Abstract
Objectives
To date no definitive cut-off value for cytomegalovirus (CMV) DNA load in bronchoalveolar lavage (BAL) fluid specimens has been established to discriminate between CMV pneumonia and pulmonary CMV DNA shedding in allogeneic hematopoietic stem cell transplant (allo-HSCT) recipients.
Methods
The current retrospective study is aimed at assessing the range of CMV DNA loads quantified in BAL fluid specimens from allo-HSCT patients with pneumonia in which different microorganisms were causally involved.
Results
A total of 144 BAL specimens from 123 patients were included. CMV DNA was detected in 56 out of 144 BAL fluid specimens and the median CMV DNA load from patients in whom CMV pneumonia was unlikely or could be tentatively ruled out was 1210 (31–68, 920) IU/ml. The frequency of CMV DNA detection and median CMV DNA loads were comparable, irrespective of the attributable cause of pneumonia. Detection of CMV DNA loads in BAL fluid specimens >500 IU/ml was independently associated with pneumonia-attributable mortality.
Conclusions
The current study highlights the difficulty in establishing universal CMV DNA load thresholds in BAL fluid specimens for distinguishing between CMV pneumonia and pulmonary CMV DNA shedding, and suggests that the presence of CMV DNA in BAL fluid specimens beyond a certain level may have a deleterious impact on patient outcome.
Keywords: Cytomegalovirus, CMV DNA in BAL, CMV pneumonia, CMV DNAemia, Pre-emptive antiviral therapy
Introduction
The implementation of effective prevention strategies has significantly reduced the incidence of cytomegalovirus (CMV) pneumonia after allogeneic hematopoietic stem transplantation (allo-HSCT)1; nevertheless, this clinical entity persists as a major clinical problem owing to poor survival, despite timely initiation of targeted antiviral therapy.1, 2 Currently, diagnosis of CMV pneumonia still remains a challenge. Proven CMV pneumonia can only be diagnosed using virological, histopathological or immunochemistry methods on biopsied lung tissue1; this specimen type, however, is rarely obtained due to potential life-threatening complications. Traditionally, culture-based bronchoalveolar lavage (BAL) testing has been the mainstay for suggesting CMV involvement ex vivo,3 although detection of viable CMV in BAL fluids only points to a probable causality.4 Moreover, conventional virological procedures have been largely abandoned in most laboratories. Qualitative detection of CMV DNA in lung specimens by nucleic acid testing assays does not allow diagnosis of CMV pneumonia, since it may be present in the absence of probable or proven disease (pulmonary CMV shedding).5, 6, 7 Recent studies suggested that quantitation of CMV DNA in BAL fluids may permit discrimination between these two conditions9 – 11; specifically, a cut-off CMV DNA level >500 IU/ml was proposed to serve that purpose, this displaying a positive predictive value of roughly 50% for probable CMV pneumonia using current prevalence rates.10 Nevertheless, validation of diagnostic viral load threshold across centers using different real-time PCR assays for CMV DNA quantitation and procedures for BAL obtention seems of paramount relevance. This task is hampered by the very low incidence of CMV pneumonia nowadays (<2%)3, 12 and the difficulty in establishing an incontrovertible diagnosis, even when lung tissue specimens are available for testing.13, 14, 15 – 16 Exploratory studies gathering information on the range of CMV DNA loads measured in BAL specimens from allo-HSCT patients with pneumonia in whom CMV causality is unlikely or can be reasonably ruled out may provide useful information and are thus warranted. Here, we report on our experience on this matter.
Patients and methods
Study population
The study cohort consisted of 123 consecutive patients who received an allo-HSCT at Hospital Clínico Universitario-HCU-(n = 61) or at Hospital Universitario Politécnico “La Fe” –HLF- (n = 62) and underwent diagnostic bronchoscopy. Clinical and microbiological data of patients and BAL fluid specimens (n = 144; HCU, n = 80; HLF, n = 64) submitted to the respective Microbiology Service between May 2012 and May 2017, respectively, were retrospectively reviewed. As per protocol, quantitative CMV PCR testing has been routinely performed on BAL specimens at both centers since 2012. All patients had clinical and radiography signs of pneumonia at the time of sampling. Bronchoscopy was performed using standard procedures according to international consensus guidelines.17 The first BAL fluid specimen per pneumonia event was taken into consideration for analysis purposes. This study was approved by the Hospital Clínico, Fundación INCLIVA ethics committee and informed consent was obtained from all patients.
Management of CMV infection
A preemptive antiviral therapy approach was used at HCU to prevent CMV end-organ disease.18 CMV DNA in plasma was quantified using the RealTime CMV PCR assay (Abbott Molecular, Des Plaines, IL, USA), which exhibits a limit of detection of approximately 31 IU/ml.19 Patients were preemptively treated with oral valganciclovir, i.v. ganciclovir or i.v. foscarnet upon detection of CMV DNA levels exceeding 1500 IU/ml or a CMV DNA doubling time ≤ 2 days, as previously reported.18, 20 Surveillance for CMV DNA detection and quantitation was conducted at least once a week within the first 100 days after allo-HSCT and this was extended beyond day 100 in patients at risk for recurrent episodes of CMV DNAemia.21 In turn, a universal prophylaxis strategy was deployed at HLF.22 Briefly, HLA-matched related allo-HSCT recipients were given oral valganciclovir (900 mg/day, three times a week) through day 90 after transplantation. Unrelated allo-HSCT recipients were treated with oral valganciclovir (900 mg/day) through day 180 after transplantation. At this center, plasma CMV DNA load was quantified using the CMV R-GENE® (Biomerieux, L'Etoile, Paris, France),23 which displays a limit of detection of 150 IU/ml, prior DNA extraction using the Virus/Pathogens Mini kit (Qiagen, Valencia, CA, USA) on the QIAsymphony DSP platform (Qiagen). Plasma CMV DNA monitoring was performed once a week within the first 100 days, fortnightly from day 100 to day 180, and every 2–4 weeks thereafter through day 365 after transplantation. Detection of any level of CMV DNA in plasma prompted the administration of antiviral therapy with (val)ganciclovir or foscarnet at the doses specified above. Anti-CMV therapy was given at the physician discretion when patients with CMV DNA detected in BAL specimens had concurrent CMV DNAemia with CMV DNA levels below the threshold for preemptive antiviral therapy.
