Abstract
There is a need to develop reliable methods to assess the safety of genetically modified and other novel foods. The aim of this study was to identify protein biomarkers of food allergy in mice exposed to ovomucoid (OVM), a major food allergen found in chicken egg white. BALB/c mice were repeatedly sensitized by gavage with OVM and cholera toxin (CT) and control mice were exposed to a mixture of amino acids with CT. At the endpoint, all mice were challenged intraperitoneally with OVM and alum. Type-1 hypersensitivity was confirmed in OVM-sensitized mice by observation of clinical signs of anaphylaxis and elevated levels of plasma histamine, OVM-specific IgE and OVM-specific IgG by ELISA. Differential protein expression was assessed in albumin-depleted plasma as well as in mesenteric lymph node, liver, spleen, and ileum by two-dimensional difference gel electrophoresis (2D-DIGE). Differentially expressed proteins were identified by liquid chromatography with tandem mass spectrometry. Plasma proteins overexpressed in OVM-sensitized mice included haptoglobin (41-fold), serum amyloid A (19-fold) and peroxiredoxin-2 (1.9-fold). Further validation of these plasma proteins in other animal models of food allergy with different food allergens is required to assess their potential as candidate biomarkers for use in evaluating the allergenicity of novel foods.
Keywords: Anaphylaxis, Food allergy, Mice, Ovomucoid, Plasma, Proteomics
Abbreviations: 2D-DIGE, two-dimensional difference gel electrophoresis; CT, cholera toxin; Hp, haptoglobin; OVM, ovomucoid; PRXII, peroxiredoxin-2; SAA2, serum amyloid A2
1. Introduction
Food allergies affect an estimated 3.7% of adults (Sampson, 2005) and 4.0% of infants (Venter et al., 2006). Adults are most often affected from eating fish or shellfish (2.3%) while infants frequently develop allergies to eggs 1.6% and milk 1.1% (Eggesbo et al., 2001a, Eggesbo et al., 2001b, Sicherer et al., 2004). Although most food allergies in humans cause mild reactions, on rare occasions they can be fatal and are now the leading cause of anaphylaxis in many countries (Sampson, 2003). Current knowledge of allergens, tolerance, effector mechanisms and predisposing conditions does not allow for accurate prediction of allergenicity in humans without food trials or prior accidental exposure. Consequently, there has been recent heightened interest to develop a validated animal model to objectively assess the allergenicity of foods for safety testing and immunotherapy development. While various models of human food allergy have been investigated none have yet been validated (Knippels and Penninks, 2005).
Serum allergen-specific IgE titers are used as an indicator of sensitization and confirmation of anaphylaxis (Dearman et al., 2003) and have been suggested as a clinical aid to predicting allergy or tolerance in children (Shek et al., 2004, Eigenmann, 2005). Antigen-specific IgE titers have excellent positive predictive value for clinical allergic reactivity in individuals with markedly increased titers but poor negative predictive value in those with low titers, necessitating food trials for these individuals (Sampson, 2005). Moreover, analysis of IgE titers and passive cutaneous skin assays require repetitive prior exposure for development of sensitization. Thus, there is a need for a rapid and accurate screening assay with a high negative predictive value of allergenicity for the valuation of allergenicity of novel foods or the effects of novel food processing methods on common foods. These biomarkers should be derived from reliable, validated, and relevant animal models and should be quantifiable and proportional to the degree of allergenicity of the food and should be detectable during both sensitization and post-challenge anaphylaxis. Newly identified biomarkers may eventually be used in an allergy screening panels with known mediators of allergy including histamine, antigen-specific IgE, and IL-4. Panels of biomarkers would then require validation with different food allergens in various animal models before applying them to the safety testing of novel foods.
Ovomucoid (OVM) is a heat stable glycoprotein comprising about 11% of chicken egg white, and is the major cause of allergic reactions in children with egg allergies (Bernhisel-Broadbent et al., 1994). OVM allergenicity has previously been characterized in a cholera-toxin-based BALB/c model (Kroghsbo et al., 2003, Adel-Patient et al., 2005) and we have previously demonstrated that OVM challenge increases plasma histamine, OVM-specific IgE and OVM-specific IgG levels and the secretion of type-2 cytokines such as IL-4 by cultured splenic lymphocytes (Rupa and Mine, 2006). BALB/c mice have often been used as animal models allergy, because they are high IgE-responders and are predisposed to developing Th2 responses more readily than other mouse strains (Jyonouchi et al., 2001).
The advent of proteomics technology has greatly facilitated profiling of plasma and tissues for disease-related biomarkers. Two-dimensional difference gel electrophoresis (2D-DIGE) is an efficient and accurate technology that efficiently separates proteins in complex mixtures and quantifies differential expression in treated and control samples (Alban et al., 2003). 2D-DIGE is very useful as a broad-based screening tool to identify differentially expressed proteins of interest, however, the technique has limited capability in resolving very small, very large, or highly basic, acidic, or hydrophobic proteins. 2D-DIGE is best applied in pH ranges approximating physiological pH and narrowing the range increases separation and resolution of protein spots.
To the best of our knowledge, proteomic profiling to detect biomarkers of food allergy has not been previously reported in mice or other animal models. The aim of this study was to identify protein biomarkers of OVM allergy in a mouse model for eventual use in the assessment of allergic potential of human foods during food safety testing.
