Abstract
Investigations of the etiologic agents of community-acquired acute respiratory illness may lead to better treatment decisions and patient outcomes. In a routine care setting, we assessed the diagnostic performance of a multiplex PCR assay with respect to conventional microbiological methods, in a continuous series of adult cases of community-acquired acute respiratory illness. We enrolled 279 adult patients hospitalised for community-acquired acute respiratory illness at Tours University Hospital during the winter of 2011–2012. Respiratory samples (mostly nasopharyngeal aspirates) were studied prospectively by indirect immunofluorescence assay and multiplex PCR, that enable detection of 8 viruses and 21 respiratory pathogens respectively. In total, 255 of the 279 (91.4%) samples had interpretable results by both methods. At least one respiratory pathogen was detected by multiplex PCR in 171 specimens (65%). Overall, 130 (76%) of the 171 positive samples were positive for only one respiratory pathogen, 37 (22%) samples were positive for two pathogens and four (2%) were positive for three pathogens. With indirect immunofluorescence assay, a respiratory virus was detected in 27 of the 255 (11%) specimens. Indirect immunofluorescence assay detected some of the influenza virus A (15/51, 29%) infections identified by multiplex PCR and some (7/15, 47%) human metapneumovirus and (5/12, 42%) respiratory syncytial virus infections, but it did not detect all the adenovirus infections. Thus, access to multiplex molecular assays improves the diagnostic spectrum and accuracy over conventional methods, increasing the frequency of identification of the respiratory pathogens involved in community-acquired acute respiratory illness.
Keywords: Respiratory infection, Etiology, Diagnostic methods, Indirect immunofluorescence, Serology
Résumé
La prise en charge des patients présentant une infection respiratoire aiguë communautaire peut être améliorée lorsque l’agent étiologique est identifié. Les performances diagnostiques d’une PCR multiplex ont été comparées aux méthodes microbiologiques conventionnelles sur une série continue d’adultes présentant une infection respiratoire aiguë communautaire. Durant l’hiver 2011–2012, 279 adultes avec une infection respiratoire aiguë communautaire et hospitalisés au centre hospitalier universitaire de Tours ont été inclus. Les prélèvements respiratoires, principalement des aspirations naso-pharyngées, ont été analysés prospectivement avec une technique d’immunofluorescence indirecte et de PCR multiplex, permettant de détecter respectivement 8 virus et 21 pathogènes respiratoires. Au total, pour 255 des 279 (91,4 %) échantillons, les résultats étaient interprétables avec les deux techniques. Au moins un pathogène respiratoire a été détecté grâce à la PCR multiplex dans 171 échantillons (65 %). Parmi ces échantillons, 130 (76 %), 37 (22 %) et 4 (2 %) échantillons étaient respectivement positifs pour un, deux et trois pathogènes. Avec la technique d’immunofluorescence indirecte, un pathogène respiratoire a été détecté dans 27 des 255 échantillons (11 %). Seulement certaines infections détectées par la PCR multiplex ont été identifées avec la technique d’immunofluorescence indirecte : virus influenza A (15/51, 29 %), metapneumovirus (7/15, 47 %) et virus respiratoire syncytial (5/12, 42 %). Aucune des infections à adenovirus n’a été détectée avec cette technique. Comparé aux méthodes de diagnostic conventionnelles, l’accès à une technique de PCR multiplex augmente la fréquence de détection de pathogènes respiratoires impliqués dans les infections respiratoires aiguës communautaires.
