Abstract
Mitochondria have a central role in regulating a range of cellular activities and host responses upon bacterial infection. Multiple pathogens affect mitochondria dynamics and functions to influence their intracellular survival or evade host immunity. On the other side, major host responses elicited against infections are directly dependent on mitochondrial functions, thus placing mitochondria centrally in maintaining homeostasis upon infection. In this review, we summarize how different bacteria and viruses impact morphological and functional changes in host mitochondria and how this manipulation can influence microbial pathogenesis as well as the host cell metabolism and immune responses.
Keywords: mitochondrial morphology, mitochondrial metabolism, bacteria, viruses, cell death, innate immunity
Highlights
Bacteria and viruses have evolved specific ways of targeting mitochondria to perturb mitochondrial function that can prove to be beneficial for these microbes.
Many bacteria and viruses use specific virulence mechanisms to modulate mitochondrial dynamics, leading to either mitochondrial fusion or fission.
Mitochondrial metabolism can also be impacted by bacterial and viral infections.
While in some cases bacteria and viruses induce the mitochondrial cell death pathway, in others cell death is inhibited promoting intracellular bacterial and viral proliferation.
Mitochondria regulate different innate immune signaling pathways induced upon bacterial or viral infections.
Mitochondrial Morphology
Mitochondria are important organelles in the cell involved in a plethora of different functions, from energy production and fatty acid oxidation to regulating cell cycle and cell death. Mitochondria originated from an endosymbiotic relationship between bacteria and ancestral eukaryotic cells billions of years ago [1]. During evolution, mitochondria evolved to perform significant functions fostering the endosymbiotic relationship within the eukaryotic cells while still retaining their own DNA (mtDNA), and transcription and translation machineries. mtDNA encodes only 13 proteins and ~99% of mitochondrial proteins are nuclear encoded.
Mitochondrial networks are dynamic in nature, undergoing cycles of fission and fusion that are mediated by a dedicated set of dynamin-related GTPases. Mitofusin 1 and 2 (MFN1 and MFN2) and optic atrophy 1 (OPA1) coordinate their functions to bring about mitochondrial fusion. Mitochondrial fusion essentially involves fusion of the outer mitochondrial membrane (OMM) and the inner mitochondrial membrane (IMM). MFN1 and MFN2, which are OMM proteins, interact with each other in a homo- and heterotypic manner involving GTP hydrolysis, which eventually leads to the fusion of the OMM. Fusion of the IMM is mediated by OPA1, which is a GTPase-protein anchored in the IMM. OPA1 is proteolytically cleaved into different fragments, including a long (L) form and a short (S) form, in the intermembrane space (IMS) of the mitochondria by two membrane-bound metalloproteases OMA1 and YME1L. L-OPA1 has been associated with mitochondrial fusion [2,3]. Loss of these mitochondrial fusion-mediating proteins (MFN1, MFN2, and OPA1) leads to mitochondrial fragmentation [4,5]. Mitochondrial fusion is an essential cellular process that helps to merge fragments of mitochondria, mediating the exchange of mtDNA, proteins, and metabolites [6]. Mitochondria can also break their network down into smaller fragments in a process termed ‘mitochondrial fission’. The most critical factor regulating this process is the cytosolic GTPase-protein dynamin-related protein 1 (DRP1), also called dynamin-1-like protein (DNM1L). Under conditions of mitochondrial fragmentation, DRP1 is recruited from the cytosol to the OMM, where it forms oligomeric structures wrapping around the mitochondrial scission site and, eventually through GTP hydrolysis, undergoes a conformational change that results in membrane constriction and scission [7]. On the OMM, DRP1 interacts with mitochondrial fission factor (MFF) and mitochondrial dynamics proteins (MiD49 and MiD51), which serve as receptors for its recruitment [8,9]. Consistent with their role in mitochondrial fission, loss of MFF, MiD49, and MiD50 leads to mitochondrial hyperfusion and defected DRP1 recruitment to the mitochondria [10., 11., 12.]. There is no known regulator of IMM fission, and it is not understood how the IMM and OMM coordinate in the process of fission.
Mitochondrial fission is required for removing damaged parts of mitochondria, which get eventually cleared by mitophagy. Mitochondrial fission also has a critical role in replicating mitochondria during cell cycle. There are also specific sites on mitochondria that serve as designated sites for mitochondrial fission. These sites are often where the tubules of endoplasmic reticulum (ER) physically contact mitochondria. These ER–mitochondria contact sites induce a constriction, where DRP1 is eventually recruited [13]. Furthermore, the receptors of DRP1, MiD49, and MiD51 are also present at the ER–mitochondria contact sites, suggesting that these sites are involved in the recruitment of DRP1 and subsequent mitochondrial fission [13,14]. It is not clearly understood how much of mitochondrial fission is driven by ER and whether the role of ER in mediating mitochondrial fission depends on specific stimuli or if it is a general phenomenon under all conditions that promote mitochondrial fission.
Given the central role of mitochondria in regulating a range of cellular activities, their role in regulating host response to an infection is plausible. Multiple studies have reported the subversion of mitochondrial functions upon microbial infections. Different pathogens have evolved ways of targeting the mitochondria to influence their intracellular survival, dissemination by mediating mitochondria-induced cell death, or evading host immunity. Research into the microbial strategies of impacting mitochondria has yielded a wealth of knowledge about their pathogenesis mechanisms. Additionally, pathogens could serve as essential tools in understanding the missing links in mitochondrial dynamics and function. Historically, studying bacterial internalization and their intracellular life-cycles elucidated mechanistic details of endocytic trafficking and vesicular biology. It will be fascinating to understand using similar strategies mitochondrial fission–fusion cycles and the key players involved in regulating the processes that are as yet unknown. In this review, we explore how bacteria and viruses manipulate the mitochondria and how this manipulation can influence microbial pathogenesis. Furthermore, we discuss how mitochondria influence the host immune response against infections and other stimuli.