2. Materials and methods
2.1. Materials
Immobilized pH gradient strips 13 cm pH 4–7 and pH 3–11, Cy2, Cy3, Cy5, 2D-Clean Up Kit, pharmalytes 3–10, Nuclease mix, and lower molecular weight markers were purchased from GE Healthcare (Montreal, QC). Urea, thiourea, ASB 14, DMF, trifluoroacetic acid, goat anti-mouse-IgG, and pNPP were acquired from Sigma (St. Louis, MO); iodacetamide, CHAPS and DTT from UBS (Cleveland, OH); Qproteome Murine Albumin Depletion Kit from Qiagen (Mississauga, ON); Biorad protein assay, BSA protein standard, 40% acrylamide, and Sypro Ruby stain; Bio-Scale S5 column from Bio-Rad Laboratories (Hercules, CA); 2% alhydrogel from Superfos Biosector (Denmark); histamine ELISA assay kit from Neogen Corporation (Lexington, KY); acetonitrile and water from Fisher Scientific (Ottawa, ON); formic acid from VWR (Mississauga, ON); rat anti-mouse-IgE and purified mouse IgE from BD Pharmingen (San Diego, CA); and purified mouse IgG from AbD Serotec (Raleigh, NC).
2.2. Animal treatments and sample collection
Thirty, 6–8 week old, female, BALB/c mice were purchased from Charles River (Montreal, QC, Canada) randomly divided into two groups (15 control and 15 OVM-sensitized) and conventionally housed 3 per cage at the Central Animal Facility at the University of Guelph. Mice were fed, exclusively, ad libitum, acidified bottled water along with 2014 Teklad Global 14% Protein Rodent Maintenance Diet which has a fixed formula and is free of all animal protein including egg and fish meal. Animal studies were conducted in accordance with the guidelines of the Canadian Council on Animal Care.
Whole OVM antigen consisting of all three domains was isolated from chicken egg white according to an earlier described procedure (Fredericq and Deutsch, 1949) and further purified using HPLC ion-exchange chromatography. Fifty milligram of crude OVM was dissolved in 5 ml of 20 mM sodium acetate buffer, pH 4.0, and applied to a Bio-Scale S5 column equilibrated with the same buffer. The column was eluted with a linear gradient of 0–1.0 M NaCl in 20 mM sodium acetate buffer, pH 4.0, at a flow rate of 1.0 ml/min using a Bio-Rad Biologic HPLC system.
Mice were gavaged twice weekly for 5 weeks with OVM (1 mg/mouse/gavage) whereas control mice were gavaged with a mixture of amino acids (1 mg) in a ratio similar to OVM. Both gavages contained cholera toxin (10 μg) as a mucosal adjuvant in sterile water for a total volume of 100 μL/mouse/gavage. At week 6, all mice were challenged with an intraperitoneal (IP) injection of OVM (1 mg) in sterile water (50 μl) and aluminum hydroxide adjuvant (50 μl, 2% alhydrogel) to induce an immediate type-1 hypersensitivity reaction. Clinical responses were scored for anaphylaxis (see below) and at 40 min post-challenge, mice were euthanized with CO2, whole blood was collected by cardiac puncture into citrated tubes and samples of liver, spleen, ileum, and mesenteric lymph node were flash frozen in liquid nitrogen and stored at −70 °C. Plasma was obtained by centrifugation at 2000g for 15 min, pooled by cage (3 mice/cage/group) and stored at −70 °C. Additional liver, ileum and spleen samples were fixed in 10% v/v buffered formalin, paraffin-embedded, and 5 μm sections were stained with hematoxylin and eosin and evaluated in a blinded fashion by two independent pathologists.
3. Immunologic analysis of OVM allergy
3.1. Anaphylaxis scoring
Clinical responses in all mice were assessed from 0 to 40 min post IP challenge by three independent investigators. Clinical responses of the mice were categorized as follows: no reaction = 0; persistent scratching or rubbing the face, ears, or head = 1; increased respiratory rate or facial edema = 2; dyspnea or cyanosis of the face or tail = 3, tremors and convulsion = 4; and death = 5 according to previously published criteria (Li et al., 1999).
3.2. Immunologic assays for plasma histamine and OVM-specific IgE, and IgG
Plasma histamine levels were determined by competitive direct ELISA using a commercial histamine ELISA kit (Neogen Corporation; Lexington, KY) and the concentration of histamine was calculated by comparison to a standard curve according to the manufacturer’s instructions.
OVM-specific IgE and IgG levels in the plasma of all the mice were determined by ELISA according to a similar previously described method (Rupa and Mine, 2006). This approach is commonly accepted for making relative rather than quantitative comparisons of antigen-specific antibody levels between control and treated groups of animals by assessing differences in optical absorbance. Absolute quantification of OVM-specific IgE and IgG was not performed as OVM-specific monoclonal antibodies are not available commercially for use in standard curves or as positive controls. Microtiter plates were coated with 100 μl (5 μg/ml) of purified OVM in 50 mM sodium bicarbonate buffer, pH 9.6, overnight at 4 °C. After three washes with PBST (phosphate-buffered saline containing 0.05% Tween 20), the plates were incubated with blocking buffer (2% BSA in PBS) for 1 h at 37 °C. The plates were further washed three times with PBST and 100 μl/well of diluted sera (1:50 for IgG; 1:10 for IgE) were added and incubated at 37 °C for 2 h. After washing three times with PBST, plates were then incubated with either alkaline phosphatase-conjugated goat anti-mouse-IgG (1:15,000 for 1 h at 37 °C) or with biotinylated rat anti-mouse-IgE monoclonal antibody (1:1000 for 2 h at 37 °C). The plates were then washed again and incubated with extr-avidin–alkaline phosphatase (1:3000) for 1 h at 37 °C. After a final washing the reaction was visualized with p-nitrophenol phosphatase (pNPP) (1 mg/ml) and the absorbance measured at 405 nm. ELISAs used to detect total IgG and IgE (data not shown) were validated in standard curves using commercially available recombinant mouse IgG and IgE as per the manufacturer’s instructions (BD Pharmingen). The average background absorbance was determined to be <0.1 OD using serum-free PBS applied to OVM-coated wells, as a control to verify absence of non-specific binding.