Plasma specimens obtained within 72 h of BAL sampling were available from all patients for CMV DNA quantitation.
Quantitation of CMV DNA in bronchoalveolar lavages
All BAL fluid specimens underwent CMV PCR testing within 24–48 h. upon reception. Samples were kept at 4 °C until processed. BAL fluid specimens obtained from different locations (when available) were collected in sterile containers, pooled and vortexed for 30 s after the addition of sterile glass beads. Two-hundred µl of each undiluted BAL fluid samples were subjected to nucleic acid extraction using the m2000 SP system (Abbott Diagnostics) or the QIAamp DNA Blood Mini Kit (Qiagen) on either the QIA Symphony or the EZ-1 platforms (Qiagen). CMV DNA quantitation was performed using the RealTime CMV PCR assay (Abbott Molecular) at HCU or the CMV R-GENE® assay (Biomerieux) at HLF. Commutability of the CMV WHO International Standard24 for CMV DNA quantitation in BAL specimens was assessed. The CMV WHO standard was reconstituted in 1 ml of deionized water as recommended by the manufacturer.24 A pool of BAL specimens free of CMV DNA, as determined by the Abbott PCR assay was made and spiked with the WHO International Standard with a predefined CMV DNA concentration to achieve nominal values of 102 103, 104 and 105 IU/ml. The reference materials were assayed in triplicate in a single run. CMV DNA levels measured in BAL fluid specimens closely matched those expected (the mean standard deviation for both PCR assays was <0.2 log10 IU/ml for each CMV WHO standard concentration tested).
Microbiological methods for the diagnosis of pneumonia
Gram stain, fungal stain and acid-fast bacilli stain were routinely performed. Cytology examination (BAL fluid cytospins) was performed systematically in patients attended at HCU, and non-routinely at HLF. In all, BAL fluid cytospin data were available from 83 patients. In addition, BAL fluid specimens were examined for the presence of respiratory viruses (RVs) using either the Luminex xTAG RVP Fast assay (Luminex Molecular Diagnostics, Austin, TX,USA) at HCU, which allows detection of 19 RVs, or the CLART® PneumoVir assay (Genomica, Coslada, Spain)-at both centers- that makes it possible to detect simultaneously 17 RVs, as previously reported.25 Quantitative cultures of BAL specimens for bacterial isolation were performed on conventional media as recommended26; bacterial loads >104 CFU/mL were deemed to be clinically relevant.26 BAL specimens were cultured on BCYE-alpha agar, BD (becston Dickinson) MGIT® (Mycobacteria Growth Indicator Tube)/Lowenstein-Jensen agar slants and Sabouraud agar for recovery of Legionella pneumophila, Mycobacterium spp., and fungal organisms, respectively. The Platelia™ Aspergillus Ag Kit (Bio-Rad, Hercules, CA, USA) was used for quantitation of Aspergillus spp. galactomannan in BAL fluid and serum specimens. Calcofluor white, blue toluidine or direct immunofluorescence staining procedures were used for detection of Pneumocystis jiroveci. 26 Lung biopsy tissue specimens were not obtained. Likewise, neither shell vial and conventional viral cultures for CMV detection or recovery nor direct fluorescent antibody testing for CMV detection were performed.
Antimicrobial prophylaxis
Acyclovir/valacyclovir prophylaxis against Herpes simplex and Varicella Zoster viruses was given to all patients as previously described.18, 20 – 22 All patients received standard antibacterial and antifungal prophylaxis.18, 20 – 22
Definitions
CMV DNAemia and pulmonary CMV DNA shedding were defined as the detection of CMV DNA (at any level) in one or more plasma or BAL fluid specimens, respectively. Proven CMV pneumonia categorization required histopathological evidence (i.e., viral inclusions and immunohistochemical staining) in biopsy (autopsy) lung tissue. CMV pneumonia diagnosis was reasonably excluded if CMV-induced cytopathogenetic effect was not observed in post-mortem (autopsy) lung tissue (data were available from 23 patients) or BAL fluid cytospins (data available from 83 patients), there was a lack of chest X ray and computed tomography (CT) evidence consistent with CMV pneumonia (ie. reticulonodular infiltrates, the presence of bilateral ground-glass opacities, air-space opacities, small centrilobular nodules <1 cm, and absence of larger nodules – see27, for review –) and alternative diagnoses that could account for the signs and symptoms, particularly when the presence of other significant bacterial, virus or fungal pathogens was demonstrated. The clinical and radiographic response to targeted antimicrobial therapy (for bacterial, fungal and non-CMV viral infections) was also considered to be against the involvement of CMV. The probable CMV pneumonia category, according to recent criteria4 was not considered herein, since culture-based BAL fluid testing was not performed and no diagnostic CMV DNA cut-off level has been definitely established. Diagnosis of proven, probable, and possible invasive fungal infection and Pneumocystis jiroveci pneumonia was achieved following consensus criteria.28, 29 Acute graft versus host disease (aGvHD) was diagnosed and graded according to standard criteria.30
Statistical analysis
Frequencies were compared using the χ 2 test (Fisher exact test) for categorical variables. Differences between medians were compared using the Mann–Whitney U test (for two independent variables) or the Kruskal–Wallis test (for more than two independent variables). Cumulative incidence plots of mortality from pneumonia were generated with the GraphPad Prism Software (La Jolla, CA, USA) and the curves were compared by using the Gehan–Wilcoxon test. Cox proportional hazards models were used to evaluate unadjusted and adjusted hazard ratios (aHRs) for mortality attributable to pneumonia, as previously reported.21 For multivariate analyses, only variables with parameter estimates showing a P value ≤ 0.10 in the univariate analyses were included. Two-sided exact P values are reported and P values ≤ 0.05 were considered statistically significant. The data were analyzed with the SPSS (version 20.0) statistical package.
