline. Immediately following the injection, the sites of injection were subjected to electroporation using the Derma Vax™, commercial DNA vaccine delivery system (Cyto Pulse Sciences Inc. MD, USA) [20]. The IDA-4-6-2 needle array (Cyto Pulse Sciences Inc.) with 6 needles per row was used for all electroporations and the pulse protocol used was two pulses of 1125 V/cm, 50 μs duration plus eight pulses of 275 V/cm and 10 ms duration [20]. Control animals were injected with 2 × 100 μl of saline by the same immunization procedure.
2.3. Toxicological evaluation: general health status, clinical chemistry and histopathology
Ten male and 10 female BALB/c mice were immunized with the plasmid vaccine and 10 males and 10 females were injected with saline. Animals were immunized at four occasions (weeks 0, 2, 6 and 12) as described above. Animals were regularly weighed and observed for changes in behaviour and fur quality. Two weeks after the last vaccine administration, animals were anaesthetised and blood was obtained through the orbital plexus for analysis with regards to white and red blood cell count (WBC and RBC, respectively), haemoglobin (HGB), platelet count (PLT), hematocrit (HCT). Sera were analyzed with regards to Aspartate transaminase (AST), Alanine transaminase (ALT), bilirubin, albumine, creatinine, lactase dehydrogenase (LDH), Gamma-glutamyltransferase (GGT), potassium, sodium and glucose. For the histopathological investigation the following organs were analysed: brain, lungs, ileum, liver, heart, spleen, kidneys, testes or ovaries, mesenteric lymph nodes, inguinal lymph nodes, non-injected skin and skin from injection sites.
2.4. Biodistribution and quantification of plasmid in tissue
For the biodistribution experiment, female BALB/c mice were immunized once and at various time-points following immunization, one animal was sacrificed by cervical dislocation. Animals were carefully dissected using a separate set of tools for each individual organ in order to avoid contamination and organs were snap-frozen after the collection until further use. Still frozen, the organs were sliced with scalpel and extraction of total DNA was performed on frozen samples using the DNAeasy Blood and Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Quantitative polymerase chain reaction (qPCR) was performed using a TaqMan-based assay on organs from the animals using the ABI Prism 7900 HT Fast real-time PCR system (Applied Biosystems). The primers and probe used in the qPCR reactions were designed to react with the plasmid (pKCMV) backbone, thus the analyses is independent of the inserted HIV-1 genes. The level of detection was determined to 10 copies of plasmid/μg of total DNA and the limit of quantification was determined to 100 copies of plasmid/μg of DNA. To establish the level of inhibition for each tissue type, 104 copies of plasmid were added to extracted DNA from each tissue type prior to PCR reaction. The samples containing inhibitory factors were diluted in water to the extent where the PCR reaction was no longer inhibited. Each following PCR sample containing extracted DNA from a tissue type that was shown to inhibit the PCR reaction was diluted to the appropriate level.
2.5. Association of plasmid DNA with high molecular weight (HMW) DNA
Female BALB/c mice were purchased from Charles River breeding facility in Germany. The study included 32 immunized mice and 16 untreated control mice. Animals were housed 8 per cage in clean rooms in which no plasmid vaccine had been handled prior to the study. The animals were injected once with 200 μg of DNA according to the procedure described above and 60 days after the injections the animals were killed by cervical dislocation and shaved thoroughly by the use of disposable single-use razors. The skin of the lower part of the back was removed from each animal using scissors and forceps. One set of instruments was used for each mouse to avoid contamination between animals. The samples were snap-frozen in liquid nitrogen and kept there until analysis of association of plasmid DNA with high molecular weight DNA. The analysis was performed at Althea Technologies, Inc. (CA, USA). DNA was subsequently extracted from the skin (site of injection) from both immunized and saline injected mice. Amount of DNA was determined by UV spectrophotometry and DNA from 4 or 8 mice were pooled from the saline controls or immunized mice, respectively, in order to obtain enough DNA for the subsequent gel-separation of heavy and light DNA. The DNA was analysed using qPCR with the same parameters as described above for the biodistribution analysis. If the plasmid content was above 10 copies/μg of DNA the material was subjected to EagI restriction digestion and subsequently field inversion gel electrophoresis (FIGE) for 3–5 h with an inversion ratio of 3:1. Upon completion of the FIGE, high molecular weight DNA (∼17KB) was excised from the gel and analysed by qPCR. In case detectable levels of plasmid remained in the HMW fraction after a first round of separation, the FIGE and subsequent qPCR were repeated.