4. Proteomic analysis
4.1. 2D-DIGE analysis of plasma
Total protein was quantified in four pooled control and four pooled OVM-sensitized plasma samples using the Biorad protein assay. Plasma (1.3 mg of protein) was loaded into an Qproteome mouse albumin removal column in phosphate-buffered saline (PBS) equal to three times the sample volume, mixed by rocking for 10 min, then briefly centrifuged, washed with PBS, and then centrifuged again. The combined eluate was then concentrated and impurities removed using the 2D Clean Up Kit. The final precipitated pellets were resolubilized in 60 μl of lysis buffer (8 M urea, 4% CHAPS, 30 mM Tris–Cl, pH 8.5) and total protein was quantified again and stored on ice prior to subsequent Cy dye labeling.
Each sample was labeled with 200 pmol (1 μl) of Cy dye per 50 μg of protein, incubated on ice for 30 min in the dark and quenched with 1 μl of 10 mM lysine and then incubated on ice for 10 min in the dark, according to the manufactures protocol. Four control and four OVM-sensitized samples (each 50 μg of protein) were labeled separately with either Cy3 or Cy5, and the internal standard (200 μg of protein comprising 25 μg from each of the eight samples), was labeled with Cy2. One control, OVM-sensitized and standard sample forming a set of Cy2, Cy3, and Cy5 labeled samples was combined for each of four gels and mixed with rehydration buffer (8 M urea, 1% CHAPS, 0.5% ASB14, 65 mM DTT, 0.5% v/v pharmalytes pH 3–10, 0.005% bromophenol blue) to a total volume of 250 μl per gel. The first dimension separation was performed on an IPGphor isoelectric focusing (IEF) unit (GE Healthcare) by applying combined samples to an immobilized pH gradient (IPG) 13 cm pH 4–7 strips with rehydration at 30 V for 10 h followed by isoelectric focusing at 500 V step for 2 h, 1000 V gradient for 1 h, 8000 V gradient for 2.5 h, then 8000 V step for 16,000 V h, for a total of 7.5 h and 29.4 kV h. Strips were each immediately equilibrated in a 5 ml solution of 6 M urea, 30% glycerol, 2% SDS, 75 mM Tris–Cl (pH 8.8), 0.005% bromophenol blue, for 15 min with 1% (w/v) DTT and subsequently 15 min with 2.5% (w/v) iodoacetamide. A molecular weight marker (14–97 kDa) was added to one gel for reference. For the second dimension, strips were applied directly to 12% SDS-polyacrylamide gels and four gels were run simultaneously on a DALT 6 (GE Healthcare) electrophoresis unit at 20 °C at 0.25 W/gel for 1 h, 0.5 W/gel for 1 h and then 17 W/gel for 5.5 h. A picking gel was created using 600 μg of total protein pooled from each of the four OVM-sensitized plasma samples analyzed and run under conditions similar to analytical gels except it was unlabeled (non-DIGE) and the IEF was extended to a total of 37.6 kV h. Picking gels were fixed in 10% methanol and 7% acetic acid for 2 h, stained with Sypro Ruby overnight, destained in fixative for 2 h, and scanned with Typhoon 9410 at 532 nm with a 610 nm filter.
4.2. 2D-DIGE analysis of liver, spleen, mesenteric lymph node, and ileum
Tissue samples flash frozen in liquid nitrogen following the initial collection, were thawed and briefly homogenized in 1.5 ml glass tubes. Lysis buffer, consisting of 7 M urea, 2 M thiourea with 4% w/v CHAPS, 1% v/v nuclease mix, 30 mM Tris–Cl (pH 8.5), was added in a volume equal to 20 or 25 times the tissue mass depending on the tissue type. Tissues were further solubilized by sonification and stored on ice for subsequent Cy dye labeling. Liver and ileum samples were combined with rehydration buffer containing 8 M urea, 2% CHAPS, 18 mM DTT, 0.5% v/v pharmalytes pH 3–10, 0.005% bromophenol blue while spleen and lymph node samples combined with 8 M urea, 2% CHAPS, 65 mM DTT, 0.5% v/v pharmalytes pH 3–10, 0.005% bromophenol blue, before being applied to 13 cm pH 3–11 IPG strips (all tissues) and pH 4–7 IPG strips (liver, spleen, lymph node). As stated for plasma above, strips were rehydrated at 30 V for 10–12 h followed by IEF to 24–30 kV h. Equilibration, PAGE, gel scanning, post-staining, and analysis were identical to plasma.
4.3. Quantification of protein overexpression
Immediately following 2D-DIGE, gels were scanned with a Typhoon 9410 imager using an excitation/emission filter of 488 nm/520 nm for Cy2, 532 nm/580 nm for Cy3, and 633 nm/670 nm for Cy5 to generate multiplexed DIGE image files. Statistical and quantitative analyses of spot changes on images were completed using DeCyder 6.5 software (GE Healthcare). Measurements of spot abundance were normalized to the internal standard sample, which was common to all four gels in the experiment. The ratio of normalized control protein abundance to normalized OVM-sensitized protein abundance was calculated for paired control–OVM-sensitized samples on each gel and the average of all four gels was calculated to represent the change in protein abundance (average ratio).