Results
Table 1 shows relevant demographic and clinical characteristics of the 123 patients in the cohort. BAL fluid samples were obtained at a median of 172.5 days after allo-HSCT (range 3 days to five years).
Table 1.
Demographic and clinical characteristics of patients.
| Parameter | No. (%) | HLF (n = 62) | HCU (n = 61) |
|---|---|---|---|
| Sex (male /female) | 76 (61.8)/47 (38.2) | 36 (58.1)/26 (41.9) | 40 (65.6)/21 (34.4) |
| Age ≥65/<65 years | 10 (8.1)/113 (91.9) | 2 (3.2)/60 (96.8) | 8 (13.1)/53 (86.9) |
| Underlying disease | |||
| NHL/CLL | 41 (33.3) | 11 (17.7) | 30 (49.2) |
| AL/MDS | 65 (52.8) | 31 (50) | 18 (29.5) |
| HD | 6 (9.8) | 0 (0) | 6 (9.8) |
| MM | 2 (3.3) | 2 (3.2) | 0 (0) |
| MPS | 5 (8.2) | 2 (3.2) | 3 (4.9) |
| Others | 4 (6.6) | 2 (3.2) | 2(3.2) |
| Number of transplants | |||
| 1 | 89 (72.4) | 49 (79) | 40 (65.6) |
| ≥2 | 34 (27.6) | 13 (20.9) | 21 (34.4) |
| Donor source and type of transplant | |||
| Umbilical cord blood | 26 (21.1) | 22 (35.5) | 4 (6.6) |
| Peripheral blood (adult unrelated) | 28 (22.8) | 19 (31.1) | 9 (14.5) |
| Peripheral blood (haploidentical) | 33 (26.8) | 20 (32.8) | 13 (21) |
| Peripheral blood (HLA-identical sibling) | 36 (29.3) | 18 (29.5) | 18 (29) |
| Human leukocyte antigen matching | |||
| Matched | 58 (47.2) | 26 (41.9) | 32 (52.5) |
| Mismatched | 65 (52.8) | 36 (58.1) | 29 (47.5) |
| Donor (D) and recipient (R) CMV serostatus | |||
| D+/R+ | 57 (46.3) | 24 (38.7) | 33 (54.1) |
| D-/R+ | 39 (31.7) | 24 (38.7) | 15 (24.6) |
| D+/R- | 14 (11.4) | 9 (14.5) | 5 (8.2) |
| D-/R- | 13 (10.6) | 5 (8.1) | 8 (13.1) |
| Use of ATG in the conditioning regimen | 37 (30.1) | 27 (43.5) | 10 (16.4) |
| Myeloablative vs. reduced-intensity conditioning | 46 (37.4)/77 (62.6) | 37 (59.7)/25 (40.3) | 9 (14.8)/52 (85.2) |
| Acute Graft vs. Host disease prophylaxis | |||
| CSA/MMF±Cy | 38 (30.9) | 20 (32.3) | 18 (29.5) |
| Tacrolimus/Cy | 4 (3.3) | 0 (0) | 4 (6.6) |
| Tacrolimus/Sirolimus | 23 (18.7) | 3 (4.8) | 20 (32.8) |
| CSA/MTX | 33 (26.8) | 18 (29) | 15 (24.6) |
| CSA/PDN | 25 (20.3) | 21 (33.9) | 4 (6.6) |
| Acute Graft vs. Host disease | |||
| No | 54 (48) | 31 (50) | 28 (45.9) |
| Yes | 69 (52) | 31 (50) | 33 (54.1) |
| Grade III-IV | 23 (32.8) | 11 (17.7) | 12 (19.7) |
| Chronic Graft vs. Host disease | 29 (23.6) | 15 (24.2) | 14 (23) |
| Treatment with corticosteroids | 66 (54) | 46 (74) | 20 (33) |
| Immunodeficiency score indexa | |||
| High (7–12) | 25 (20) | 16 (26) | 9 (15) |
| Moderate (3–6) | 61 (50) | 38 (61) | 23 (38) |
| Low (0–2) | 37 (30) | 8 (13) | 29 (48) |
AL, acute leukemia; CLL, chronic lymphocytic leukemia; CMV, cytomegalovirus; CSA, cyclosporine A, Cy, cyclophosphamide; HCU, Hospital Clínico Universitario; HD, Hodgkin lymphoma; HLF, Hospital Universitario Politécnico “La Fe”; MDS, myelodisplastic syndrome; MM, multiple mieloma; MMF, micophenolate mofetil; MPS, myeloproliferative syndrome; MTX, methotrexate; NHL, non-Hodgkin lymphoma; PDN, prednisone.