Mots clés: Infection respiratoire, Étiologie, Méthodes diagnostiques, Immunofluorescence indirecte, Sérologie
1. Introduction
Acute respiratory tract infections are a major cause of morbidity and mortality worldwide. They are caused by a diverse range of viruses and bacteria. Establishing a rapid etiological diagnosis of community-acquired acute respiratory illness (CARTI) may improve treatment decisions and patient outcomes. Selection of the most appropriate testing method and of the pathogens to be investigated remains a challenge [1]. A combination of several microbiological methods is often required: bacterial or viral culture, bacterial or viral antigen detection, nucleic acid amplification techniques and serology. The investigation of all potential causal agents rapidly becomes too time-consuming and costly. Choices as to the diagnostic methods to be used must therefore be made, and these choices are often driven by clinical presentation [2]. However, in clinical practice, symptoms, such as fever and myalgia, and even lung imaging findings are frequently misleading. The H1N1pdm09 flu epidemic showed that hospitalised cases of influenza-like illness were frequently attributed to a wide range of respiratory viruses and bacteria [3]. The confusion of these infections with true influenza hampered preventive and treatment measures and resulted in the unnecessary occupation of hospital beds by patients that did not actually require hospitalisation.
In recent years, multiplex RT-PCR methods have been developed, with the aim of detecting a large panel of respiratory pathogens in a single sample [4]. These techniques have been shown to be the least costly strategy, generating significant savings for hospitals [5]. In this study, we compared, in a routine care setting, the diagnostic efficacies of a multiplex PCR assay and conventional microbiological methods, in a continuous series of adult cases of CARTI.
2. Materials and methods
2.1. Ethics statement
We carried out a non-interventional study, with no addition to the usual procedures. Biological material and clinical data were obtained only for standard bacterial and viral diagnosis in accordance with doctors’ prescriptions (no specific sampling, no modification of the sampling protocol, no additional questions). Data analyses were carried out with an anonymized database. According to French Public Health laws (CSP Art L1121-1.1), such a protocol does not require the approval of an ethics committee and is exempted from informed consent application.
2.2. Patients and samples
We enrolled a continuous series of 279 adult patients hospitalised for CARTI at Tours University Hospital during the winter of 2011–2012 (from week 47 in 2011 to week 18 in 2012). The mean age of the patients was 61 years (range: 15–95 years). The sex ratio (M/F) was 1.30. In total, 167 patients were hospitalised in the intensive care unit (ICU) and 112 in medical wards. Most of the samples studied were nasopharyngeal aspirates (n= 235). For some patients, only bronchoalveolar lavages (n= 42) or sputum samples (n= 2) were obtained.
2.3. Detection of respiratory viruses
2.3.1. Antigen detection by indirect immunofluorescence assay
Bronchial cells obtained from nasopharyngeal aspirates by centrifugation were suspended in buffer and spotted onto slides, then air-dried, fixed in acetone and incubated for 15 minutes with a specific mouse monoclonal antibody (Argène bioMérieux, France). The slides were washed and incubated with goat anti-mouse fluorescein-conjugated monoclonal antibodies for 15 minutes. The slides were then washed again and examined under a fluorescence microscope. We analysed only samples containing at least 20 bronchial cells per spot. All samples were tested for influenza virus A (INF A), influenza virus B (INF B), adenovirus (AdV), human metapneumovirus (HMPV), parainfluenza viruses (PIV) 1 to 3 and respiratory syncytial virus (RSV).
2.3.2. Multiplex real-time PCR
Total nucleic acids were extracted with an EZ1 Advanced XL automatic extractor (Qiagen, France), according to the manufacturer's instructions, beginning with 200 μl of each respiratory specimen. The final elution volume was 90 μl.
Samples were analysed with Respifinder22, (Pathofinder, The Netherlands), a multiplex molecular assay for the detection of 18 respiratory viruses (AdV, human bocavirus (HBoV) human coronaviruses (HCoV) NL63, OC43, 229E, HKU1, HMPV, INF A, INF B, INF A-H1N1pdm 2009, PIV 1 to 4, RSV A, RSV B, rhinovirus/enterovirus (HRV/EV) and four bacteria (Bordetella pertussis, Chlamydophila pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila). Assays were performed on a LightCycler 480 (Roche) according to the manufacturer's instructions.
2.3.3. M. pneumoniae serology
IgG and IgM antibodies were detected with the alphaWell M. pneumoniae ELISA kit (Mikrogen Diagnostik, Germany), according to the manufacturer's instructions.