4.4. Identification of overexpressed proteins
Plasma protein spots of interest identified by DeCyder and 2D-DIGE were manually excised from Sypro Ruby-stained picking gels and sent to the Advanced Protein Technology Center, Hospital for Sick Children (Toronto, Ontario, Canada) where tryptic digests were analyzed by mass spectrometry. Peptide sequencing was determined by on-line liquid chromatography tandem mass spectrometry (LC/MS/MS) using a QSTAR XL electrospray ionization QTOF mass spectrometer (Applied Biosystems/MDS Sciex, Concord, ON, Canada) coupled with an Agilent 1100 NanoLC system (Santa Clara, CA). Samples (5 μL) were first loaded into a precolumn (100 μm i.d. × 5 cm), and then eluted to an analytical column (75 μm i.d. × 10 cm) for further separation. Both columns were packed with Pursuit C18 (5 μm particle size, 200 Å pore size, by Varian Inc., Palo Alto, CA). For the reverse phase chromatography, a 125 min gradient elution from water to acetonitrile, each containing 0.1% formic acid and 0.02% trifluoroacetic acid, was performed at a flow of 200 nL/min. Tandem mass spectra were extracted, charge state deconvoluted and deisotoped by BioWorks version 2.0. All MS/MS spectra were analyzed using Mascot (Matrix Science, London, UK; version 2.1.03) and X! Tandem (http://www.thegpm.org; version 2006.04.01.2), using the NCBInr_20061106 database (Mus musculus), with trypsin digestion and a fragment ion mass tolerance of 0.20 Da and a parent ion tolerance of 0.20 Da, iodoacetamide derivative of cysteine was specified as a fixed modification while variable modifications were specified as Pyro-glu from E of the n-terminus, s-carbamoylmethylcysteine cyclization of the n-terminus, deamidation of asparagine and glutamine, oxidation of methionine and acetylation of the n-terminus. Scaffold (Proteome Software Inc., Portland, OR, version-01_06_04) was used to validate MS/MS-based peptide and protein identifications. Protein identifications were accepted if they could be established at greater than 80.0% probability and contained at least three identified peptides.
In some cases, matrix-assisted laser desorption/ionization top-of-flight mass spectrometry (MALDI-TOF-MS) was used to confirm the identity of isomer protein spots initially identified by LC–MS/MS. MALDI-TOF-MS has much lower specificity compared to LC–MS/MS and as a result it is generally accepted that the number of theoretical protein modifications chosen during a MALDI search should be held to a minimum, to maximize specificity even though there is some loss in sensitivity. The number of modifications allowed during the MASCOT search was minimized to improve the specificity of matching MALDI-TOF-MS data. Proteins were analyzed by MALDI-TOF-MS using an Applied Biosystems/MDS Sciex API QSTAR XL Pulsar MALDI-QqTOF amd protein identification was determined using the Mascot search engine. Significant measured peptide masses in the spectrum were matched to peptides in the NCBI database for mus musculus with no more than one missed cut, complete carbamidomethyl (C), partial oxidation (M), 0.2 Da peptide tolerance, one charge state (MH+) and Mascot scores greater than 63 (p ⩽ 0.05). The number of modifications allowed during the Mascot search was minimized to maximize the specificity of matching MALDI-TOF-MS data, because it is less specific than MS/MS data.
4.5. Stastistical analysis
A Student’s t-test was performed on histamine, IgE and IgG data for comparison between OVM-sensitized and control groups and data are presented as the mean ± standard error of the mean with p values less than 0.05 considered to be statistically significant. Differences in protein spots were analyzed for statistical significance by comparing the control (n = 4 pools) and OVM-sensitized (n = 4 pools) sample populations using the Student’s t-test in DeCyder 6.5. Proteins of interest were initially selected based on an average ratio of abundance greater than 2.0 with p < 0.05. Additional proteins with changes approaching these criteria were also considered as proteins of interest based on biological relevance.
5. Results
5.1. Clinical assessment of anaphylaxis
Clinical signs of anaphylaxis were monitored for 40 min from the time of challenge to the end point. OVM-sensitized mice had an average anaphylaxis score of 3/5 demonstrating variable levels of face scratching, hunched posture, dyspnea, and tail cyanosis, while there were no observable clinical signs of anaphylaxis in control mice. In addition, gross examination of the viscera during sample collection revealed mildly enlarged Peyer’s patches on the small intestinal serosa of the OVM-sensitized mice. There were no microscopic lesions evident on histologic analysis of H&E-stained sections of liver, spleen, and ileum in control and OVM-sensitized mice.
5.2. Clinical pathology assessment of anaphylaxis
To assess the induction of an IgE-mediated response to the OVM challenge, histamine, and OVM-specific IgE levels were assayed in plasma 40 min post-challenge. In addition, OVM-specific IgG was assayed to differentiate an immunogenic response from general inflammation. The OVM-sensitized mice demonstrated an 8-fold increase in plasma histamine levels (Fig. 1 a, p = 0.01), a 1.5-fold increase in OVM-specific IgE (p = 0.05, Fig. 1b), and a 6-fold increase in OVM-specific IgG levels (Fig. 1c, p = 0.05) relative to the unsensitized control mice, confirming the occurrence of a type-1 hypersensitivity response.
Fig. 1.
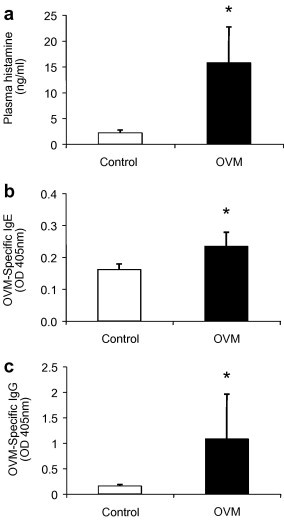
Histamine, OVM-specific IgE and IgG are elevated in OVM-challenged mice. (a) Histamine; (b) OVM-specific IgE; and (c) OVM-specific IgG, were measured in pooled plasma samples (n = 5 pools with 3 mice/pool) of amino acid control mice and OVM-sensitized mice. Plasma was collected 40 min following IP challenge with OVM and assayed by ELISA. Results represent the mean ± SEM. *Significantly different from control (p ⩽ 0.05).