According to [39].
Microbiological yield of bronchoalveolar lavage specimens and attributable cause of pneumonia
Specific details on the microbiological yield of BAL fluid specimens are shown in Table 2 . Proven CMV pneumonia was diagnosed in 2 patients (1.6%) on the basis of histopathological and immunohistochemistry findings in lung autopsy tissue. In the remaining 142 episodes, pneumonia was attributable to bacteria in 18 cases (12.5%), and to one or more viruses in 37 (25.7%). There were 22 invasive aspergillosis infection cases (13.3%), these being categorized as possible (n = 11), probable (n = 7), or proven (n = 4). Mixed infections were documented in 42 patients (29.2%). The individual pathogenetic contribution of each detected or cultured microbial agent to pneumonia was not attempted to be settled. Twenty-three pneumonia cases (15.9%) were deemed not to have an infectious cause on the basis of clinical and radiographic data and the lack of detection of any potential pathogenic microorganism (other than CMV) in BAL fluid specimens.
Table 2.
Pneumonia attributable etiology and microbiological findings in bronchoalveolar lavage fluid specimens.
| Etiology | No. (%) |
|---|---|
| Bacterial | 18 (12.5%) |
| Pseudomonas aeruginosa | 4 (22.2%) |
| Multi-resistant Klebsiella pneumoniae | 1 (5.6%) |
| Enterococcus faecalis | 2 (11.1%) |
| Stenotrophomonas maltophilia | 1 (5.6%) |
| Streptococcus pneumoniae | 1 (5.6%) |
| Streptococcus mitis | 1(5.6%) |
| Enterococcus faecalis | 2 (11.1%) |
| Enterococcus faecium | 1 (5.6%) |
| Meticillin- resistant Staphylococcus aureus | 1 (5.6%) |
| Escherichia coli | 1(5.6%) |
| Nocardia asteroides complex | 1 (5.6%) |
| Not documenteda | 4 (22.2%) |
| Viral | 37 (25.7%) |
| One virus | 26 (70.3%) |
| Enterovirus/Rhinovirus | 10 (38.5%) |
| Parainfluenza 3 virus | 5 (19.2%) |
| Respiratory syncytial virus | 4 (16.7%) |
| Metapneumovirus | 3 (11.5%) |
| Influenza A H1N1 | 1 (3.8%) |
| Influenza A H3N2 | 1 (3.8%) |
| Influenza B | 1 (3.8%) |
| Parainfluenza 1 virus | 1 (3.8%) |
| Two viruses | 6 (16.2%) |
| Metapneumovirus/Respiratory syncytial virus | 2 (33.3%) |
| Enterovirus/Rhinovirus/Parainfluenza 1 virus | 1 (16.7%) |
| Influenza A H1N1/Respiratory syncytial virus | 1 (16.7%) |
| Influenza A H3N2/Respiratory syncytial virus | 1 (16.7%) |
| Influenza B/ Respiratory syncytial virus | 1 (16.7%) |
| Three viruses | |
| Adenovirus/Coronavirus NL63/Parainfluenza 3 virus) | 1 (2.7%) |
| Not detectedb | 4 (10.8%) |
| Invasive Fungal Infection (Aspergillus spp.)c | 22 (15.3%) |
| Possible | 11 (50%) |
| Probable | 7 (35%) |
| Proven | 4 (18.2%) |
| Polymicrobial | 42 (29.2%) |
| Bacteria/virus | 18 (42.9%) |
| Bacteria/fungus (Aspergillus spp.) | 10 (23.8%) |
| Bacteria/virus/fungus (Aspergillus spp.) | 4 (9.5%) |
| Virus/fungus (Aspergillus spp.) | 2 (4.8%) |
| Mycobacterium tuberculosis/virus | 2 (4.8%) |
| Mycobacterium tuberculosis/fungus (Aspergillus spp.) | 2 (4.8%) |
| Pneumocystis jiroveci/virus | 2 (4.8%) |
| Bacteria/virus/fungus (Aspergillus spp.)/ Mycobacterium tuberculosis | 1 (2.4%) |
| Pneumocystis jiroveci/virus/bacteria | 1 (2.4%) |
| Proven CMV Pneumoniad | 2 (1.4%) |
| Non-infectious etiologye | 23 (15.9%) |
Suspected etiology based on clinical and radiographic data.
Suspected etiology based on clinical (i.e. pneumonia preceded by an upper-respiratory tract infection) and radiographic data.
According to [28].
According to [4].
Including idiopathic pneumonia syndrome, bronchiolitis obliterans and cryptogenic organizing pneumonia among other causes.
CMV DNA detection in BAL fluid and plasma specimens
CMV DNA was detected in 56 out of 144 BAL fluid specimens (38.9%): 49 from patients with pneumonia episodes in which potential respiratory pathogens (one or more) were either cultured or detected, and 7 from cases deemed not to have an infectious origin. In these latter cases, the involvement of CMV as the causative agent was reasonably ruled out on clinical and radiographic grounds. CMV DNA was present in BAL fluid specimens from the two patients with proven CMV pneumonia; no other microorganisms were concurrently detected/cultured in these two specimens.