3. Results
Sixteen of the 279 (6%) samples could not be studied with the multiplex assay because they contained RT-PCR inhibitors. At least one respiratory pathogen was detected by multiplex PCR in 171 of the 263 (65%) specimens analysed. Multiple infections were observed in 41 samples (37 with two pathogens and four with three pathogens). Thus, in total, 195 viruses and 21 bacteria were detected (Table 1 ). The percentage of specimens testing positive varied from 74% (January 2012) to 44% (March 2012); peak incidence was at week 8, when 90% of samples tested positive. INF A (n = 50) was the most common etiologic agent detected, followed by HRV/EV (n = 35) and HBoV (n = 34). Eight pathogens accounted for more than 5% of the total (AdV, HBoV, HCoV OC43, HMPV, INF A, HRV/EV, RSV A and B, M. pneumoniae). AdV, HBoV and HRV/EV were observed throughout the study period. The incidence of influenza virus infections peaked during weeks 8 to 11. HCoV OC43, HMPV and RSV and Mycoplasma infections were more evenly distributed over the winter months (Fig. 1 ). We observed no difference in the distribution of pathogens between ICUs and medical wards or between male and female patients (data not shown). Only M. pneumoniae infections were significantly more frequent in younger patients. The mean age of the patients infected with M. pneumoniae was 47 years, whereas the mean age of patients infected with other agents was 61 years (P < 0.05; Wilcoxon test).
Table 1.
Respiratory pathogens detected by multiplex real-time PCR during the winter 2011–2012.
| ICUa | Medical ward | Total (%) | |
|---|---|---|---|
| AdVb | 15 | 6 | 21 (9.7) |
| HBoV | 20 | 14 | 34 (15.7) |
| HCoV 229E | 1 | 0 | 1 (0.4) |
| HCoV NL63 | 4 | 3 | 7 (3.2) |
| HCoV OC43 | 8 | 4 | 12 (5.5) |
| HCoV HKU1 | 0 | 0 | 0 |
| HMPV | 9 | 6 | 15 (6.9) |
| INF A | 34 | 16 | 50 (23.1) |
| INF A (H1N1) pdm 2009 | 1 | 0 | 1 (0.4) |
| INF B | 0 | 2 | 2 (0.9) |
| PIV 1 | 1 | 0 | 1 (0.4) |
| PIV 2 | 0 | 0 | 0 |
| PIV 3 | 1 | 0 | 1 (0.4) |
| PIV 4 | 2 | 1 | 3 (1.4) |
| HRV/EV | 24 | 11 | 35 (16.2) |
| RSV A | 2 | 2 | 4 (1.8) |
| RSV B | 2 | 6 | 8 (3.7) |
| Bor | 1 | 2 | 3 (1.4) |
| C. pn | 0 | 0 | 0 |
| L. pn | 1 | 0 | 1 (0.4) |
| M. pn | 9 | 8 | 17 (7.9) |
| Total | 135 | 81 | 216 (100.0) |
Virus: AdV: adenovirus; HBoV: human bocavirus; HCoV: human coronavirus; HMPV: human metapneumovirus; HRV/EV: rhinovirus/enterovirus; INF A: influenza virus A; INF B: influenza virus B; PIV: parainfluenza virus; RSV: respiratory syncytial virus. Bacteria: Bor: Bordetella pertussis; L. pn: Legionella pneumophila; M. pn: Mycoplasma pneumoniae.
Intensive care unit.
In bold typeface, pathogens accounting for more than 5% of the total.
Fig. 1.
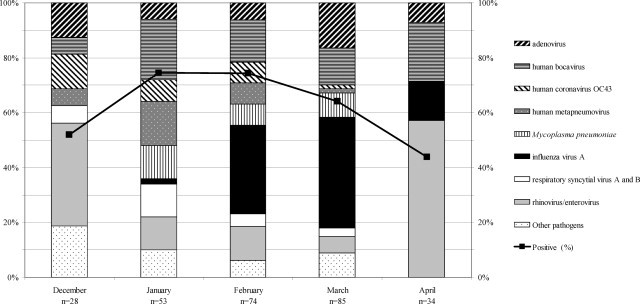
Temporal distribution of the prevalent pathogens detected by multiplex PCR between December 2011 and April 2012. The pathogen distribution for November and May is not shown because of the small number of patients tested during these two months (November: n = 4; May: n = 1).