5.3. 2D-DIGE analysis of differentially expressed proteins
Serum proteins that were differentially expressed in OVM-sensitized mice were detected by 2D-DIGE analysis. An example of a three-channel 2D-DIGE overlay image (Fig. 2 ) illustrates several proteins (red, Cy5) that were significantly overexpressed in the plasma of OVM-sensitized mice compared to proteins (green, Cy3) from control mice. The majority of remaining protein spots were expressed equally in plasma of both control and OVM-sensitized mice and appeared as white spots on DIGE gels. Grayscale images representing individual scans of OVM-sensitized or control plasma proteins (e.g. Fig. 3 ) were analyzed using Decyder 6.5 software, to quantify the increased expression of the seven protein spots of interest (Table 1 ). Haptoglobin (Hp), peroxiredoxin-2 (PRXII), and serum amyloid A2 (SAA2) were overexpressed proteins that were subsequently identified by LC–MS/MS of tryptic digests of gel plugs (Table 2 ) whereas additional isomers of Hp were identified by MALDI-TOF-MS (Table 3 ). Spot 1928 contained minor amounts of hemoglobin-alpha likely representing a contaminant of the more abundant SAA2 (Table 3).
Fig. 2.
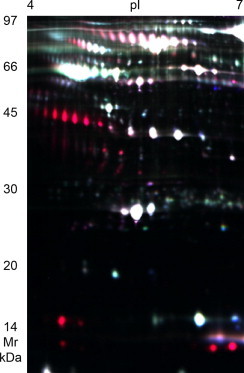
2D-DIGE overlay image of differentially expressed plasma proteins in OVM-challenged mice. 2D-DIGE overlay image of protein spots comparing albumin-depleted plasma from unsensitized control mice labeled with Cy3 (green) to OVM-sensitized mice labeled with Cy5 (red), and an internal standard sample common to all gels labeled with Cy2 (blue). Proteins with similar expression levels in control and OVM-sensitized mice appear white in color. Plasma was collected 40 min following intraperitoneal challenge with ovomucoid. This image is representative of one of four gels analyzed.
Fig. 3.
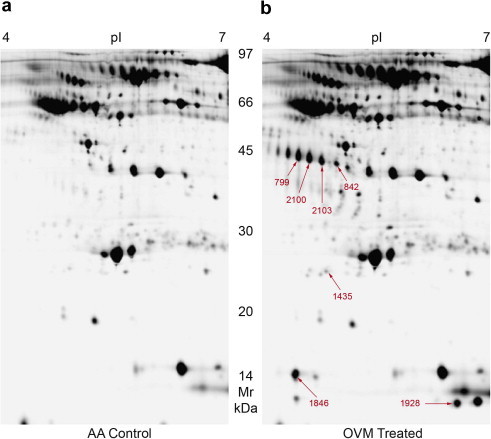
2D-DIGE gray scale image of plasma protein expression in control and OVM-challenged mice. Analytical 2D-DIGE gray scale images of samples depicted in Fig. 2 used to quantify changes in protein spot abundance in albumin-depleted plasma from (a) unsensitized control mice labeled with Cy3 and (b) OVM-sensitized mice labeled with Cy5, electrophoresed within the same gel. Protein spots that were significantly increased (change ⩾ 1.9-fold, p ⩽ 0.06) in OVM-sensitized mouse plasma are indicated. These images of individually labeled samples are representative of one the four gels analyzed.
Table 1.
DIGE analysis of plasma proteins overexpressed in OVM-sensitized mice
| Spot # | Protein | Average ratio of abundance T vs C | Significance t-test |
|---|---|---|---|
| 2103 | Haptoglobin | 41.2 | 0.020 |
| 2100 | Haptoglobin | 27.0 | 0.029 |
| 799 | Haptoglobin | 20.0 | 0.052 |
| 842 | Haptoglobin | 14.7 | 0.033 |
| 1846 | Haptoglobin | 61.6 | 0.019 |
| 1435 | Peroxiredoxin-2 | 1.9 | 0.004 |
| 1928 | Serum amyloid A2 | 19.3 | 0.060 |
Results of analysis 2D-DIGE images of albumin-depleted plasma from OVM-sensitized and control mice using Decyder 6.5 indicating the average ratio of abundance of protein spots and statistical significance (Student’s t-test). Positive ratios indicate increased expression of proteins in the plasma from OVM-sensitized mice relative to control mice.
Table 2.
LC–MS/MS analysis and identification of plasma proteins overexpressed in OVM-exposed mice
| Protein | Spot # | Accession # | Database MW/pI | Scaffold % probability protein identity | # Unique peptides | % Coverage | # Amino acids/total |
|---|---|---|---|---|---|---|---|
| Haptoglobin | 2100 | gi|8850219 | 38,752/5.9 | 100 | 28 | 41 | 143/347 |
| Haptoglobin | 1846 | gi|8850219 | 38,752/5.9 | 89 | 3 | 12 | 41/347 |
| Peroxiredoxin-2 | 1435 | gi|2499469 | 21,779/5.2 | 100 | 4 | 44 | 87/198 |
| Serum amyloid A2 | 1928 | gi|6755394 | 13,623 | 100 | 9 | 48 | 58/122 |
| Hemoglobin-alpha | 1928 | gi|6680175 | 15,085 | 100 | 4 | 31 | 44/142 |
Proteins of interest were excised from 2D-DIGE gels, digested with trypsin and analyzed by LC–MS/MS. Proteins were identified by matching unique spectra in the NCBI database for mus musculus using Scaffold software with dual search engines Mascot and X! Tandem.
Table 3.