Modulation of Mitochondrial Dynamics upon Infection
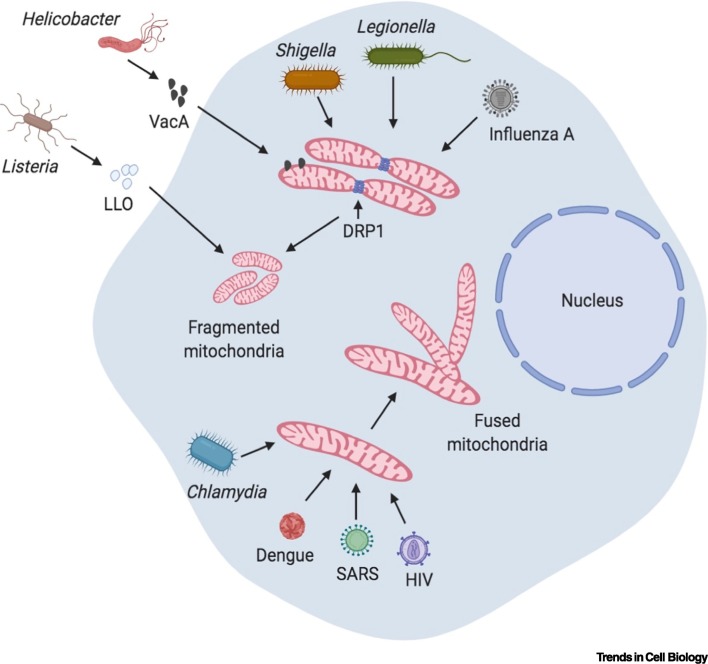
Several bacteria have developed strategies to subvert mitochondrial dynamics to benefit their intracellular niche. An effective way of subverting the mitochondrial function can be beneficial for intracellular bacteria since mitochondria can produce reactive oxygen species (ROS), which can be detrimental for the survival and proliferation of intracellular bacteria. A common theme among bacteria that target mitochondrial dynamics is that they form a niche to proliferate inside host cells. However, there are clear exceptions to this rule, as exemplified by Helicobacter pylori, an extracellular bacterium that targets mitochondria (discussed later). Moreover, most of the bacteria targeting mitochondrial morphology generally cause fragmentation of the mitochondria, thereby perturbing their function significantly. An interesting example is the intracellular Gram-positive bacterium Listeria monocytogenes. Infection with L. monocytogenes leads to rapid mitochondrial fission, causing fragmentation of the mitochondrial network, with effects that are dependent on its pore-forming toxin listeriolysin O [15]. Perturbation of mitochondrial dynamics by knocking down components of the fusion (MFN1 and MFN2) and fission (DRP1) machineries impacts the intracellular survival of L. monocytogenes, suggesting that the bacterium actively deploys listeriolysin O to fragment mitochondria for its sustenance inside the cell [15]. Interestingly, mitochondrial fission caused by L. monocytogenes is independent of DRP1 [16], pointing towards other mechanisms of mitochondrial fission that are as yet unknown. Similar to Listeria, H. pylori also targets mitochondria via its secreted vacuolating cytotoxin VacA, which localizes to mitochondria [17]. VacA induces fragmentation of the mitochondrial network and subsequent release of cytochrome C, which are both dependent on the activity of DRP1 [18]. DRP1-dependent mitochondrial fragmentation is also observed in Shigella flexneri infections [19]. Knockdown of the DRP1 gene by small interfering (si)RNA reverses mitochondrial fragmentation, and reduces the infectious foci counts and the plaque size, suggesting a decrease in cell-to-cell spreading of bacteria [19]. Furthermore, Legionella pneumophila secretes an effector called MitF through its type IV secretion system, which induces DRP1-dependent mitochondrial fragmentation in macrophages [20]. Blocking mitochondrial fragmentation leads to a decrease in intracellular replication of L. pneumophila [20]. By contrast, Chlamydia trachomatis preserves the elongated mitochondrial network for its intracellular proliferation. C. trachomatis upregulates a host miRNA (miR-30c-5p), which is key in maintaining the mitochondrial structure and intracellular proliferation of the bacteria [21]. While during the early infection phase, C. trachomatis induces mitochondrial elongation, it does resort to enhancing mitochondrial fragmentation during the late phases of infection [22]. Mitochondrial elongation is associated with enhanced respiratory activity and increased ATP production, which in turn promotes bacterial proliferation, thus making mitochondria a critical focal point in the intracellular life-cycle of C. trachomatis [22] (Figure 1 ).
Figure 1.
Modulation of Mitochondrial Dynamics upon Infection.
Bacteria and viruses induce changes in mitochondrial morphology. Listeria secretes Listeriolysin O (LLO), a pore-forming toxin that induces an unconventional form of mitochondrial fragmentation that is independent of dynamin-related protein 1 (DRP1). Vacuolating cytotoxin VacA from Helicobacter localizes to mitochondria and induces mitochondrial fragmentation, the release of cytochrome C into the cytosol, and subsequent cell death. Shigella and Legionella also induce mitochondrial fragmentation in a DRP1-dependent manner. By contrast, Chlamydia infection leads to mitochondrial fusion, which is required for its intracellular proliferation. Viruses can also modulate mitochondrial dynamics. Influenza A leads to the dissipation of mitochondrial membrane potential, which ultimately causes mitochondrial fragmentation. Dengue virus inhibits DRP1 and induces mitochondrial fragmentation, which is necessary for its replication and immune evasion. Similarly, severe acute respiratory syndrome (SARS) virus and HIV cause DRP1 degradation, thereby bringing about mitochondrial fragmentation. Created with BioRender (www.BioRender.com).
All these observations invoke an intriguing question: given the bacterial origin of mitochondria, do these bacteria use similar mechanisms to attack other bacterial populations to maintain their own niche? Bacteria secrete soluble factors, including microcins and bacteriocins, to inhibit the growth of other competing bacteria. In addition, mechanisms such as contact-dependent inhibition (CDI) are in place where certain bacteria use type V or VI secretion systems to restrict the growth of other bacteria by direct contact [23]. It will be exciting to determine whether bacteria use similar mechanisms to interact with mitochondria and if this arm of defense against bacterial competition serves a dual role.
Similar to bacteria, many viruses also target mitochondrial functions to establish a proliferative niche for themselves and subsequently disseminate by killing the cells. Influenza A viral protein PB1-F2, which is a key virulence factor for the viral infection, targets mitochondria, leading to mitochondrial fragmentation due to the loss of mitochondrial membrane potential [24]. By contrast, dengue viruses inhibit DRP1, thus inducing mitochondrial fusion and elongation of the mitochondrial network via a nonstructural protein NS4B. Mitochondrial fusion is required for intracellular proliferation of the virus and evasion of the antiviral innate immune signaling [25,26]. Moreover, ORF-9b, a virulence factor of severe acute respiratory syndrome coronavirus (SARS-CoV), induces proteasomal degradation of DRP1, thereby leading to mitochondrial fusion, which eventually limits host cell interferon (IFN) responses against the virus [27]. Similarly, the HIV envelope protein gp120 causes a reduction in DRP1 levels, leading to mitochondrial hyperfusion [28]. These studies and other research [29] have clearly established a role of mitochondrial dynamics and their interplay with antiviral immunity upon a viral invasion. Since many of the viral proteins that target mitochondria are essential for viral replication, they might serve as critical drug targets for generating therapeutics against viral infections.