In total, 130 (76%) of the 171 positive samples were positive for only one respiratory pathogen, 37 (22%) samples were positive for two pathogens and four (2%) were positive for three pathogens. HBoV was most frequently found associated with other pathogens in co-infections (21/34, 62%). Co-infections were also frequent with AdV and HCoV. By contrast, INF A and HRV/EV were mostly found in single infections (22% and 29%, respectively) (Table 2 ). For each pathogen, we calculated the frequency of association with other pathogens. Differences between the expected and observed rates of co-infection may indicate preferential association or mutual exclusion. Overall, the associations observed were consistent with the prevalence of the partners in the population of pathogens, with the exception of INF A and HRV/EV, which appeared to be mutually exclusive (P = 6 × 10−4).
Table 2.
Respiratory pathogens involved in single infections or co-infections during the winter 2011–2012.
| Total | Co-infections | AdV | HBoV | Bor | HCoV | HMPV | M. pn | INF A | PIV | HRV/EV | RSV | 4 infections with 3 pathogens: a |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AdV | 21 | 11 | (52.4%) | 3 | 1 | 4 | 2 | 3 | b,c | |||||
| HBoV | 34 | 21 | (61.8%) | 3 | 1 | 5 | 2 | 6 | 2 | 3 | 2 | b,d,e | ||
| Bor | 3 | 1 | (33.3%) | 1 | ||||||||||
| HCoV | 20 | 10 | (50.0%) | 1 | 1 | 1 | 1 | 2 | 4 | 1 | e | |||
| HMPV | 15 | 6 | (40.0%) | 5 | 1 | 1 | b | |||||||
| L. pn | 1 | 0 | – | |||||||||||
| M. pn | 17 | 7 | (41.2%) | 4 | 2 | 1 | 1 | 1 | b,c | |||||
| INF A | 51 | 11 | (21.6%) | 2 | 6 | 2 | 1 | 1 | d | |||||
| INF B | 2 | 0 | – | |||||||||||
| PIV | 5 | 4 | (80.0%) | 2 | 1 | 1 | ||||||||
| HRV/EV | 35 | 10 | (28.6%) | 3 | 3 | 4 | 1 | 1 | c,e | |||||
| RSV | 12 | 5 | (41.7%) | 2 | 1 | 1 | 1 | |||||||
Virus: AdV: adenovirus; HBoV: human bocavirus; HCoV: human coronavirus; HMPV: human metapneumovirus; HRV/EV: rhinovirus/enterovirus; INF A: influenza virus A; INF B: influenza virus B; PIV: parainfluenza virus; RSV: respiratory syncytial virus. Bacteria: Bor: Bordetella pertussis; L. pn: Legionella pneumophila; M. pn: Mycoplasma pneumonia.
Four infections with three pathogens; each infection is explicited below.
Infection with AdV, HBoV and M. pn.
Infection with AdV, M. pn and RSV.
Infection with HBoV, HMPV and INF A.
Infection with HBoV, HCoV and RSV.
Note that data are mirrored across the diagonal.
We assessed the correlation between the results obtained by multiplex real-time PCR and by other conventional microbiological methods. In total, 255 of the 279 (91.4%) samples were studied by both indirect immunofluorescence assay (IFA) and multiplex PCR. We excluded 24 samples from the comparative study because less than 20 epithelial cells were observed in IFA and/or because they contained PCR inhibitors. IFA detected a respiratory virus in 27 of 255 (11%) specimens: 15 INF A, seven HMPV and five RSV. No double infections were picked up with this technique. IFA detected some of the INF A (15/51, 29%), HMPV (7/15, 47%) and RSV (5/12, 42%) infections identified by multiplex PCR but did not detect all the AdV infections. All positive results obtained by IFA were confirmed by multiplex real-time PCR (Fig. 2 ).
Fig. 2.