MALDI-TOF-MS identification of haptoglobin in plasma of OVM-sensitized mice
| Protein | Spot # | Peptides matched | Coverage (%) | Mascot score |
|---|---|---|---|---|
| Haptoglobin gi|8850219 | 799 | 10 | 22 | 95 |
| 842 | 8 | 20 | 90 | |
| 2103 | 13 | 24 | 124 |
Tryptic digests of proteins spots in immediate proximity to spots 2100 and 1846 were identified as haptoglobin using MALDI-TOF-MS by matching peptide mass fingerprints to the NCBI database for mus musculus, using the Mascot search engine. Mascot scores greater than 63 are considered significant (p < 0.05).
Analysis of liver, spleen, and mesenteric lymph node subjected to two pH ranges of isoelectric focusing, pH 4–7 and pH 3–11, and ileum to only pH 3–11, did not reveal any significant changes in protein expression.
6. Discussion
The objective of this study was to evaluate differential protein expression in plasma and tissues of mice sensitized by gavage to the egg white allergen OVM. Using 2D-DIGE, we have identified three proteins, namely Hp, SAA2, and PrxII, that are overexpressed in plasma of OVM-sensitized mice during OVM-induced anaphylaxis. There was no significant alteration in protein expression in liver, spleen, ileum and mesenteric lymph nodes associated with OVM treatment. These results indicate that increased expression of these specific acute phase and antioxidant proteins may represent important biomarkers for assessing the allergic potential of food in mice.
Acute phase proteins (APP) including Hp and SAA2, are a group of proteins whose expression in plasma varies as a component of the acute phase response immediately following tissue injury. This response is highly non-specific and species-dependent and considered to be one of the earliest markers of specific pathological processes. While individual APPs can have both pro- and anti-inflammatory effects, their ultimate purpose is to restore homeostasis by providing immediate control of inflammation until appropriate defense mechanisms can be up-regulated or initiated. Both Hp and SAA2 are positive APPs that are used as biomarkers of inflammation and infection in both human and veterinary medicine (Saini et al., 1998, Murata et al., 2004).
Hp is a serum glycoprotein synthesized predominantly in the liver in response to proinflammatory mediators but is also expressed in lung, skin, spleen, kidney, and adipocytes (Baumann and Gauldie, 1994, D’Armiento et al., 1997). Hp is commonly known for its role as a scavenger of hemoglobin, which is a proinflammatory product of hemolysis. In addition, Hp is angiogenic (Cid et al., 1993), and strongly anti-inflammatory (Arredouani et al., 2005), through modulation of macrophage function (Baseler and Burrell, 1983) and inhibition of granulocyte chemotaxis and phagocytosis (Rossbacher et al., 1999). Hp receptors have been discovered on monocytes, macrophages, granulocytes, natural killer cells, B and T lymphocytes, and mast cells, which reflects the multifaceted immunosuppressive effect of this APP (Arredouani et al., 2005). While the role of Hp in allergic reactions is unclear, Hp preferentially and strongly inhibits the release of Th2 cytokines in mice thereby promoting a Th1-dominant cellular response (Arredouani et al., 2003). Similarily, Hp−/− mice are more prone to asthma (Arredouani et al., 2005). Therefore, it is possible that BALB/c mice, with their genetic predisposition for Th2-biased responses, may release Hp to attenuate anaphylaxis. Because serum Hp is more easily assessed than highly labile cytokines, it may represent a useful indicator of anaphylaxis associated with exposure to food allergens.
Although PrxII was only increased by 1.9-fold, this change was statistically highly significant p = 0.004, and PrxII can be mechanistically linked to inflammation, hemolysis, and endothelial damage. Peroxiredoxins previously named thioredoxin peroxidases, are intracellular antioxidants which function as scavengers of hydrogen peroxide and alkyl hydroperoxides by utilizing thioredoxin. Six distinct isozymes have been detected in a variety of mammalian cells including erythrocytes, skin, endothelium, and lymphocytes (Rhee et al., 2001). The isozyme PrxII is highly expressed in erythrocytes and is believed to play a major role in protecting erythrocytes from oxidative stress (Lee et al., 2003b). In this study, a possible confounding source of PrxII may be from complement-mediated hemolysis associated with anaphylaxis. However, complement activation was not confirmed in this study and its relationship to anaphylaxis in OVM-sensitized mice is not clear. Other reported functions of PrxII include enhancement of natural killer cell activity (Shau et al., 1994), and inhibition of apoptosis (Zhang et al., 1997). PrxII has been reported to be a novel marker of neoplasia, expressed in endothelial cells of benign and malignant human vascular tumors of the skin (Lee et al., 2003a). PrxII has also been detected in the plasma of patients with severe acute respiratory syndrome (Chen et al., 2004). The role of PrxII in allergy is unclear but the results of the current study suggest that this protein may represent a potential biomarker of anaphylaxis.
SSA2 expression was markedly increased (19-fold) and was included as a potential biomarker in spite of the fact that this change did not quite reach statistical significance (p = 0.06). SSA2 is biologically relevant to this study as it is mechanistically linked to various pathophysiological factors and events (i.e. acute phase proteins, trauma, infection, and inflammation) that can be associated with anaphylaxis. SAA2 is a member of a group of small serum apolipoproteins which are predominantly expressed in the liver in response to various injuries, including trauma, infection, inflammation, and neoplasia (Uhlar and Whitehead, 1999). Extrahepatic expression has been reported in human epithelial and endothelial cells, monocytes and macrophages, cultured smooth muscle, atherosclerotic plaques, synovial tissue from patients with rheumatoid arthritis, and brains of patients with Alzheimer’s disease (Urieli-Shoval et al., 2000). SAA2 is a precursor protein causing amyloidosis, a common condition in most strains of mice but rare in BALB/c mice. In this study SAA2 was detected in combination with lesser amounts of hemoglobin-alpha (Table 2). The presence of hemoglobin in the MS analysis of SAA2 (spot 1928) may represent contamination during spot picking considering SAA2 and hemoglobin differ by approximately 2 kDa.