Modulation of Mitochondrial Metabolism upon Infection
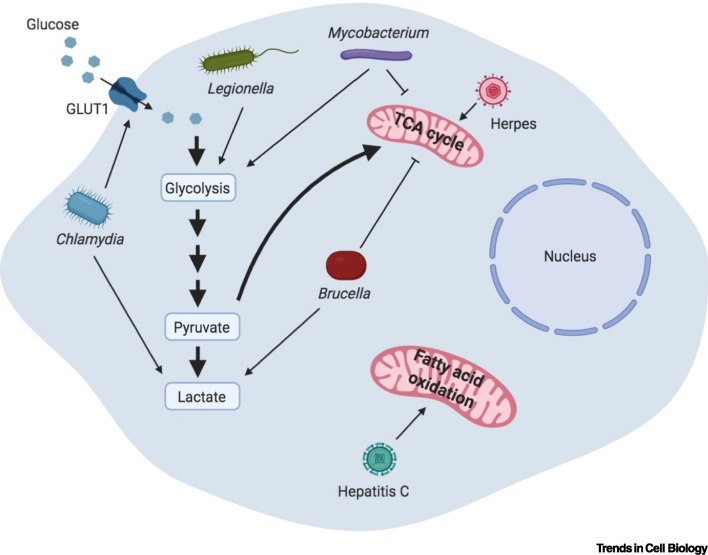
The mitochondrion is the central organelle that regulates the metabolism of macromolecules, including carbohydrates, amino acids, and fatty acids. Glucose, which is the major source of energy, is converted to pyruvate in the cytoplasm via glycolysis. Under normal conditions in most cells, pyruvate is shuttled into mitochondria, where it is oxidized via the tricarboxylic acid (TCA) cycle, eventually generating ATP through the electron transport chain of the mitochondria in a process called oxidative phosphorylation (OXPHOS). OXPHOS is an efficient process; complete oxidation of a single molecule of glucose generates 36 molecules of ATP. Even though OXPHOS is energetically efficient, it is metabolically slow. Rapidly dividing cells (e.g., cancer cells or activated immune cells) that have a high metabolism need quick energy production to maintain their activity. These cells resort to an alternative form of ATP production, termed aerobic glycolysis (also called the Warburg effect), in which pyruvate generated from glycolysis is converted to lactate in the cytoplasm, generating two ATP molecules for every glucose molecule (Figure 2 ).
Figure 2.
Modulation of Mitochondrial Metabolism upon Infection.
Many bacteria and viruses hijack cellular metabolism for their own benefit. Mycobacterium is the best-known bacterium that influences host metabolism by enhancing aerobic glycolysis. Epithelial cells and immune cells infected with Mycobacterium exhibit a reduction in the tricarboxylic acid (TCA) cycle and a corresponding increase in the glycolytic flux. Elevated aerobic glycolysis is also observed in Mycobacterium-infected lung granulomas in animal models of infection and in patients with active tuberculosis. Legionella also promotes glycolytic flux similar to Mycobacterium, thus enhancing aerobic glycolysis. Brucella infection promotes lactate production via aerobic glycolysis, while suppressing the TCA cycle. Chlamydia enhances the levels of the glucose transporter GLUT1, leading to augmented glucose uptake and increased glycolytic flux, which is necessary for its intracellular growth. Viruses also perturb metabolic pathways for their replication and subsequent dissemination. Herpes virus induces the TCA cycle, thereby promoting mitochondrial respiration, while Hepatitis C perturbs mitochondrial fatty acid oxidation. Created with BioRender (www.BioRender.com).
Many bacteria are known to modulate cellular metabolism. They can actively remodel the host cell metabolism to enhance their intracellular survival or the remodeling can be a result of the host response towards bacteria to ramp up the immune response. In either case, the end result is generally a slowdown of the TCA cycle and an induction of aerobic glycolysis. One of the best-known bacteria that perturbs metabolism is Mycobacterium tuberculosis. Macrophages and T cells infected with M. tuberculosis exhibit increased levels of glycolytic enzymes and transporters for glucose uptake and reduced levels of TCA cycle and OXPHOS enzymes, suggesting that M. tuberculosis infection induces a metabolic state similar to the Warburg effect [30]. Human alveolar macrophages also display a similar shift towards aerobic glycolysis, which is essential to curb the intracellular growth of bacteria. Inhibition of this metabolic switch leads to reduced production of proinflammatory cytokine IL-1β and enhanced growth of intracellular bacteria [31]. Similarly a switch towards aerobic glycolysis was also observed in a rabbit model of active tuberculosis (TB), in M. tuberculosis-infected lung granulomas of guinea pigs, and in lung granulomas from patients with TB [32., 33., 34.]. M. tuberculosis also enhances the levels of aerobic glycolysis in human peripheral blood mononuclear cells (PBMCs) dependent on Toll-Like Receptor 2 (TLR2) and the AKT-mTOR pathway [35]. Another Mycobacterium species, Mycobacterium avium, also induces aerobic glycolysis in infected cells, which is dependent on the presence of IFN-γ [36]. Additionally, Brucella abortus, an intracellular bacterium causing chronic human and live-stock disease, can alter host cell metabolism to benefit its own growth and proliferation in cells. The metabolic shift induced by B. abortus is characterized by reduced TCA metabolism, reduced amino acid consumption, altered mitochondrial localization, and augmented lactate production [37]. Chemical inhibition of glycolysis and lactate production attenuates the intracellular survival of B. abortus [37]. Furthermore, infection with C. trachomatis also rewires cellular metabolism. C. trachomatis infection causes upregulation of the glucose transporter Glut1 and downregulation of TIGAR, which is an inhibitor of fructose-2,6-bisphosphate, thereby altering glycolysis [38]. These findings are in line with C. trachomatis being an obligate intracellular bacterium the growth of which depends on the uptake of glucose from the host cells [39]. Similarly, Chlamydia psittaci induces metabolic perturbations and alterations of mitochondrial function characterized by increased glutamate and lactate production and accumulation of glycogen due to higher consumption of glucose by infected cells [40]. Macrophages infected with L. pneumophila also exhibit an abrupt halt of mitochondrial respiration and an increase in glycolysis, thus tipping the balance towards aerobic respiration, which is required for intracellular bacterial replication [20]. Thus, it is now clear that bacteria influence host metabolism. It will be interesting to further explore what kind of effectors and molecular mechanisms bacteria use to hijack the mitochondrial metabolism. To obtain a complete picture, it will also be important to address the outstanding mechanistic questions as to how sensing of certain bacteria, such as M. tuberculosis, leads the host cell to remodel its cellular metabolism to restrict bacterial proliferation.