The frequency of CMV DNA detection was comparable (P = 0.40), regardless of the established (or presumed) etiology of pneumonia (Table 3 ). In 41 episodes, CMV DNA BAL detection occurred in the face of an ongoing episode of CMV DNAemia (including two episodes that occurred in the setting of autopsy-proven CMV pneumonitis), and isolately in the remaining 15 episodes. Clinical and laboratory characteristics of patients (n = 33) in whom CMV DNA was co-detected in BAL and plasma specimens in the absence (presumed) of CMV pneumonitis (n = 39 episodes) merit special attention, as CMV lung disease usually occurs concomitantly with CMV DNAemia.3, 4 The data are shown in Supplementary Table 1. CMV pneumonitis was deemed to be unlikely in these patients owing to one or more of the following: (i) lack of typical findings in CTs (in all episodes); (ii) negative BAL cytospin results (in 25 episodes); (iii) negative lung histopathology at autopsy (in 7 out of 16 patients who died); (iv) survival of patients who did not undergo specific anti-CMV treatment courses with appropriate doses for CMV pneumonitis (23 episodes); Of note, no patient in this series was treated with anti-CMV drugs for CMV pneumonitis as recommended3; (v) Documentation of the presence of bacteria, viruses (other than CMV) or fungal pathogens known to cause pneumonia (in 25 episodes); (vi) Clinical response (survival) to targeted anti-bacterial, anti-viral (influenza virus) or anti-fungal therapy (in 17 episodes). Nevertheless, in particular, there were four episodes in which the causative involvement of CMV raised doubts (second episode in patient 31, and episodes in patients 35, 37 and 51-Supplementary Table 1. Despite the fact that CT scans were not suggestive of CMV pneumonitis, no alternative microbial etiology was documented and all these patients died (pneumonia was the attributable cause of death). In addition, CMV DNA was detected at high levels in both BAL (ranging from 1382 to 40,048 IU/ml) and plasma (ranging from 3510 to 54,540 IU/ml) specimens. One of these patients (patient 51) had negative BAL cytospin results.
Table 3.
Frequency of Cytomegalovirus DNA detection in bronchoalveolar lavage fluids according to the etiology of pneumonia.
| Hospital | CMV DNA detected | Etiology, no. (%) |
P value | |||||
|---|---|---|---|---|---|---|---|---|
| Bacterial | Virus | Fungal | Mixed infection | CMV | Other causesa | |||
| Overall | Yes | 7 (38.9) | 12 (32.4) | 10 (45.5) | 18 (42.9) | 2 (100) | 7 (30.4) | 0.40 |
| No | 11 (61.1) | 25 (67.7) | 12 (54.5) | 24 (57.1) | 0 (0) | 16 (69.6) | ||
| HCU | Yes | 6 (54.5) | 4 (33.3) | 7 (53.8) | 15 (42.9) | 0 (0) | 3 (33.3) | 0.74 |
| No | 5 (65.5) | 8 (66.7) | 6 (46.2) | 20 (57.1) | 0 (0) | 6 (66.6) | ||
| HLF | Yes | 1 (14.3) | 8 (32.0) | 3 (33.3) | 3 (42.9) | 2 (100) | 4 (28.6) | 0.34 |
| No | 6 (85.7) | 17 (68.0) | 6 (66.6) | 4 (57.1) | 0 (0) | 10 (71.4) | ||
CMV, cytomegalovirus; HCU, Hospital Clínico Universitario; HLF, Hospital Politécnico Universitario, “La Fe”.
Including idiopathic pneumonia syndrome, bronchiolitis obliterans, cryptogenetic organizing pneumonia, among other causes.
In turn, CMV DNAemia was documented in 33 cases in the absence of CMV DNA detection in BAL fluid samples. Recipient CMV seropositivity and treatment with corticosteroids were associated with detection of CMV DNA in BAL fluid specimens in univariate analyses (Supplementary Table 2).
CMV DNA load in BAL fluid specimens
Overall, the median CMV DNA load in BAL fluid specimens from patients categorized as not having CMV pneumonia was 1210 IU/ml (range, 31–68,920 IU/ml). This magnitude was greater (P = 0.001) in specimens analyzed at HLF (median, 1938 IU/ml; range, 180–68,920 IU/ml) than in those being tested at HCU (median, 345 IU/ml; range, 21–11,263 IU/ml) (Supplementary Fig. 1). The CMV DNA load was >500 IU/ml in 32 pneumonia episodes and <500 IU/ml in the remaining 22 (Table 4 ). The CMV DNA load in BAL fluid in the two proven CMV pneumonia cases was 1453 and 12,998 IU/ml. A trend towards higher CMV DNA loads in BAL fluid specimens was observed (P = 0.09) in episodes in which CMV DNAemia was detected concurrently (1382 IU/ml; range, 31–68,920 IU/ml vs. 289 IU/ml; range, 66–12,839 IU/ml in its absence).
Table 4.