Of the three plasma proteins detected, Hp, with multiple receptor expression on various cell types and known immune-modulating functions, is likely the best candidate for use as a biomarker of allergic reactions. If serum Hp increases during the sensitization process, then this protein would have tremendous utility as a readily accessible biomarker to qualify and potentially quantify early subclinical allergic reactions. These protein changes, although detected during anaphylaxis, could have been elevated prior to OVM challenge, in response to successive gavages during sensitization. However, we did not assess changes in protein expression during sensitization when proteins were assumed to be highly labile and of low abundance. We felt that analysis at a point immediately post-challenge, when OVM hypersensitivity is highly amplified, would provide the greatest likelihood of detecting significant changes. We are currently investigating the expression of these biomarker proteins in time-course studies in other animal models of food allergy using different allergens and methodologies that are less labor-intensive than 2D-DIGE.
While Hp, SAA2, and PrxII are clearly associated with anaphylaxis in this study, it is unclear whether they are mechanistically linked to anaphylaxis or whether they represent non-specific proteins associated with bystander injury. Further validation of serum levels of Hp, SAA2, and PrxII in other animal models of allergy to other food allergens, and at various time points during both sensitization and anaphylaxis is necessary to evaluate their sensitivity, specificity and predictive value as biomarkers of food allergy.
Acknowledgments
This research was supported by the Ontario Ministry of Agriculture, Food and Rural Affairs (OMAFRA). We thank Li Zhang at the Advanced Protein Technology Center, Hospital for Sick Children, Toronto, Ontario, Canada for his MS analysis. We gratefully thank A. Morrison, J. Bruin, P. Huber, A. MacKay, and A. Maslen, for their technical assistance, and Dr. J. Lumsden, and Dr. P. Shewen for their helpful advice and guidance.
References
- Adel-Patient K., Bernard H., Ah-Leung S., Creminon C., Wal J.M. Peanut- and cow’s milk-specific IgE, Th2 cells and local anaphylactic reaction are induced in BALB/c mice orally sensitized with cholera toxin. Allergy. 2005;60:658–664. doi: 10.1111/j.1398-9995.2005.00767.x. [DOI] [PubMed] [Google Scholar]
- Alban A., David S.O., Bjorkesten L., Andersson C., Sloge E., Lewis S., Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- Arredouani M., Matthijs P., Van Hoeyveld E., Kasran A., Baumann H., Ceuppens J.L., Stevens E. Haptoglobin directly affects T cells and suppresses T helper cell type 2 cytokine release. Immunology. 2003;108:144–151. doi: 10.1046/j.1365-2567.2003.01569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredouani M.S., Kasran A., Vanoirbeek J.A., Berger F.G., Baumann H., Ceuppens J.L. Haptoglobin dampens endotoxin-induced inflammatory effects both in vitro and in vivo. Immunology. 2005;114:263–271. doi: 10.1111/j.1365-2567.2004.02071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseler M.W., Burrell R. Purification of haptoglobin and its effects on lymphocyte and alveolar macrophage responses. Inflammation. 1983;7:387–400. doi: 10.1007/BF00916303. [DOI] [PubMed] [Google Scholar]
- Baumann H., Gauldie J. The acute phase response [see comments] Immunol. Today. 1994;15:74–80. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- Bernhisel-Broadbent J., Dintzis H.M., Dintzis R.Z., Sampson H.A. Allergenicity and antigenicity of chicken egg ovomucoid (Gal d III) compared with ovalbumin (Gal d I) in children with egg allergy and in mice. J. Allergy Clin. Immunol. 1994;93:1047–1059. doi: 10.1016/s0091-6749(94)70054-0. [DOI] [PubMed] [Google Scholar]
- Chen J.H., Chang Y.W., Yao C.W., Chiueh T.S., Huang S.C., Chien K.Y., Chen A., Chang F.Y., Wong C.H., Chen Y.J. Plasma proteome of severe acute respiratory syndrome analyzed by two-dimensional gel electrophoresis and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:17039–17044. doi: 10.1073/pnas.0407992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid M.C., Grant D.S., Hoffman G.S., Auerbach R., Fauci A.S., Kleinman H.K. Identification of haptoglobin as an angiogenic factor in sera from patients with systemic vasculitis. J. Clin. Invest. 1993;91:977–985. doi: 10.1172/JCI116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Armiento J., Dalal S.S., Chada K. Tissue, temporal and inducible expression pattern of haptoglobin in mice. Gene. 1997;195:19–27. doi: 10.1016/s0378-1119(97)00123-6. [DOI] [PubMed] [Google Scholar]
- Dearman R.J., Skinner R.A., Herouet C., Labay K., Debruyne E., Kimber I. Induction of IgE antibody responses by protein allergens: inter-laboratory comparisons. Food Chem. Toxicol. 2003;41:1509–1516. doi: 10.1016/s0278-6915(03)00167-4. [DOI] [PubMed] [Google Scholar]
- Eggesbo M., Botten G., Halvorsen R., Magnus P. The prevalence of allergy to egg: a population-based study in young children. Allergy. 2001;56:403–411. doi: 10.1034/j.1398-9995.2001.056005403.x. [DOI] [PubMed] [Google Scholar]