Viruses also tweak mitochondrial metabolism to maintain a suitable replication niche. Interestingly, viruses can induce different effects on the host metabolism, which specifically depend of the type of virus. For example, two related herpes viruses [human cytomegalovirus (HCMV) and herpes simplex virus type-1 (HSV-1)] induce different effects on host metabolism. While HCMV enhances glycolytic flux, HSV-1 leads to induction of the TCA cycle [41]. HCMV appears to directly impact metabolism by elevating mitochondrial biogenesis, which is accompanied by increased respiration, both of which are required for HCMV replication [42]. Measles virus also enhances mitochondrial functions characterized by reduced ROS levels and enhanced mitochondrial membrane potential in infected cells, and this perturbation is required for persistent infection [43]. Hepatitis C virus (HCV) infection enhances mitochondrial fatty acid oxidation [44] and, concordantly, pharmacological inhibition of the transport of long-chain fatty acids into mitochondria restricts viral replication [45]. From numerous studies, it has become apparent that different viruses target different nodes of mitochondrial metabolism, including β-oxidation and the TCA cycle. However, we are still far from a clear understanding as to what the mechanisms of mitochondrial targeting are and why there exists such a diversity of metabolic perturbations caused by different viruses.
Modulation of Mitochondria-Induced Cell Death upon Infection
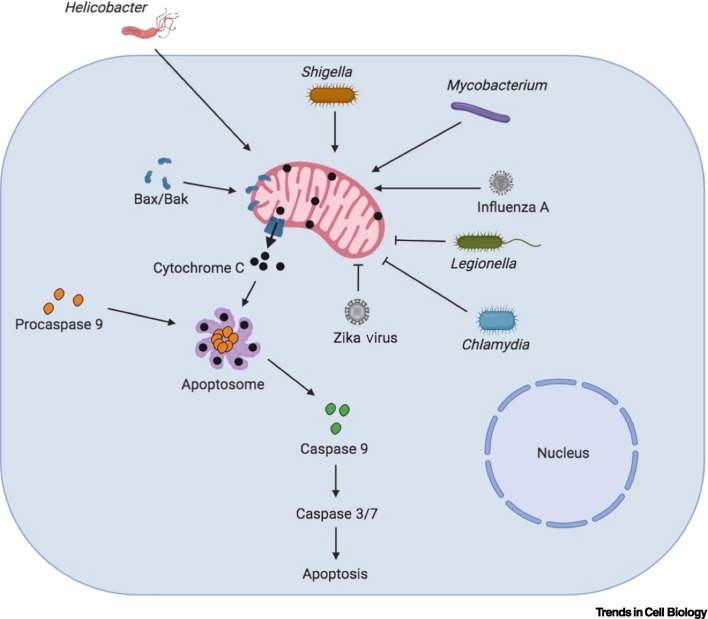
Mitochondria directly impact cell death via the intrinsic apoptosis pathway. The major players regulating cell death through this pathway belong to the B cell lymphoma 2 (Bcl-2) protein family. Some of these proteins are pro- while others are antiapoptotic in function. Major factors carrying out apoptosis include Bcl-2-associated X protein (Bax) and Bcl-2 homologous antagonist/killer (Bak), which, upon activation, localize to the OMM and induce OMM permeabilization (MOMP) [46]. MOMP leads to the release of proapoptotic factors, including cytochrome c and second mitochondrion-derived activator of caspases (SMAC/DIABLO), from the mitochondria into the cytoplasm. Cytochrome c interacts with the cytosolic protein apoptotic protease activating factor 1 (APAF1) in the cytoplasm and forms a specialized protein complex, called the apoptosome. The apoptosome cleaves procaspase-9 into active caspase-9, which subsequently cleaves and activates the executioner caspases 3/6/7, resulting in cell apoptosis [46].
Many pathogenic bacteria hijack this pathway to influence their own survival by either promoting cell death to obtain nutrients and disseminate further or suppressing cell death to enable their proliferation inside the infected cells. For example, Shigella flexneri induces the loss of mitochondrial membrane potential, leading to disruption of mitochondrial function that causes cell death in macrophages [47] and non-myeloid cells [48]. Blocking the damage to mitochondria is sufficient to reverse the cell death phenotype, suggesting that S. flexneri actively uses strategies to perturb mitochondrial function [48]. Macrophages infected with M. tuberculosis also exhibit dissipation of mitochondrial membrane potential and a MOMP phenotype [49]. Virulent M. tuberculosis induces the translocation of Bax to mitochondria, leading to MOMP, the release of cytochrome-c in the cytoplasm, and eventually cell death [49]. In addition, a virulence factor called SipB produced by Salmonella enterica and delivered via the type III secretion system (T3SS) targets mitochondria in macrophages and induces cell death [50]. Similarly, H. pylori vacuolating toxin VacA activates DRP1 and induces its localization to mitochondria, leading to mitochondrial fragmentation, MOMP, activation of Bax, and release of cytochrome c into the cytoplasm. All these effects are rescued by inhibition of mitochondrial fragmentation, suggesting that H. pylori actively induces disruption of the mitochondria that leads to cell death [18]. Additionally, Pseudomonas aeruginosa secretes an effector, called ExoT, through its T3SS, which activates the mitochondrial intrinsic pathway of cell death by inducing higher levels of the proapoptotic proteins Bax, Bid, and Bim, loss of mitochondrial membrane potential, and release of cytochrome c [51].
There are also several bacteria that inhibit cell death instead of promoting it. This is seen as a strategy to protect their niche inside the cells where they replicate. For example, L. pneumophila secretes an effector called SidF through its T4SS, which inhibits apoptosis by suppressing the effects of the Bcl2 family proapoptotic proteins BNIP3 and Bcl-rambo [52]. SdhA is another L. pneumophila T4SS effector, which prevents apoptosis of the infected macrophages. Macrophages infected with the L. pneumophila sdhA mutant display enhanced cell death, thereby hindering the intracellular proliferation of the bacteria [53]. Coxiella burnetii uses an effector, named CaeB, to limit MOMP, thereby inhibiting the intrinsic apoptosis pathway [54]. CaeB expression in HEK293 cells leads to its localization to mitochondria, suggesting that this effector manipulates mitochondrial physiology, which then modulates cell death [54]. Additionally, C. trachomatis and Chlamydia pneumoniae induce proteasomal degradation of the proapoptotic proteins Bim, Puma, and Bad without affecting the transcript levels [55,56]. Degradation of these proteins eventually leads to suppression of cytochrome c release into the cytosol and inhibition of cell death, which assists the proliferation of the bacteria in the cells. A C. trachomatis effector called CPAF is responsible for inhibiting cell death by promoting the degradation of proapoptotic proteins [57]. While there are many different pathways that induce the demise of a cell, the mitochondrial pathway is widely targeted by bacteria to manipulate the host. It requires further investigation to understand whether there is crosstalk between cell death pathways when cells are challenged with a bacterial infection or whether bacteria also use similar effectors and mechanisms to target other forms of cell death. Bacterial modulation of the mitochondrial cell death pathway appears logical in terms of keeping the immune system silent, since it triggers apoptosis, which is an anti-inflammatory form of cell death (Figure 3 ).