2.6. Cytokine ELISpot assay
Lymphocytes derived from the spleen (referred to as splenocytes) were isolated from 5 male and 5 female animals from the toxicological experiment. Ficoll-paque plus (Amersham Biosciences Europe, Uppsala, Sweden) was used to purify the splenocytes, which were subsequently suspended in RPMI 1640 medium (Sigma–Aldrich, Stockholm, Sweden) supplemented with penicillin/streptomycin (PEST; Invitrogen Corporation, Carlsbad, CA) and 10% fetal calf serum (Sigma–Aldrich, Stockholm, Sweden). Cells were distributed in anti-IFN-γ antibody-precoated 96-well polyvinylidene fluoride-bottomed plates (Mabtech AB, Nacka, Sweden). To stimulate the splenocytes, 15-mer peptides (Thermo Fisher Scientific, Waltham, MA) overlapping by 10 amino acids covering the HIV-1 proteins p24Gag and gp120Env (both of subtype B) were used at a final concentration of 2.5 μg of each peptide/ml. A control peptide library consisting of 18 peptides derived from tick-borne encephalitis virus and medium alone were used as controls. Concanavalin A (1 μg per well) was used as a positive control to confirm cell viability. The plates were then developed as instructed by the manufacturer (Mabtech AB, Nacka, Sweden). Results are given as cytokine-producing spot-forming cells (SFCs) per 106 splenocytes, and responding animals were defined as having more than 50 SFCs per 106 plated cells and twice the number of SFCs for unstimulated cells from the same animal.
3. Results
3.1. Toxicology of GMP-produced 7 plasmid vaccine delivered in combination with intradermal electroporation
Female and male mice were immunized intradermally using the Biojector 2000 with saline or the 7 plasmid vaccine at 4 occasions (week 0, 2, 6 and 12). Saline or plasmids (200 μg total DNA) were injected at a volume of 2 × 100 μl and immediately thereafter, both injection sites were electroporated with the Derma Vax™ DNA vaccine delivery system.
The general health status (change in food intake, activity and fur quality) of the immunized animals did not deviate from the saline-injected control animals throughout the study. The body weights of the animals were recorded 3 times/week and at the end of the study no significant differences in body weights were found between the groups of females. However, the male mice in the vaccine group had statistically significant higher body weights than male control animals (mean difference 2.4 g) but no obvious reasons for the different weight gain between groups could be identified.
No clinical chemistry parameters were significantly different between the groups of female mice. In males, statistically significant differences between groups were found in levels of lactase dehydrogenase (LDH) and sodium. The LDH values were, for unknown reasons, unusually high in the control group, both among males and females, and suggests that this observation is not related to the treatment. The differences in sodium levels between groups were small and not considered to be of any biological significance. No other clinical chemistry parameters analyzed were significantly different between the groups of males.
No histopathological changes were found in any of following tissues that could be suspected to be the result of the treatment: brain, lungs, ileum, liver, heart, spleen, kidneys, testes or ovaries, mesenteric lymph nodes, inguinal lymph nodes and non-injected skin. In samples of skin from the injection site, signs of tissue reaction were occasionally found reflecting healing of an inflammatory response to a previous injection procedure. The reactions included increased numbers of inflammatory cells in dermis and subcutis, remnants of small epidermal “crusts” containing keratin and dead cells and sometimes disrupted epidermis.
It was concluded that repeated injections of the 7 plasmid HIV vaccine followed by electroporation was well tolerated by the animals in this study, which was performed according to good laboratory practice (GLP) at Visionar Preclinical AB, Uppsala, Sweden.
3.2. Biodistribution
In order to investigate the biodistribution of plasmids following immunization, female BALB/c mice were injected with 200 μg (total DNA) of the 7 plasmid mixture according to procedure described above. Control animals were injected with 2 × 100 μl of saline by the same procedure. Animals were immunized and housed at Visionar Preclinical AB, Uppsala, according to GLP. Major organs were collected after which DNA was extracted and subjected to quantitative PCR (qPCR) analysis using primer and probe designed to detect the plasmid (pKCMV) backbone with a sensitivity of 10 copies of plasmid/μg of total extracted DNA.