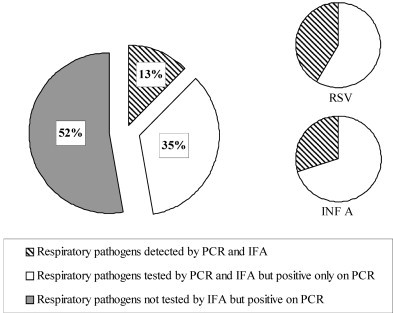
Correlation between multiplex real-time PCR and indirect immunofluorescence assay results. All the detected pathogens are shown on the first pie chart (n = 216); RSV: respiratory syncytial virus (n = 12); INF A: influenza virus A (n = 51).
Multiplex real-time PCR identified 17 patients with M. pneumoniae infection. At least one serum sample was available for each patient. The first serum sample was obtained within one week of illness onset for eight patients. Only one of these samples was tested positive for anti-Mycoplasma IgM. Four other patients were tested for the first time during the second week of illness: three tested negative and the fourth tested positive for anti-Mycoplasma IgM. Samples were collected from five patients after the second week of illness: three tested positive for anti-Mycoplasma IgM. A second serum sample was available for only four patients: one patient displayed IgG seroconversion (second serum sample collected during the third week of illness); one patient displayed IgM seroconversion (second serum sample on day 133) and serological tests remained negative for the other two patients (second serum sample collected on day 13 for one patient and day 40 for the other).
4. Discussion
It was widely thought that severe CARTI in infants is caused by a relatively limited number of viral pathogens, including predominantly paramyxovirus. This led to a diagnostic strategy based on immunofluorescence assays exploring a dozen viruses, together with single immunochromatography assays for RSV infections [6]. Viruses highly prevalent in infancy, such as HMPV and RSV, are also detected in adults, together with a diverse range of other bacterial and viral pathogens. Given this broad spectrum of etiologies for CARTI in adults, the routine use of a broad panel of single time-consuming techniques is far from ideal.
The introduction of multiplex PCR has revolutionized the management of CARTI in adults. In a continuous series of 279 patients hospitalised for CARTI, the putative causal agent was identified in 171 cases (61%): 195 viruses and 21 bacteria were identified in 130 single infections and 41 co-infections. The clinical significance of these results is unclear. Some of the infections detected may be bona fide infections directly responsible for the symptoms observed. Others may be viral infections unassociated with the symptoms, detected fortuitously in the patients carrying them. The H1N1pdm09 flu epidemic illustrates the paradigm shift resulting from the introduction of multiplex PCR. A large number of patients were hospitalised for a severe flu-like syndrome and were tested by multiple PCRs or multiplex PCR; many of these patients were found to be infected with pathogens unsuspected in this clinical context [3]. These findings suggest that the roles and distributions of the different viruses in CARTI should be reconsidered. For instance, rhinovirus infections are very common throughout the year in temperate areas. A given individual may suffer several episodes of rhinovirus infection due to the large number of genotypes and the lack of cross-protection. However, there are several lines of evidence suggesting that rhinoviruses play a significant role in CARTI in adults: asymptomatic rhinovirus carriage is seldom encountered in this population (0.1 to 2%) [7] and these viruses have been shown to have a significant pathogenic impact in vivo during experimental infections in adults [8], [9]. Rhinoviruses are also thought to interfere with other potential viral respiratory pathogens, preventing co-infection [10], [11]. Consistent with these findings, rhinovirus and influenza virus were the most prevalent pathogens in our study, and they appeared to be mutually exclusive. In the 2009–2010 H1N1pdm09 epidemic, large numbers of patients were hospitalised for severe CARTI and subjected to drastic prevention measures to prevent the spread of the epidemic flu virus, but were eventually found to be infected with rhinovirus [3]. Testing for rhinovirus has thus become paramount in the evaluation of respiratory tract infections. The position of HBoV remains much less clear. These viruses are highly prevalent, with almost 100% of the population having antibodies directed against them by the age of six years. The incidence of HBoV-associated CARTI is low in adults, at 1 to 5% [12]. In our study, HBoV were found in 16% of samples. They were associated with another bacterial or viral pathogen in 21 cases, accounting for 52% of all co-infections, and three of the four triple infections observed involved HBoV. These observations argue against HBoV playing a key role in CARTI. However, conflicting observations have been made in infants, in whom HBoV are more strongly associated with lower respiratory tract infection [13], the severity of the disease being correlated with viral load [14]. The inclusion or removal of HBoV probes from multiplex PCR assays has been much debated. For instance, the most recent format of the Film array™ Respiratory panel (BioFire Diagnostics Inc, United States) does not contain HBoV probes whereas previous versions did. HCoV are recognized to be responsible for some cases of self-resolving common cold syndrome. They have also been associated with severe pneumonia in immunocompromised patients [15]. They appear to vary considerably between years with seasonal sporadic outbreaks [16]. HCoV OC43 was the most frequent of these viruses, detected in 12 of 20 cases in our study. It was frequently found in mixed infections, co-infections with coronavirus accounting for 50% of our cases.