Cytomegalovirus DNA load in bronchoalveolar lavage fluids in patients with non-proven CMV pneumonia at each participating center.
| CMV DNA load (IU/ml)a | Center, no. of cases (%) |
Total number | |
|---|---|---|---|
| HCU | HLF | ||
| Not detected | 45 (56.3) | 43 (69.4) | 88 (62.0) |
| <500 | 19 (23.8) | 3 (4.8) | 22 (15.5) |
| 500-999 | 3 (3.8) | 5 (8.1) | 8 (5.6) |
| 1000-5000 | 10 (12.5) | 3 (4.8) | 13 (9.2) |
| >5000 | 3 (3.8) | 8 (12.9) | 11 (7.7) |
CMV, cytomegalovirus; HCU, Hospital Clínico Universitario; HLF, Hospiatl Universitario Politécnico “La Fe”.
a The RealTime CMV PCR assay (Abbott Molecular, Des Plaines, IL, USA) was used at HCU, and the CMV R-GENE® (Biomerieux, L'Etoile, Paris, France) was used at HLF.
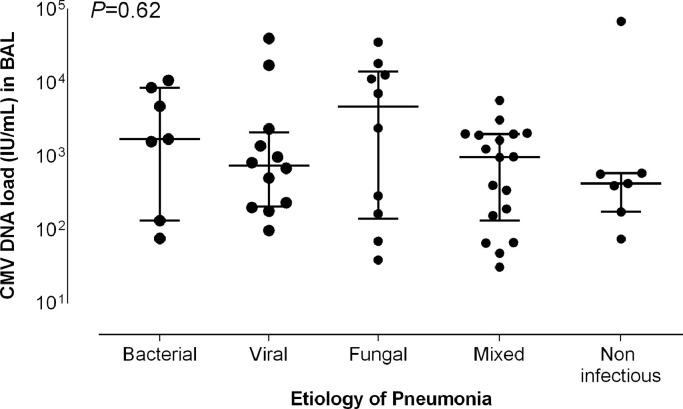
CMV DNA loads in BAL fluid specimens were comparable (P = 0.62), irrespective of the etiology (attributable) of pneumonia (Fig. 1 , and Supplementary Table 3).
Fig. 1.
CMV DNA loads in bronchoalveolar lavage fluid specimens in patients with non-proven CMV pneumonia according to the etiological diagnosis. Bars indicate median values (and 95% CI values). The P values for comparisons across groups (Kruskal–Wallis test) are shown.
Overall, there was a trend towards an inverse correlation between the level of immunosuppression, as inferred by the immunodeficiency index (ISI) – see footnotes in Table 1 –, and the CMV DNA load quantified in BAL specimens (Rho,−0.15; P = 0.08).
Anti-CMV therapy at the time of BAL fluid sampling
Overall, 72 patients (50%) were under anti-CMV therapy at the time of BAL sampling (prophylaxis, n = 14; preemptive therapy, n = 58). Among those with CMV DNA detectable in BAL fluids, 37 (66%) were receiving anti-CMV therapy (preemptive therapy, n = 35, and antiviral prophylaxis, n = 2). The CMV DNA load in BAL was significantly higher in patients who were under antiviral therapy (median, 1585 IU/ml; range, 31–68,920 IU/ml vs. median, 345 IU/ml; range, 39–7159 IU/ml; P = 0.04 in non-treated patients). Two out of the 19 patients with CMV DNA detected in BAL fluids and no concurrent CMV DNAemia were treated with i.v. ganciclovir.
CMV DNA in BAL fluids and mortality from pneumonia
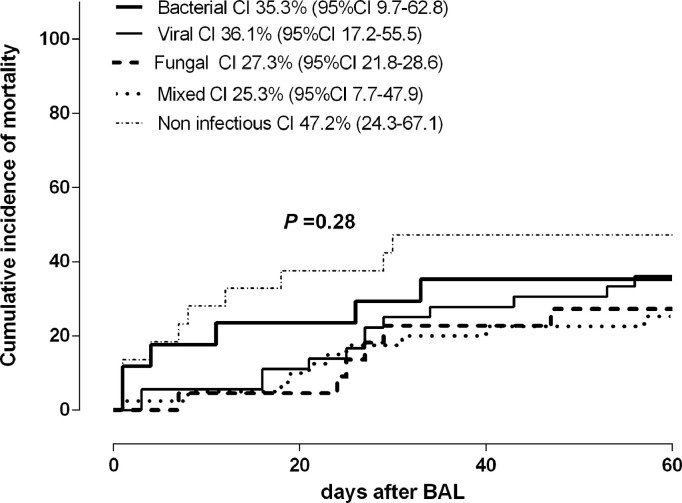
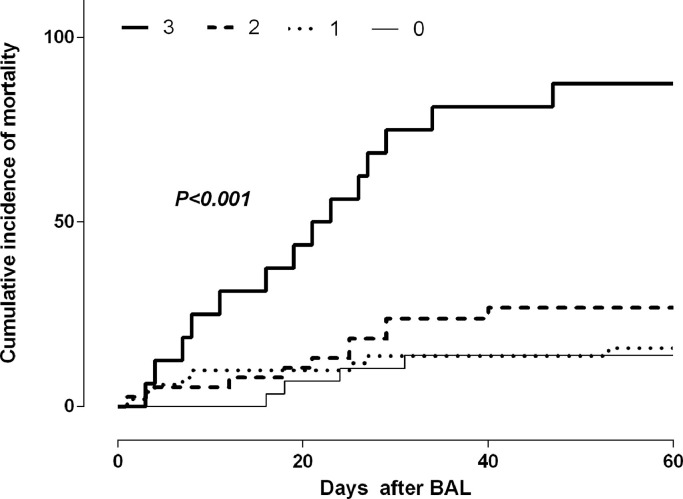
Forty-six patients (37%) died within 60 days after BAL sampling, including one patient with proven CMV pneumonia. The cause of death was deemed to be related to the pneumonia episode in 40 patients (87%). The cumulative incidence of pneumonia-attributable mortality was comparable across the different etiological categories (Fig. 2 ). We investigated whether the presence of CMV DNA in BAL fluid specimens was associated with increased mortality, and found this not to be the case. Nevertheless, ROC curve analysis enabled to determine a cut-off CMV DNA level in BAL fluid identifying those patients with increased mortality; this threshold was 500 IU/ml (sensitivity: 84.2%; 95% CI 62.4–94.5%; specificity, 53.1%; 95% CI, 36.4−69.1%) (Supplementary Fig. 2). Adjusted Cox models including in addition to this variable a number of others that could have had an impact on pneumonia-attributable mortality identified the detection of a CMV DNA load in BAL fluid specimens at levels >500 IU/ml, treatment with corticosteroids (at any dose) and lymphocyte counts <0.7 × 109/L, the latter two at the time of BAL sampling, as the parameters that were independently associated with pneumonia-attributable mortality (Table 5 ). Moreover, as shown in Fig. 3, these factors appeared to exert a synergistic impact on mortality.