With the exception of the skin of the injection site and in the underlying muscle, vaccine plasmids were readily cleared from the majority of organs (Table 2 ). At day 40 we detected plasmid in the spleen of the analyzed animal and by day 60 post immunization we could detect low levels of plasmid in the lungs of the analyzed animal (Table 2). At day 120 post immunization the plasmid DNA vaccine had been cleared from all organs except skin of the injection sites (Table 2, Fig. 2 ). The level of plasmids at the injection site decreased substantially over time but at the end of the experiment, at day 120 post immunization, several thousand copies of plasmid/μg of extracted DNA still remained.
Table 2.
Biodistribution, pKCMV quantitative PCR results (copies of pKCMV per microgram total DNA).
| Sample | No. of plasmids/μg DNA |
|||
|---|---|---|---|---|
| 14 days | 40 days | 60 days | 120 days | |
| Brain | – | – | – | – |
| Heart | – | – | – | – |
| Liver | – | – | – | – |
| Lung | – | – | 98 | – |
| Spleen | – | 2382 | – | – |
| Kidney | – | – | – | – |
| Ovaries | 103 | – | – | – |
| Intestine | – | – | – | – |
| Musclea | 4370 | 4760 | 606 | – |
| Skinb | 4,984,841 | 117,700 | 138,548 | 3096 |
(−) <10 copies of plasmid/μg of total DNA.
Muscle located directly beneath the site of injection.
Skin from the site of injection.
Fig. 2.
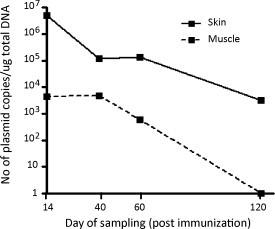
Presence of plasmid in skin at the injection site and underlying muscle. DNA was extracted and subjected to qPCR-analysis detecting the plasmid backbone. Results are shown as number of copies of plasmid/μg of extracted DNA at the indicated time points after immunization. Exact plasmid copy number for each time-point can be found in Table 2.
3.3. Plasmid persistence and integration analyses
In order to investigate the potential risk of integration of plasmid DNA into the host genome, we performed a study designed to quantify the amount of plasmid DNA that was associated with high molecular weight DNA (genomic DNA) at the site of injection, 60 days after immunization with the plasmid vaccine. The mice were immunized once with DNA or saline as described in the materials and methods section. FIGE was used to separate high molecular weight genomic DNA from extrachromosomal plasmid DNA and following the separation, a qPCR assay was used to detect potential genomic integration of the plasmid vector [21]. The primers used in the qPCR analysis are the same as used in the biodistribution assay and are designed to detect the plasmid backbone, independently of the inserted vaccine gene.
The number of copies of pKCMV plasmid detected in genomic DNA of vaccine-treated skin ranged from 3.68 × 105 to 8.81 × 105 per μg of DNA prior to gel separation (Table 3 ). Following restriction enzyme digestion and gel electrophoresis (primary FIGE), the number of copies of pKCMV detected in each pool ranged from 385 to 807 copies per microgram of HMW DNA. The HMW DNA isolated from the gel was then subjected to a second round of FIGE (secondary FIGE) and all samples tested below the assay limit of quantification, indicating minimal to no risk of plasmid DNA integration into host genomic DNA.
Table 3.
pKCMV quantitative PCR results before and after FIGE (Copies of pKCMV per microgram total DNA).
| Pool no. | Animal Nos. | Extracted gDNAa | Primary FIGEb | Secondary FIGEc |
|---|---|---|---|---|
| 1 (Untreated controls) | 1–8 | BLD | N/A | N/A |
| 2 (Untreated controls) | 9–16 | NQ | N/A | N/A |
| 3 (Immunized) | 17–24 | 881,316 | 419 | NQ |
| 4 (Immunized) | 25–32 | 546,352 | 385 | BLD |
| 5 (Immunized) | 33–40 | 664,524 | 554 | NQ |
| 6 (Immunized) | 41–48 | 367,767 | 807 | BLD |
| Mean value groups 3–6 | 615,989 ± 215,000 | 541 ± 191 | <10 | |
| Median value groups 3–6 | 605,438 | 487 |
BLD – below the limit of detection of the assay (<10 copies/μg DNA). NQ – below level of quantification (<100 copies/μg DNA). N/A – not applicable; separation not performed; specimen tested BLD or NQ for the pKCMV assay.