Figure 3.
Modulation of Mitochondrial Apoptosis Pathway upon Infection.
B cell lymphoma 2 (Bcl-2) family proteins, including include Bcl-2-associated X protein (Bax) and Bcl-2 homologous antagonist/killer (Bak), regulate the mitochondrial cell death pathway. Bax and Bak are proapoptotic proteins that localize to mitochondria and induce outer mitochondrial membrane permeabilization (MOMP), leading to the release of proapoptotic factors, such as cytochrome C, into the cytosol, which induce cell death by activating Caspases 9, 3, and 7 via the apoptosome complex. Bacteria and viruses influence this pathway and modulate the host response. Helicobacter, Shigella, and Mycobacterium infections lead to mitochondrial disruption that stimulates the mitochondrial apoptotic machinery and causes cell death. By contrast, bacteria such as Chlamydia and Legionella block the mitochondrial cell death pathway to promote their intracellular proliferation. Different viruses also exert varied effects on the mitochondrial cell death pathway. Influenza A enhances cell death, which helps in its dissemination, while Zika virus blocks cell death. Created with BioRender (www.BioRender.com).
Since many pathogenic bacteria modulate the mitochondrial cell death pathway, it is plausible that bacteria induce damage to the epithelial barrier by inducing cell death, thereby disseminating to other organs and ultimately leading to sepsis. Indeed, perturbed mitochondrial morphology, a dysfunctional mitochondrial electron transport chain, and enhanced oxidative stress have been reported in sepsis [58]. However, specific strategies to restore mitochondrial homeostasis in sepsis have not yielded promising results in humans [59], even though results from animal experiments were encouraging [60]. This suggests that bacteria use multiple strategies to disseminate and evade the immune system, of which mitochondrial targeting appears to be only one.
Similar to bacteria, viruses can also modulate cell death for their own replication and dissemination. For example, Influenza A utilizes the mitochondrial cell death pathway by targeting its virulence factor PB1-F2 to the IMM space, where it disrupts mitochondrial organization, thereby inducing cell death [61]. Similarly, an HIV protein, called the viral protein R, translocates to OMM upon viral infection, inhibits Mfn2, and disrupts mitochondrial membrane potential, leading ultimately to cell death induction [62,63]. Interestingly, in contrast to HIV and Influenza A, some viruses, including Dengue [64], Zika [65], and Chikungunya [66] viruses, promote autophagy in the host, which inhibits cell death and promotes viral replication and dissemination. Similarly, measles virus enhances mitophagy in host cells, which limits apoptosis, but induces necrotic cell death in the later stages due to reduced levels of ATP [67]. Studies of viral infections have highlighted the involvement of autophagy and its interplay in different forms of cell death. These interesting studies have also revealed how certain aspects of bacterial infections match viral strategies, while others differ, in the context of modulating cell death.
Mitochondria and Innate Immunity
Multiple studies have established a crosstalk between signaling via the innate immune receptors and the mitochondria. Innate immune receptors, also known as pathogen recognition receptors (PRRs), are either cytosolic or membrane bound. The most-studied PRRs include the membrane bound TLRs and the cytosolic nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs). Ligand-activated TLRs or NLRs lead to the induction of downstream signaling cascades, causing the activation of major immune-regulatory transcription factors, such as AP-1 and NF-κB, ultimately leading to the production of cytokines and chemokines. Here, we describe the role of mitochondria in regulating innate immune responses.
Mitochondria and TLR Signaling upon Infection
TLRs are highly conserved transmembrane receptors that bind to their ligands, called microbe-associated molecular patterns (MAMPs) or danger-associated molecular patterns (DAMPs). Ligand binding leads to activation of TLRs and downstream signaling, resulting in a variety of cellular responses, including the activation and nuclear localization of the transcription factor NF-κB and the subsequent production of proinflammatory cytokines and interferons. Ten human (TLR1–10) and 12 mouse TLRs (TLR1–9, TLR11, 12, and 13) have been identified and characterized so far. In humans, TLR1, 2, 4, 5, 6, and 10 are localized to the plasma membrane and TLRs 3, 7, 8, and 9 are localized intracellularly in the endosomal membrane. While TLRs are activated significantly by bacterial or viral components, several studies have highlighted the role of mitochondrial components in modulating TLR signaling. mtDNA is similar to bacterial DNA in that both share hypomethylated CpG motifs that activate TLR signaling. Purified human and murine mtDNA injected intra-articularly led to arthritis in mice, while this effect was not observed with nuclear DNA, revealing the inflammatory nature of mtDNA [68]. In addition to mitochondrial components being proinflammatory, mitochondria themselves regulate TLR signaling in response to MAMPs. Activation of TLRs 1, 2, and 4 leads to the recruitment of mitochondria to macrophage phagosome and enhances mitochondrial ROS production. After TLR activation, the TLR adaptor protein TRAF6 interacts with, and ubiquitinates, a mitochondrial respiratory chain assembly factor, ECSIT. Ubiquitination of ECSIT leads to enhanced mitochondrial ROS production, which augments intracellular bacterial killing [69]. ECSIT interacts with TRAF6 and the serine/threonine kinase TAK1 upon lipopolysaccharide (LPS) stimulation, which activates TLR4 signaling. These protein interactions are deemed necessary for NF-κB activation upon LPS stimulation [70]. Activation of TLR signaling upon viral infections is also modulated by mitochondria. The OMM protein MARCH5 positively regulates TLR7 signaling by poly-ubiquitinating TANK (a TRAF-binding protein) and impairing its ability to bind to TRAF6 [71]. Additionally, the transcription factor NFAT1 is known to translocate into mitochondria in murine microglia with LPS stimulation, inhibition of which leads to reduced levels of cytokines and suppression in ROS production, suggesting a link with mitochondrial regulation of TLR signaling in microglia [72]. Bacterial infection also upregulates mitochondrial biogenesis through the upregulation of the PGC family of transcriptional co-activators [73., 74., 75.]. This induction of mitochondrial biogenesis is also dependent on TLR2 and TLR4, signifying that the innate immune function feeds into the regulation of mitochondrial function [73]. Moreover, it was recently reported that methicillin-resistant Staphylococcus aureus (MRSA) induces the generation of mitochondria-derived vesicles that contain ROS and that help in the clearance of intracellular bacteria dependent on TLR signaling [76]. Even though there is substantial evidence for the involvement of mitochondria in TLR signaling, there are significant gaps that need to be addressed, in particular how mitochondria sense activation of TLRs and how they play into regulating downstream signaling. It will also be of interest to determine whether different TLRs follow similar mechanisms of mitochondrial involvement and whether the loss of mitochondrial functionality completely dampens TLR responses phenotypically. Ongoing and future work will address these conundrums.