Fig. 2.
Cumulative incidence of mortality attributable to pneumonia complications by day 60 after bronchoalveolar lavage fluid testing according to the etiology. Cumulative incidence plots were generated with the GraphPad Prism Software (La Jolla, CA, USA) and the curves were compared by using the Gehan–Wilcoxon test (the P value is shown).
Table 5.
Multivariate Cox models of risk factors for mortality attributable to pneumonia by day 60 after bronchoalveolar lavage fluid sampling.
| Variable | Univariate |
Multivariate |
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95%CI) | P value | |
| Sex (male vs. female) | 0.87 (0.46-1.63) | NS | ||
| Age ≥65 years | 1.30 (0.46-3.64) | NS | ||
| Donor type | ||||
| Umbilical cord blood | 3.07 (1.33-7.10) | 0.009 | NS | |
| Unrelated | 1.24 (0.51-3.06) | NS | ||
| Haploidentical | 0.99 (0.37-2.65) | NS | ||
| HLA-identical sibling | 1 | |||
| CMV serostatus | ||||
| D+/R+ | 0.48 (0.19-1.20) | NS | ||
| D-/R+ | 0.55 (0.21-1.43) | NS | ||
| D+/R- | 0.45 (0.11-1.78) | NS | ||
| D-/R- | 1 | |||
| ATG as a part of conditioning | 2.15 (1.15-4.03) | 0.02 | NS | |
| aGvHD grades III-IVa | 1.95 (0.95-3.99) | 0.068 | NS | |
| cGvHD | 1.02 (0.49-2.15) | NS | ||
| Etiology of pneumonia | ||||
| Non-infectious | 1.63 (0.55-4.88) | NS | ||
| Viral | 0.93 (0.32-2.67) | NS | ||
| Fungal | 0.66 (0.19-2.28) | NS | ||
| Mixed infection | 0.67 (0.22-1.99) | NS | ||
| Bacterial | 1 | |||
| Presence of CMV DNA in BAL vs. absence | 1.70 (0.91-3.18) | 0.10 | ||
| CMV DNA load in BAL (IU/mL) | ||||
| >500 IU/mL | 2.31 (1.24-4.30) | 0.009 | 2.21 (1.13-4.32) | 0.02 |
| <500 IU/mL | 0.68 (0.24-1.97) | NS | ||
| No CMV | 1 | 1 | ||
| High (7-12) | 3.71 (1.35-10.22) | 0.003 | NS | |
| Moderate (3-6) | 3.39 (1.39-8.30) | 0.008 | NS | |
| Low (0–2) | 1 | |||
| Treatment with corticosteroids | 2.80 (1-39-5.62) | 0.004 | 2.13 (1.04-4.33) | 0.04 |
| Absolute lymphocyte countb< 0.7 × 109/L | 4.88 (2.05-11.63) | <0.001 | 4.16 (1.72-10.04) | 0.002 |
aGvHD, acute graft versus host disease; ATG, anti-thymocyte globulin; BAL, bronchoalveolar lavage fluid; cGvHD, chronic graft versus host disease; CMV, cytomegalovirus; D, donor; NS, not significant; R, recipient.
According to [30].
The cut-off was established by ROC analysis (not shown).
Fig. 3.
Cumulative incidence of mortality attributable to pneumonia complications by day 60 after bronchoalveolar lavage fluid testing according to the presence of one or more factors associated with mortality in multivariate Cox models (CMV DNA load in BAL fluid 500 IU/ml, treatment with corticosteroids and total lymphocyte counts < 0.7 × 109/L). Cumulative incidence plots were generated with the GraphPad Prism Software (La Jolla, CA, USA) and the curves were compared by using the Gehan–Wilcoxon test (the P values for comparisons are shown.