Template is gDNA.
Template is HMW DNA derived from restriction enzyme digested genomic DNA separated from plasmid DNA by field inversion gel electrophoresis.
Template is HMW DNA derived from primary FIGE HMW DNA subjected to successive round of field inversion gel electrophoresis.
3.4. Immunogenicity in mice
The readout of successful gene-transfer and expression, resulting in subsequent vaccine-specific immune responses, was measured as cellular immune responses. From the set of animals in the toxicological study that were assessed for blood chemistry parameters, spleens were collected from 5 female and 5 male animals from each group where after splenocytes were isolated and analyzed for vaccine-specific immune responses (Fig. 3 ). The vaccine-specific immune responses were analysed with IFN-γ ELISpot (Mabtech AB, Nacka, Sweden) and the splenocytes were stimulated with overlapping peptides covering either p24Gag or gp120Env, both of subtype B. All immunized mice, both female and male, responded against both antigens, Env and Gag. The strongest immune response could be detected in the female mice and was directed to Gag (Fig. 3).
Fig. 3.
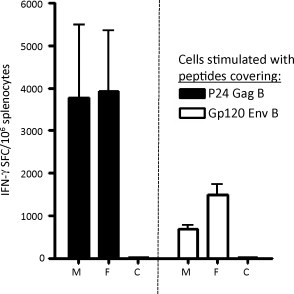
ELISpot analysis of IFN-γ secretion by splenocytes from mice immunized with the 7 plasmid vaccine and stimulated with peptides from HIV-1 Gag or Env, respectively. The animals were sacrificed 2 weeks after the fourth immunization. Error bars show standard deviation. M: male, F: female, C: saline injected animals (both female and male mice).
4. Discussion
Plasmid vaccines have been developed and evaluated in clinical trials for the prevention against, or treatment of, many different infectious diseases and cancers. Plasmid vaccines have shown an excellent safety profile and more than ten years have passed since the first clinical trials including genetic vaccines were concluded [22], [23]. However, plasmid vaccines have so far only induced moderate immune responses in man and the best responses have been obtained after boosting with recombinant microbial vectors or recombinant proteins. However, the application of in vivo electroporation increases the transfection efficacy and the immunogenicity of plasmid vaccines in animal models several fold [24], [25], [26], [27]. Currently, several clinical trials are ongoing in order to determine if the benefits of this methodology can be translated into successful use in humans. However, the significant increase in transfection efficacy observed after electroporation has raised concerns of prolonged persistence of plasmid in the target tissue and increased risk of integration of plasmid DNA into the host genome. Although electroporation is unlikely to influence the distribution of plasmids to other organs it is reasonable to assume that the more efficient transfection could affect the local persistence of plasmid at the site of injection [28], [29]. As shown in the present study, the plasmid DNA is indeed rapidly cleared from all organs except the site of the injection and the underlying muscle. At days 40 and 60 very low amounts of plasmid could be found in spleen and lungs, respectively. However, by day 120 no plasmid could be detected in any of the analyzed organs beside the skin at the site of injection. The presence of plasmid in the spleen at day 40 could potentially be a result of immune cells obtaining plasmid at the site of injection followed by trafficking to the spleen but the reason for the low, but detectable, levels of plasmid observed in the lung at day 60 is unclear. Possible explanations for the detected plasmid in lung might be that the mice inhaled plasmid during grooming (as there are high levels of plasmid in the skin at day 40 and 60) or that plasmids temporarily get retained in the fine mesh of capillaries surrounding the alveoli in the lung. As the analyses showed that plasmid was retained at the site of injection up to 120 days, we performed an extensive integration study of the skin from the injection site. The qPCR used for quantifying plasmids in extracted DNA allows for detection of as low as 10 plasmid copies/μg of total DNA, corresponding to a sensitivity of 10 plasmids/150,000 diploid cells. The results from integration experiments confirmed the persistence of plasmid in the injection site skin at day 60 but showed that there was no association of plasmid DNA with the host chromosomal DNA (Table 3). A previous study has shown that the transfected and antigen-expressing cells are concentrated in the deepest layer of the skin but also found in the hypodermis as well as in the dermis and epidermis [30]. It is unknown in what compartment the majority of plasmids are retained, but one can assume that both intra- and extracellular localization is involved.