Our assay investigated only four bacterial pathogens: B. pertussis, C. pneumoniae, M. pneumoniae and L. pneumophila. The culture of these pathogens requires specific growth media and is slow, fastidious and costly. In a clinical setting, it is not possible to perform such cultures routinely, and the choice of culture technique depends on the clinical presentation. The introduction of primers for these respiratory pathogens into multiplex PCR assays would therefore facilitate diagnosis. M. pneumoniae infection was diagnosed in 17 cases (8%), and was associated with other pathogens in seven of these cases. Serological tests were used to assess the significance of positive PCR results for Mycoplasma, because the long-term carriage of Mycoplasma has been observed [17]. In five patients, IgM antibodies directed against Mycoplasma were detected in the month following PCR. For the other patients, the absence of IgM, particularly during the period shortly after infection, would have hindered interpretation [18]. Blood samples for serological tests were generally collected at admission and the taking of a second sample was not systematic in our setting. We have since recommended a change in practice, with the taking of a second sample some time after admission in cases in which the interval between the onset of symptoms and hospital admission is less than two weeks.
Other bacterial pathogens, such as Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis also commonly cause CARTI. It is difficult to distinguish between colonization and infection, as the bacterial involved are commensal microorganisms of the oropharyngeal flora. The use of quantitative PCR assay seems to overcome this problem [19], but culture remains necessary, for the testing of antimicrobial drug susceptibility.
This study shows that multiplex molecular assays provide greater diagnostic accuracy than conventional methods for patients hospitalised for CARTI. With molecular detection, 144 more samples (of 279 in total) than for conventional techniques were tested positive for at least one pathogen. These results are consistent with previous findings [20]. However, the clinical significance of these findings remains unclear, particularly as concerns the multiple infections. Commercial multiplex assays are not appropriate for all situations. For example, they are not suitable for the detection of emerging pathogens, for which an additional specific, sensitive test, such as monoplex PCR, is required. In the last 10 years, there have been several epidemics of acute respiratory tract infections due to emerging pathogens. These epidemics caused considerable logistic problems, because it was not possible to isolate patients (SRAS outbreak) or to apply specific therapeutic and preventive measures (H1N1pdm09 outbreak) in the absence of specific diagnosis. Highly automated laboratories making use of combinations of multiple assays are therefore required and must be capable of producing sensitive new tests to enable us to face the unknown.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
References
- 1.Brittain-Long R., Westin J., Olofsson S., Lindh M., Andersson L.M. Access to a polymerase chain reaction assay method targeting 13 respiratory viruses can reduce antibiotics: a randomised, controlled trial. BMC Med. 2011;9:44. doi: 10.1186/1741-7015-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johansson N., Kalin M., Tiveljung-Lindell A., Giske C.G., Hedlund J. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50:202–209. doi: 10.1086/648678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnepf N., Resche-Rigon M., Chaillon A., Scemla A., Gras G., Semoun O. High burden of non-influenza viruses in influenza-like illness in the early weeks of H1N1 v epidemic in France. PloS One. 2011;6:e23514. doi: 10.1371/journal.pone.0023514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21:716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahony J.B., Blackhouse G., Babwah J., Smieja M., Buracond S., Chong S. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol. 2009;47:2812–2817. doi: 10.1128/JCM.00556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Hajje M.J., Lambe C., Moulin F., Suremain Nd, Pons-Catalano C., Chalumeau M. The burden of respiratory viral disease in hospitalized children in Paris. Eur J Pediatr. 2008;167:435–436. doi: 10.1007/s00431-007-0529-5. [DOI] [PubMed] [Google Scholar]