- Eggesbo M., Botten G., Halvorsen R., Magnus P. The prevalence of CMA/CMPI in young children: the validity of parentally perceived reactions in a population-based study. Allergy. 2001;56:393–402. doi: 10.1034/j.1398-9995.2001.056005393.x. [DOI] [PubMed] [Google Scholar]
- Eigenmann P.A. Are specific immunoglobulin E titres reliable for prediction of food allergy? Clin. Exp. Allergy. 2005;35:247–249. doi: 10.1111/j.1365-2222.2005.02183.x. [DOI] [PubMed] [Google Scholar]
- Fredericq E., Deutsch H.F. Studies on ovomucoid. J. Biol. Chem. 1949;181:499–510. [PubMed] [Google Scholar]
- Jyonouchi H., Sun S., Winship T., Kuchan M.J. Dietary ribonucleotides modulate type 1 and type 2 T-helper cell responses against ovalbumin in young BALB/cJ mice. J. Nutr. 2001;131:1165–1170. doi: 10.1093/jn/131.4.1165. [DOI] [PubMed] [Google Scholar]
- Knippels L.M., Penninks A.H. Recent advances using rodent models for predicting human allergenicity. Toxicol. Appl. Pharmacol. 2005;207:157–160. doi: 10.1016/j.taap.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Kroghsbo S., Christensen H.R., Frokiaer H. Experimental parameters differentially affect the humoral response of the cholera-toxin-based murine model of food allergy. Int. Arch. Allergy Immunol. 2003;131:256–263. doi: 10.1159/000072137. [DOI] [PubMed] [Google Scholar]
- Lee S.C., Na Y.P., Lee J.B. Expression of peroxiredoxin II in vascular tumors of the skin: a novel vascular marker of endothelial cells. J. Am. Acad. Dermatol. 2003;49:487–491. doi: 10.1067/s0190-9622(03)01485-3. [DOI] [PubMed] [Google Scholar]
- Lee T.H., Kim S.U., Yu S.L., Kim S.H. Peroxiredoxin II is essential for sustaining life span of erythrocytes in mice. Blood. 2003;101:5033–5038. doi: 10.1182/blood-2002-08-2548. [DOI] [PubMed] [Google Scholar]
- Li X., Huang C.K., Schofield B.H., Burks A.W., Bannon G.A., Kim K.H., Huang S.K., Sampson H.A. Strain-dependent induction of allergic sensitization caused by peanut allergen DNA immunization in mice. J. Immunol. 1999;162:3045–3052. [PubMed] [Google Scholar]
- Murata H., Shimada N., Yoshioka M. Current research on acute phase proteins in veterinary diagnosis: an overview. Vet. J. 2004;168:28–40. doi: 10.1016/S1090-0233(03)00119-9. [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Kang S.W., Chang T.S., Jeong W., Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- Rossbacher J., Wagner L., Pasternack M.S. Inhibitory effect of haptoglobin on granulocyte chemotaxis, phagocytosis and bactericidal activity. Scand. J. Immunol. 1999;50:399–404. doi: 10.1046/j.1365-3083.1999.00609.x. [DOI] [PubMed] [Google Scholar]
- Rupa P., Mine Y. Engineered recombinant ovomucoid third domain can desensitize BALB/c mice of egg allergy. Allergy. 2006;61:836–842. doi: 10.1111/j.1398-9995.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Saini P.K., Riaz M., Webert D.W., Eckersall P.D., Young C.R., Stanker L.H., Chakrabarti E., Judkins J.C. Development of a simple enzyme immunoassay for blood haptoglobin concentration in cattle and its application in improving food safety. Am. J. Vet. Res. 1998;59:1101–1107. [PubMed] [Google Scholar]
- Sampson H.A. Anaphylaxis and emergency treatment. Pediatrics. 2003;111:1601–1608. [PubMed] [Google Scholar]
- Sampson H.A. Food allergy – accurately identifying clinical reactivity. Allergy. 2005;60(Suppl. 79):19–24. doi: 10.1111/j.1398-9995.2005.00853.x. [DOI] [PubMed] [Google Scholar]
- Shau H., Butterfield L.H., Chiu R., Kim A. Cloning and sequence analysis of candidate human natural killer-enhancing factor genes. Immunogenetics. 1994;40:129–134. doi: 10.1007/BF00188176. [DOI] [PubMed] [Google Scholar]
- Shek L.P., Soderstrom L., Ahlstedt S., Beyer K., Sampson H.A. Determination of food specific IgE levels over time can predict the development of tolerance in cow’s milk and hen’s egg allergy. J. Allergy Clin. Immunol. 2004;114:387–391. doi: 10.1016/j.jaci.2004.04.032. [DOI] [PubMed] [Google Scholar]
- Sicherer S.H., Munoz-Furlong A., Sampson H.A. Prevalence of seafood allergy in the United States determined by a random telephone survey. J. Allergy Clin. Immunol. 2004;114:159–165. doi: 10.1016/j.jaci.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Uhlar C.M., Whitehead A.S. Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- Urieli-Shoval S., Linke R.P., Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr. Opin. Hematol. 2000;7:64–69. doi: 10.1097/00062752-200001000-00012. [DOI] [PubMed] [Google Scholar]
- Venter C., Pereira B., Grundy J., Clayton C.B., Roberts G., Higgins B., Dean T. Incidence of parentally reported and clinically diagnosed food hypersensitivity in the first year of life. J. Allergy Clin. Immunol. 2006;117:1118–1124. doi: 10.1016/j.jaci.2005.12.1352. [DOI] [PubMed] [Google Scholar]
- Zhang P., Liu B., Kang S.W., Seo M.S., Rhee S.G., Obeid L.M. Thioredoxin peroxidase is a novel inhibitor of apoptosis with a mechanism distinct from that of Bcl-2. J. Biol. Chem. 1997;272:30615–30618. doi: 10.1074/jbc.272.49.30615. [DOI] [PubMed] [Google Scholar]