Mitochondria as an Activation Platform for NLR Signaling
In addition to the membrane-bound TLRs, PRRs exist in the cytosol that detect cytosolic PAMPs and DAMPs. The most widely studied cytosolic PRRs are the NLRs. NLRs activate inflammasomes, which are multiprotein complexes that act as platforms for the activation of proinflammatory caspase-1. Active caspase-1 then proteolytically cleaves pro-IL-1β, pro-IL-18, and pro-IL-33 to produce mature cytokines, which are secreted out of the cell [77,78]. Caspase-1 also cleaves gasdermin D, which shuttles to the cell membrane, where it causes pore formation that ultimately leads to pyroptosis, an inflammatory form of cell death [77,78]. Some inflammasomes, such as NLRP3 and NLRC4, are well studied, while the mechanistic and functional details of others, such as NLRP6, NLRP7, and NLRP12, remain lacking. Functional links between mitochondria and the NLR signaling have been reported, as described later.
One of the first seminal studies describing the link between mitochondrial function and NLR signaling reported that the NLRP3 inflammasome and its adaptor protein ASC localize to the mitochondria upon activation [79]. The study revealed that a block in mitophagy led to an accumulation of damaged ROS-producing mitochondria, which induced the activation of the NLRP3 inflammasome [79,80]. Subsequently, the mitochondrial-associated adaptor protein MAVS was identified as an interaction partner important for the association of NLRP3 with mitochondria [81,82]. MAVS also promoted NLRP3-induced production of mature IL-1β, revealing an important role of mitochondria in mediating NLRP3 function [81,82]. Another mitochondrial protein, MAPL (also known as MUL1), localizes to the OMM and regulates NLRP3 activity. SUMOylation of NLRP3 by MAPL results in suppression of its activity [83]. Furthermore ,mtDNA [84,85] and the mitochondrial lipid cardioliopin [86] act as potent activators of the NLRP3 inflammasome. By contrast, NLRP3 inducers also bring about aberrant perturbation of mitochondrial homeostasis by diminishing the concentration of NAD+, which consequently leads to accumulation of acetylated α-tubulin and subsequent microtubule remodeling, which is also necessary for NLRP3 localization to mitochondria [87]. Moreover, another member of the NLR family, NLRX1, interacts with MAVS in the OMM, and this interaction downregulates IFN-β production upon viral infection [88]. Loss of NLRX1 promotes virus-induced type I IFN production and decreases viral replication [88]. Overactivation of NLRX1 causes enhanced ROS production [89., 90., 91.]. NLRX1 has also been reported to target the mitochondrial matrix and interact with UQCRC2, a matrix protein of the respiratory chain complex III. This interaction is required for ROS production [90]. Besides ROS production, NLRX1 was recently shown to function as a mitophagy receptor in L. monocytogenes infection [92]. Additionally, activation of the NLRC4 inflammasome has been associated with mitochondrial damage. P. aeruginosa infection leads to increased production of ROS and release of mtDNA into the cytoplasm, which activates the NLRC4 inflammasome [93]. Thus, it will be interesting to determine how the less understood inflammasomes, such as NLRP6, NLRP7, and NLRP12, react upon mitochondrial perturbations and whether conditions that activate these inflammasomes also influence mitochondrial physiology.
Mitochondria and Other Innate Immune Pathways
Mitochondria also have an essential role in regulating the function of other innate immune receptors. The mitochondrial protein MAVS influences the function of retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), which are specialized PRRs involved in the recognition of viruses by the innate immune system [94]. The RLR family members RIG-I, melanoma differentiation-associated 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) sense viral RNA in the cytoplasm and lead to downstream activation of a potent immune response, which activates the transcription factors IRF3, IRF7, and NF-κB, culminating in the production of type I IFNs and other proinflammatory cytokines. Viral infection induces aggregation of MAVS on the mitochondrial membrane, which is mediated by RIG-I, and these MAVS aggregates are capable of activating IRF3 to generate an antiviral immune response [95]. The mitochondrial fusion protein MFN2 also regulates RLR signaling via MAVS. MFN2 directly interacts with MAVS; overexpression of MFN2 leads to a decrease in RIG-I and MDA5, thereby reducing antiviral immunity [96]. Interestingly, MAVS is also located on peroxisomes, and peroxisomal and mitochondrial MAVS work in conjunction to promote IFN production and antiviral immunity [97,98].
Finally, the cGAS-STING pathway has also been reported to have a mitochondrial dimension. This pathway senses cytosolic DNA and generates a downstream immune response driving type I IFNs and other proinflammatory cytokines. mtDNA present in the cytosol is sensed by cGAS and the cGAS-STING pathway is turned on. mtDNA stress induced by a deficiency in the transcription factor TFAM, leads to an escape of mtDNA into the cytosol, where it engages the DNA sensor cGAS, promoting STING-IRF3 signaling and ultimately culminating in a type I IFN response [99]. Mechanistic studies on the regulation of cell death and inflammation governed by mitochondria revealed that the absence of proapoptotic caspases promotes the cells to induce the cGAS-STING pathway and antiviral immunity upon MOMP [100,101]. Even though MOMP induced by Bax/Bak proteins has been associated with the escape of mtDNA into the cytoplasm [102], the exact mechanism whereby mtDNA can escape to the cytosol remains unknown. Further investigations are needed to ascertain whether additional mechanisms exist that release mtDNA into the cytoplasm to induce cGAS-STING signaling.
Concluding Remarks
Given the importance of mitochondria as a highly versatile player in governing different aspects of host responses from bacterial infections to innate immunity, it is understandable that various intracellular bacteria have evolved specific tactics to hijack mitochondrial functions to create a viable niche for themselves. Similarly, extracellular bacteria have devised ways of targeting mitochondria to induce cell death to avail themselves with nutrients. Even viruses have dedicated virulence factors that modulate mitochondrial function to influence their replication and dissemination. Downstream of an infection, it is fascinating that a plethora of immune responses, be it against bacteria or viruses or LPS stimulation, are strongly impacted by mitochondria (see Outstanding Questions). Thus, it will be exciting to extrapolate this current understanding and delve into how these fundamental processes governed by mitochondria can be translated into active therapeutics to boost immunity against pathogens or to keep overt immune responses under control in the case of inflammatory disorders.