The risk of integration after electroporation has been reported in a single study [13]. The level of integration demonstrated in the study was very low and would in a “worst case scenario” result in a level of integration similar to the rate of spontaneous gene-inactivating mutations [31]. Contrasting these results there are reports indicating that the increased immune responses induced by electroporation results in a more efficient clearing of plasmid/antigen as a consequence of a more potent cytotoxic response killing transfected antigen-expressing cells [20], [24], [32] Moreover, experiments conducted with the aim to incorporate genetic material into the host chromosome indeed confirms that plasmid vectors are highly inefficient for this purpose [33] and that viral vectors are much more powerful for purposes of integrative gene therapy. These studies thus serve as additional safety data for plasmid DNA vaccine applications, emphasizing the difficulties in achieving integration using plasmid DNA injection.
The results from the toxicological evaluation showed that the vaccine delivered by jet injection and electroporation was well tolerated and non-toxic in mice. The safety of the vaccine is further strengthened by the fact that the amount of DNA used in the mice, 200 μg, corresponds to roughly 10 mg of plasmid DNA/kg body weight and this is substantially higher than the 8–10 μg of plasmid DNA/kg body weight that is planned for the clinical testing of this vaccine strategy. Furthermore, the skin delivery, in contrast to intramuscular delivery, allows for the possibility to visually inspect the injection site, thus making it easier to detect any potentially malignant changes. In conclusion, a high dose of highly immunogenic vaccine plasmids delivered intradermally by jet injection followed by electroporation was shown to be safe and did not result in integration of plasmid DNA into the host genome. The vaccine and delivery method presented here were recently approved by the Swedish Medical Products Agency and will shortly enter a phase I clinical trial in Stockholm involving healthy non-HIV infected volunteers.
Acknowledgements
This work was funded by the Swedish international development cooperation agency and through the EU programs EUROPRISE (LSHP-CT-2006-037611) and NGIN (Health-F3-2008-201433)
References
- 1.Bacardi D., Amador-Canizares Y., Cosme K., Urquiza D., Suarez J., Marante J. Toxicology and biodistribution study of CIGB-230, a DNA vaccine against hepatitis C virus. Hum Exp Toxicol. 2009;28(August (8)):479–491. doi: 10.1177/0960327109106438. [DOI] [PubMed] [Google Scholar]
- 2.Gravier R., Dory D., Laurentie M., Bougeard S., Cariolet R., Jestin A. In vivo tissue distribution and kinetics of a pseudorabies virus plasmid DNA vaccine after intramuscular injection in swine. Vaccine. 2007;25(September (39–40)):6930–6938. doi: 10.1016/j.vaccine.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Hanke T., McMichael A.J., Samuel R.V., Powell L.A., McLoughlin L., Crome S.J. Lack of toxicity and persistence in the mouse associated with administration of candidate DNA- and modified vaccinia virus Ankara (MVA)-based HIV vaccines for Kenya. Vaccine. 2002;21(November (1–2)):108–114. doi: 10.1016/s0264-410x(02)00403-6. [DOI] [PubMed] [Google Scholar]
- 4.Kim B.M., Lee D.S., Choi J.H., Kim C.Y., Son M., Suh Y.S. In vivo kinetics and biodistribution of a HIV-1 DNA vaccine after administration in mice. Arch Pharm Res. 2003;26(June (6)):493–498. doi: 10.1007/BF02976869. [DOI] [PubMed] [Google Scholar]
- 5.Manam S., Ledwith B.J., Barnum A.B., Troilo P.J., Pauley C.J., Harper L.B. Plasmid DNA vaccines: tissue distribution and effects of DNA sequence, adjuvants and delivery method on integration into host DNA. Intervirology. 2000;43(4–6):273–281. doi: 10.1159/000053994. [DOI] [PubMed] [Google Scholar]