- 7.Fry A.M., Lu X., Olsen S.J., Chittaganpitch M., Sawatwong P., Chantra S. Human rhinovirus infections in rural Thailand: epidemiological evidence for rhinovirus as both pathogen and bystander. PloS One. 2011;6:e17780. doi: 10.1371/journal.pone.0017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutman J.A., Peck A.J., Kuypers J., Boeckh M. Rhinovirus as a cause of fatal lower respiratory tract infection in adult stem cell transplantation patients: a report of two cases. Bone Marrow Transplant. 2007;40:809–811. doi: 10.1038/sj.bmt.1705827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadopoulos N.G., Bates P.J., Bardin P.G., Papi A., Leir S.H., Fraenkel D.J. Rhinoviruses infect the lower airways. J Infect Dis. 2000;181:1875–1884. doi: 10.1086/315513. [DOI] [PubMed] [Google Scholar]
- 10.Greer R.M., McErlean P., Arden K.E., Faux C.E., Nitsche A., Lambert S.B. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J Clin Virol. 2009;45:10–15. doi: 10.1016/j.jcv.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner H., Boxall E., Osman H. Respiratory viral infections during the 2009–2010 winter season in Central England UK: incidence and patterns of multiple virus co-infections. Eur J Clin Microbiol Infect Dis. 2012;31:3001–3006. doi: 10.1007/s10096-012-1653-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longtin J., Bastien M., Gilca R., Leblanc E., de Serres G., Bergeron M.G. Human bocavirus infections in hospitalized children and adults. Emerg Infect Dis. 2008;14:217–221. doi: 10.3201/eid1402.070851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fry A.M., Lu X., Chittaganpitch M., Peret T., Fischer J., Dowell S.F. Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J Infect Dis. 2007;195:1038–1045. doi: 10.1086/512163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B., Yu X., Wang C., Teng Z., Wang C., Shen J. High human bocavirus viral load is associated with disease severity in children under five years of age. PloS One. 2013;8:e62318. doi: 10.1371/journal.pone.0062318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milano F., Campbell A.P., Guthrie K.A., Kuypers J., Englund J.A., Corey L. Human rhinovirus and coronavirus detection among allogeneic hematopoietic stem cell transplantation recipients. Blood. 2010;115:2088–2094. doi: 10.1182/blood-2009-09-244152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spuesens E.B., Fraaij P.L., Visser E.G., Hoogenboezem T., Hop W.C., van Adrichem L.N. Carriage of Mycoplasma pneumoniae in the upper respiratory tract of symptomatic and asymptomatic children: an observational study. PLoS Med. 2013;10:e1001444. doi: 10.1371/journal.pmed.1001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson A.C., Björkman P., Persson K. Polymerase chain reaction is superior to serology for the diagnosis of acute Mycoplasma pneumoniae infection and reveals a high rate of persistent infection. BMC Microbiol. 2008;8:93. doi: 10.1186/1471-2180-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kais M., Spindler C., Kalin M., Ortqvist A., Giske C.G. Quantitative detection of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in lower respiratory tract samples by real-time PCR. Diagn Microbiol Infect Dis. 2006;55:169–178. doi: 10.1016/j.diagmicrobio.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Perotin J.-M., Dury S., Renois F., Deslee G., Wolak A., Duval V. Detection of multiple viral and bacterial infections in acute exacerbation of chronic obstructive pulmonary disease: a pilot prospective study. J Med Virol. 2013;85:866–873. doi: 10.1002/jmv.23495. [DOI] [PMC free article] [PubMed] [Google Scholar]