Outstanding Questions.
Can microbes that manipulate mitochondrial morphology be used to uncover the missing mechanistic links of mitochondrial fission and fusion?
How do pathogens perturb mitochondrial metabolism and what benefits does that confer on them?
How does altered mitochondrial metabolism upon infection affect cellular immune responses?
Can therapeutics targeting mitochondria bolster immune responses against microbial infections and limit the spread of infection?
Alt-text: Outstanding Questions
References
- 1.Roger A.J. The origin and diversification of mitochondria. Curr. Biol. 2017;27:R1177–R1192. doi: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Tondera D. SlP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;278:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand R. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cipolat S. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longo D.L., Archer S.L. Mitochondrial dynamics-mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 2013;369:2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 7.Mears J.A. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer C.S. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loson O.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otera H. Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J. Cell Biol. 2016;212:531–544. doi: 10.1083/jcb.201508099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otera H. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osellame L.D. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J. Cell Sci. 2016;129:2170–2181. doi: 10.1242/jcs.185165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman J.R. ER tubules mark sites of mitochondrial division. Science (80-. ) 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elgass K.D. Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J. Cell Sci. 2015;128:2795–2804. doi: 10.1242/jcs.169136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stavru F. Listeria monocytogenes transiently alters mitochondrial dynamics during infection. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3612–3617. doi: 10.1073/pnas.1100126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stavru F. Atypical mitochondrial fission upon bacterial infection. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16003–16008. doi: 10.1073/pnas.1315784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galmiche A. The N-terminal 34 kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J. 2002;19:6361–6370. doi: 10.1093/emboj/19.23.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain P. Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16032–16037. doi: 10.1073/pnas.1105175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lum M., Morona R. Dynamin-related protein Drp1 and mitochondria are important for Shigella flexneri infection. Int. J. Med. Microbiol. 2014;304:530–541. doi: 10.1016/j.ijmm.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Escoll P. Legionella pneumophila modulates mitochondrial dynamics to trigger metabolic repurposing of infected macrophages. Cell Host Microbe. 2017;22:302–316. doi: 10.1016/j.chom.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 21.Chowdhury S.R. Chlamydia preserves the mitochondrial network necessary for replication via microRNA-dependent inhibition of fission. J. Cell Biol. 2017;216:1071–1089. doi: 10.1083/jcb.201608063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurihara Y. Chlamydia trachomatis targets mitochondrial dynamics to promote intracellular survival and proliferation. Cell. Microbiol. 2019;21 doi: 10.1111/cmi.12962. [DOI] [PubMed] [Google Scholar]
- 23.Ruhe Z.C. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshizumi T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat. Commun. 2014;5:4713. doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]
- 25.Barbier V. Dengue virus induces mitochondrial elongation through impairment of Drp1-triggered mitochondrial fission. Virology. 2017;500:149–160. doi: 10.1016/j.virol.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatel-Chaix L. Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe. 2016;20:342–356. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi C.-S. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields J.A. HIV alters neuronal mitochondrial fission/fusion in the brain during HIV-associated neurocognitive disorders. Neurobiol. Dis. 2016;86:154–169. doi: 10.1016/j.nbd.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S.J. The essential role of mitochondrial dynamics in antiviral immunity. Mitochondrion. 2018;41:21–27. doi: 10.1016/j.mito.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi L. Infection with Mycobacterium tuberculosis induces the Warburg effect in mouse lungs. Sci. Rep. 2015;5:18176. doi: 10.1038/srep18176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gleeson L.E. Cutting Edge: Mycobacterium tuberculosis induces aerobic glycolysis in human alveolar macrophages that is required for control of intracellular bacillary replication. J. Immunol. 2016;196:2444–2449. doi: 10.4049/jimmunol.1501612. [DOI] [PubMed] [Google Scholar]
- 32.Shi L. Biphasic dynamics of macrophage immunometabolism during Mycobacterium tuberculosis infection. mBio. 2019;10:e02550-18. doi: 10.1128/mbio.02550-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L. Immunometabolism in tuberculosis. Front. Immunol. 2016;7:150. doi: 10.3389/fimmu.2016.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somashekar B.S. Metabolic profiling of lung granuloma in Mycobacterium tuberculosis infected guinea pigs: ex vivo1H magic angle spinning NMR studies. J. Proteome Res. 2011;10:4186–4195. doi: 10.1021/pr2003352. [DOI] [PubMed] [Google Scholar]
- 35.Lachmandas E. Rewiring cellular metabolism via the AKT/mTOR pathway contributes to host defence against Mycobacterium tuberculosis in human and murine cells. Eur. J. Immunol. 2016;46:2574–2586. doi: 10.1002/eji.201546259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Appelberg R. The Warburg effect in mycobacterial granulomas is dependent on the recruitment and activation of macrophages by interferon-γ. Immunology. 2015;145:498–507. doi: 10.1111/imm.12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czyz D.M. Brucella abortus induces a Warburg shift in host metabolism that is linked to enhanced intracellular survival of the pathogen. J. Bacteriol. 2017;199 doi: 10.1128/JB.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegl C. Tumor suppressor p53 alters host cell metabolism to limit Chlamydia trachomatis infection. Cell Rep. 2014;9:918–929. doi: 10.1016/j.celrep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Omsland A. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojcius D.M. Enhancement of ATP levels and glucose metabolism during an infection by Chlamydia: NMR studies of living cells. J. Biol. Chem. 1998;273:7052–7058. doi: 10.1074/jbc.273.12.7052. [DOI] [PubMed] [Google Scholar]
- 41.Vastag L. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaarbø M. Human cytomegalovirus infection increases mitochondrial biogenesis. Mitochondrion. 2011;11:935–945. doi: 10.1016/j.mito.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi M. Increased mitochondrial functions in human glioblastoma cells persistently infected with measles virus. Antivir. Res. 2013;99:238–244. doi: 10.1016/j.antiviral.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 44.Diamond D.L. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasmussen A.L. Systems virology identifies a mitochondrial fatty acid oxidation enzyme, dodecenoyl coenzyme A delta isomerase, required for Hepatitis C virus replication and likely pathogenesis. J. Virol. 2011;85:11646–11654. doi: 10.1128/JVI.05605-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kale J. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koterski J.F. Virulent Shigella flexneri causes damage to mitochondria and triggers necrosis in infected human monocyte-derived macrophages. Infect. Immun. 2005;73:504–513. doi: 10.1128/IAI.73.1.504-513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carneiro L.A.M. Shigella induces mitochondrial dysfunction and cell death in nonmyleoid cells. Cell Host Microbe. 2009;5:123–136. doi: 10.1016/j.chom.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 49.Chen M. A mechanism of virulence: virulent Mycobacterium tuberculosis strain H37Rv, but not attenuated H37Ra, causes significant mitochondrial inner membrane disruption in macrophages leading to necrosis. J. Immunol. 2006;176:3707–3716. doi: 10.4049/jimmunol.176.6.3707. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez L.D. A Salmonella protein causes macrophage cell death by inducing autophagy. J. Cell Biol. 2003;163:1123–1131. doi: 10.1083/jcb.200309161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood S.J. Pseudomonas aeruginosa ExoT induces mitochondrial apoptosis in target host cells in a manner that depends on its GTPase-activating protein (GAP) domain activity. J. Biol. Chem. 2015;290:29063–29073. doi: 10.1074/jbc.M115.689950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banga S. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5121–5126. doi: 10.1073/pnas.0611030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laguna R.K. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klingenbeck L. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell. Microbiol. 2013;15:675–687. doi: 10.1111/cmi.12066. [DOI] [PubMed] [Google Scholar]