- 6.Parker S.E., Borellini F., Wenk M.L., Hobart P., Hoffman S.L., Hedstrom R. Plasmid DNA malaria vaccine: tissue distribution and safety studies in mice and rabbits. Hum Gene Ther. 1999;10(March (5)):741–758. doi: 10.1089/10430349950018508. [DOI] [PubMed] [Google Scholar]
- 7.Quezada A., Larson J., French M., Ponce R., Perrard J., Durland R. Biodistribution and safety studies of hDel-1 plasmid-based gene therapy in mouse and rabbit models. J Pharm Pharmacol. 2004;56(February (2)):177–185. doi: 10.1211/0022357022584. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez K., Barry E.M., Ulmer J., Stout R., Szabo J., Manetz S. Preclinical safety and biodistribution of Sindbis virus measles DNA vaccines administered as a single dose or followed by live attenuated measles vaccine in a heterologous prime-boost regimen. Hum Gene Ther. 2008;19(May (5)):522–531. doi: 10.1089/hum.2007.172. [DOI] [PubMed] [Google Scholar]
- 9.Sheets R.L., Stein J., Manetz T.S., Duffy C., Nason M., Andrews C. Biodistribution of DNA plasmid vaccines against HIV-1, Ebola, Severe Acute Respiratory Syndrome, or West Nile virus is similar, without integration, despite differing plasmid backbones or gene inserts. Toxicol Sci. 2006;91(June (2)):610–619. doi: 10.1093/toxsci/kfj169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winegar R.A., Monforte J.A., Suing K.D., O’Loughlin K.G., Rudd C.J., Macgregor J.T. Determination of tissue distribution of an intramuscular plasmid vaccine using PCR and in situ DNA hybridization. Hum Gene Ther. 1996;7(November (17)):2185–2194. doi: 10.1089/hum.1996.7.17-2185. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H.Y., Sun S.H., Guo Y.J., Chen Z.H., Huang L., Gao Y.J. Tissue distribution of a plasmid DNA containing epitopes of foot-and-mouth disease virus in mice. Vaccine. 2005;23(December (48–49)):5632–5640. doi: 10.1016/j.vaccine.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 12.Schalk J.A., Mooi F.R., Berbers G.A., van Aerts L.A., Ovelgonne H., Kimman T.G. Preclinical and clinical safety studies on DNA vaccines. Hum Vaccin. 2006;2(March–April (2)):45–53. doi: 10.4161/hv.2.2.2620. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z., Troilo P.J., Wang X., Griffiths T.G., Pacchione S.J., Barnum A.B. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11(April (8)):711–721. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- 14.Brave A., Ljungberg K., Boberg A., Rollman E., Isaguliants M., Lundgren B. Multigene/multisubtype HIV-1 vaccine induces potent cellular and humoral immune responses by needle-free intradermal delivery. Mol Ther. 2005;12(December (6)):1197–1205. doi: 10.1016/j.ymthe.2005.06.473. [DOI] [PubMed] [Google Scholar]
- 15.Sandstrom E., Nilsson C., Hejdeman B., Brave A., Bratt G., Robb M. Broad immunogenicity of a multigene, multiclade HIV-1 DNA vaccine boosted with heterologous HIV-1 recombinant modified vaccinia virus Ankara. J Infect Dis. 2008;198(November (10)):1482–1490. doi: 10.1086/592507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakari M. A low dose multigene, multiclade HIV DNA given intradermally induces strong and broad immune responses after boosting with heterologous HIV MVA. AIDS Vaccine conference, 2009; Paris; 2009. [Google Scholar]
- 17.Ljungberg K., Rollman E., Eriksson L., Hinkula J., Wahren B. Enhanced immune responses after DNA vaccination with combined envelope genes from different HIV-1 subtypes. Virology. 2002;302(October (1)):44–57. doi: 10.1006/viro.2002.1547. [DOI] [PubMed] [Google Scholar]
- 18.Rollman E., Brave A., Boberg A., Gudmundsdotter L., Engstrom G., Isaguliants M. The rationale behind a vaccine based on multiple HIV antigens. Microbes Infect. 2005;7(November (14)):1414–1423. doi: 10.1016/j.micinf.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 19.Brave A., Ljungberg K., Boberg A., Rollman E., Engstrom G., Hinkula J. Reduced cellular immune responses following immunization with a multi-gene HIV-1 vaccine. Vaccine. 2006;24(May (21)):4524–4526. doi: 10.1016/j.vaccine.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Roos A.K., Eriksson F., Walters D.C., Pisa P., King A.D. Optimization of skin electroporation in mice to increase tolerability of DNA vaccine delivery to patients. Mol Ther. 2009;17(September (9)):1637–1642. doi: 10.1038/mt.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin T., Parker S.E., Hedstrom R., Le T., Hoffman S.L., Norman J. Plasmid DNA malaria vaccine: the potential for genomic integration after intramuscular injection. Hum Gene Ther. 1999;10(March (5)):759–768. doi: 10.1089/10430349950018517. [DOI] [PubMed] [Google Scholar]
- 22.Calarota S., Bratt G., Nordlund S., Hinkula J., Leandersson A.C., Sandstrom E. Cellular cytotoxic response induced by DNA vaccination in HIV-1-infected patients. Lancet. 1998;351(May (9112)):1320–1325. doi: 10.1016/S0140-6736(97)09440-3. [DOI] [PubMed] [Google Scholar]
- 23.MacGregor R.R., Boyer J.D., Ugen K.E., Lacy K.E., Gluckman S.J., Bagarazzi M.L. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J Infect Dis. 1998;178(July (1)):92–100. doi: 10.1086/515613. [DOI] [PubMed] [Google Scholar]
- 24.Luckay A., Sidhu M.K., Kjeken R., Megati S., Chong S.Y., Roopchand V. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in rhesus macaques. J Virol. 2007;81(May (10)):5257–5269. doi: 10.1128/JVI.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mir L.M., Bureau M.F., Gehl J., Rangara R., Rouy D., Caillaud J.M. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc Natl Acad Sci USA. 1999;96(April (8)):4262–4267. doi: 10.1073/pnas.96.8.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otten G., Schaefer M., Doe B., Liu H., Srivastava I., zur Megede J. Enhancement of DNA vaccine potency in rhesus macaques by electroporation. Vaccine. 2004;22(June (19)):2489–2493. doi: 10.1016/j.vaccine.2003.11.073. [DOI] [PubMed] [Google Scholar]
- 27.Widera G., Austin M., Rabussay D., Goldbeck C., Barnett S.W., Chen M. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J Immunol. 2000;164(May (9)):4635–4640. doi: 10.4049/jimmunol.164.9.4635. [DOI] [PubMed] [Google Scholar]
- 28.Long Y.C., Jaichandran S., Ho L.P., Tien S.L., Tan S.Y., Kon O.L. FVIII gene delivery by muscle electroporation corrects murine hemophilia A. J Gene Med. 2005;7(April (4)):494–505. doi: 10.1002/jgm.683. [DOI] [PubMed] [Google Scholar]
- 29.Luxembourg A., Evans C.F., Hannaman D. Electroporation-based DNA immunisation: translation to the clinic. Expert Opin Biol Ther. 2007;7(November (11)):1647–1664. doi: 10.1517/14712598.7.11.1647. [DOI] [PubMed] [Google Scholar]
- 30.Roos A.K., Eriksson F., Timmons J.A., Gerhardt J., Nyman U., Gudmundsdotter L. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS ONE. 2009;4(9):e7226. doi: 10.1371/journal.pone.0007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ledwith B.J., Manam S., Troilo P.J., Barnum A.B., Pauley C.J., Griffiths T.G., 2 Plasmid DNA vaccines: investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology. 2000;43(4–6):258–272. doi: 10.1159/000053993. [DOI] [PubMed] [Google Scholar]
- 32.Ahlen G., Soderholm J., Tjelle T., Kjeken R., Frelin L., Hoglund U. In vivo electroporation enhances the immunogenicity of hepatitis C virus nonstructural 3/4A DNA by increased local DNA uptake, protein expression, inflammation, and infiltration of CD3+ T cells. J Immunol. 2007;179(October (7)):4741–4753. doi: 10.4049/jimmunol.179.7.4741. [DOI] [PubMed] [Google Scholar]
- 33.Chalberg T.W., Genise H.L., Vollrath D. Calos MP. phiC31 integrase confers genomic integration and long-term transgene expression in rat retina. Invest Ophthalmol Vis Sci. 2005;46(June (6)):2140–2146. doi: 10.1167/iovs.04-1252. [DOI] [PubMed] [Google Scholar]