- 55.Dong F. Degradation of the proapoptotic proteins Bik, Puma, and Bim with Bcl-2 domain 3 homology in Chlamydia trachomatis-infected cells. Infect. Immun. 2005;73:1861–1864. doi: 10.1128/IAI.73.3.1861-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischer S.F. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J. Exp. Med. 2004;200:905–916. doi: 10.1084/jem.20040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pirbhai M. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J. Biol. Chem. 2006;281:31495–31501. doi: 10.1074/jbc.M602796200. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H. Potential therapy strategy: targeting mitochondrial dysfunction in sepsis. Mil. Med. Res. 2018;5:41. doi: 10.1186/s40779-018-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Donnino M.W. Ubiquinol (reduced coenzyme Q10) in patients with severe sepsis or septic shock: a randomized, double-blind, placebo-controlled, pilot trial. Crit. Care. 2015;19:275. doi: 10.1186/s13054-015-0989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chopra M. Modulation of myocardial mitochondrial mechanisms during severe polymicrobial sepsis in the rat. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zamarin D. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005;1 doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang C.Y. HIV-1 Vpr triggers mitochondrial destruction by impairing Mfn2-mediated ER-mitochondria interaction. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Macreadie I.G. HIV-1 protein Vpr causes gross mitochondrial dysfunction in the yeast Saccharomyces cerevisiae. FEBS Lett. 1997;410:145–149. doi: 10.1016/s0014-5793(97)00542-5. [DOI] [PubMed] [Google Scholar]
- 64.Datan E. Dengue-induced autophagy, virus replication and protection from cell death require ER stress (PERK) pathway activation. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2015.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang Q. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell. 2016;19:663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joubert P.E. Chikungunya virus-induced autophagy delays caspase-dependent cell death. J. Exp. Med. 2012;209:1029–1047. doi: 10.1084/jem.20110996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia M. Mitophagy switches cell death from apoptosis to necrosis in NSCLC cells treated with oncolytic measles virus. Oncotarget. 2014;5:3907–3918. doi: 10.18632/oncotarget.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Collins L.V. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J. Leukoc. Biol. 2004;75:995–1000. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- 69.West A.P. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wi S.M. TAK1-ECSIT-TRAF6 complex plays a key role in the TLR4 signal to activate NF-κB. J. Biol. Chem. 2014;289:35205–35214. doi: 10.1074/jbc.M114.597187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi H.X. Mitochondrial ubiquitin ligase MARCH5 promotes TLR7 signaling by attenuating TANK action. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma B. Toll-like receptors promote mitochondrial translocation of nuclear transcription factor nuclear factor of activated T-cells in prolonged microglial activation. J. Neurosci. 2015;35:10799–10814. doi: 10.1523/JNEUROSCI.2455-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sweeney T.E. Differential regulation of the PGC family of genes in a mouse model of Staphylococcus aureus sepsis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kantrow S.P. Oxidative metabolism in rat hepatocytes and mitochondria during sepsis. Arch. Biochem. Biophys. 1997;345:278–288. doi: 10.1006/abbi.1997.0264. [DOI] [PubMed] [Google Scholar]
- 75.Haden D.W. Mitochondrial biogenesis restores oxidative metabolism during Staphylococcus aureus sepsis. Am. J. Respir. Crit. Care Med. 2007;176:768–777. doi: 10.1164/rccm.200701-161OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abuaita B.H. Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe. 2018;24:625–636. doi: 10.1016/j.chom.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vladimer G.I. Inflammasomes and host defenses against bacterial infections. Curr. Opin. Microbiol. 2013;16:23–31. doi: 10.1016/j.mib.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Broz P., Dixit V.M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 79.Zhou R. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 80.Lupfer C. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subramanian N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park S. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates Its inflammasome activity. J. Immunol. 2013;191:4358–4366. doi: 10.4049/jimmunol.1301170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barry R. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat. Commun. 2018;9:3001. doi: 10.1038/s41467-018-05321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakahira K. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shimada K. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iyer S.S. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Misawa T. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 88.Moore C.B. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573-57. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 89.Tattoli I. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arnoult D. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J. Cell Sci. 2009;122:3161–3168. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abdul-Sater A.A. Enhancement of reactive oxygen species production and chlamydial infection by the mitochondrial Nod-like family member NLRX1. J. Biol. Chem. 2010;285:41637–41645. doi: 10.1074/jbc.M110.137885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat. Immunol. 2019;20:433–446. doi: 10.1038/s41590-019-0324-2. [DOI] [PubMed] [Google Scholar]
- 93.Jabir M.S. Mitochondrial damage contributes to Pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy. 2015;11:166–182. doi: 10.4161/15548627.2014.981915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seth R.B. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 95.Hou F. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yasukawa K. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 97.Bender S. Activation of type I and III interferon response by mitochondrial and peroxisomal MAVS and inhibition by Hepatitis C virus. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dixit E. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;20 doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.West A.P. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rongvaux A. Apoptotic caspases prevent the induction of type i interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.White M.J. Apoptotic caspases suppress mtDNA-induced STING-mediated type i IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McArthur K. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science (80-. ) 2018;359 doi: 10.1126/science.aao6047. [DOI] [PubMed] [Google Scholar]