Discussion
The definitive abandonment of traditional culture-based procedures for CMV testing in most laboratories and the non-availability of lung biopsy specimens for histopathological and immunohistochemistry examination make the diagnosis of proven or probable CMV pneumonia4 elusive nowadays. Moreover, no cut-off value for CMV DNA load in BAL fluid specimens discriminating between CMV pneumonia and pulmonary CMV DNA shedding in allo-HSCT recipients has been established at the present time.4 The assessment of the performance of quantitative CMV DNA PCR testing for the diagnosis of CMV pneumonia faces several difficulties including: (i) the low incidence of this clinical event, (ii) the lack of a normalized procedure for CMV DNA PCR testing on BAL fluids, (iii) the non-negligible possibility of miscategorization of pneumonia cases as either being causally linked or unrelated to CMV when using BAL fluid specimens, or even lung tissue material, for CMV diagnosis,8 (iv) the persistence of a large variability of CMV DNA loads provided by different real-time PCR assays,31, 32 – 33 despite their calibration to the WHO International Standard for CMV DNA24 and, as already mentioned, (v) the relegation of virological procedures for CMV detection in clinical specimens. Inevitably, all these drawbacks were encountered in this study, so that we aimed not to establish a diagnostic cut-off, but rather to assess the range of CMV DNA loads quantifiable in BAL fluid specimens from patients in whom the causal involvement of CMV was highly unlikely or could be reasonably discarded; this, having in mind that we surely incurred CMV pneumonia infradiagnosis. Nevertheless, we perceived this limitation as not being an insoslayable obstacle to our purpose as: (i) CMV is unlikely to be the etiological agent of more than 10% of pneumonia cases among allo-HSCT recipients who undergo routine bronchoscopy,10 and (ii) survival of patients with CMV pneumonia who do not undergo specific anti-CMV treatment with appropriate doses of (val)ganciclovir or foscarnet is highly unlikely, provided the high rate of CMV pneumonia-associated mortality.4 We retrospectively reviewed clinical and microbiological data from patients who underwent routine quantitative CMV PCR testing on BAL fluid specimens at two transplant centers in our city. We deliberately chose not to include archived BAL specimens in our series because of the lack of data on the impact of cryopreservation on CMV DNA load quantitation in this sample type.
Several findings arose from the present study. First, we confirmed previous observations8 – 10 indicating that detection of CMV DNA in BAL fluid specimens using highly-sensitive PCR assays is a very common finding in allo-HSCT patients with pneumonia, irrespective of the definitive etiological diagnosis. In our series, CMV DNA was detected in more than one third of BAL fluid samples, the frequency of detection varying between 30% in patients seemingly with not having an infectious pneumonia and 45% in patients with mixed infections. In our series, recipient CMV seropositivity and treatment with corticosteroids were associated with detection of CMV in BAL fluid specimens. Of interest, the two patients with proven CMV pneumonia had CMV DNA detectable in BAL fluid.
Second, we found a wide range of CMV DNA loads measured in BAL fluid specimens from patients with pneumonia in whom CMV causality was unlikely or reasonably ruled out attending to clinical (including therapeutic response to nonanti-CMV drugs), radiographic, lung autopsy histopathology or BAL fluid cytology (in some patients) and microbiological criteria. Interestingly, median CMV DNA loads were comparable irrespective of the nature and the number of co-detected microorganisms (at both participating centers), and were overall higher in the presence of concurrent DNAemia. As for the latter observation, in agreement with Boeckh et al.,10 we found this not to be due to pulmonary hemorrhage (not shown). Overall, CMV DNA loads in BAL fluid specimens processed at HLF were of greater magnitude than those analyzed at HCU. Since CMV DNA loads produced by the Argene PCR assay are slightly lower than those measured by the Abbott PCR assay (not shown), differences in the net state of immunosuppression of patients at the time of BAL sampling across centers, as reflected by the immunodeficiency score index (higher for HLF patients), may account for this observation. In fact, a trend towards an inverse correlation was found between the ISI score and the CMV DNA load quantified in BAL specimens.
In a recent study,10 a CMV DNA load cut-off of 500 IU/ml in BAL fluid was found to have a positive predictive value of ∼50% for the presence of probable CMV pneumonia (considering a prevalence of this event of 10% among patients at risk and undergoing BAL testing). Tan et al.,8 in contrast, found CMV DNA levels in BAL fluid samples to have a limited value to distinguish between CMV and non-CMV pneumonia cases. It is pertinent to mention here that the above threshold is between one and two log10 lower than those tentatively proposed for diagnosing CMV pneumonia in lung transplant recipients.34, 35, 36, 37 – 38 Interestingly, control patients with non-CMV pneumonia in the Boeckh study10 showed a median CMV DNA load of 0 log10 IU/ml (IQR, 0–1.6 IU/mL), with CMV DNA levels between 100 and 500 IU/ml in roughly 64% of cases and >500 IU/ml in 36% of them. Here, the opposite was true, with nearly 60% of BAL fluid samples from patients with non-proven CMV pneumonia having CMV viral loads >500 IU/ml, of which 75% had >1000 IU/ml. It is worth highlighting that these figures were comparable at both participating centers, despite the above-referred differences in CMV DNA loads provided by PCR assays used at each center. To gauge the potential relevance of these data, it must be taken into consideration that in nearly 70% of pneumonia episodes in our cohort, BAL sampling was performed while patients were under anti-CMV therapy (>3 days), whereas in the aforementioned study,10 only 24% of patients with CMV pneumonia and 35% of patients with non-CMV pneumonia had been treated with antivirals for at least two days. Despite this fact, higher CMV DNA loads in BAL fluid specimens were quantified in the current study, likely reflecting major differences regarding the DNAemia cut-off triggering the inception of antiviral therapy between these studies (much higher in the current cohort). In fact, in this series, the median CMV DNA load in BAL was significantly higher in patients who were under antiviral therapy than in non-treated patients.
As stated above, the limited number of proven CMV pneumonia cases in our series precluded any attempt to establish a diagnostic CMV DNA load cutoff; nevertheless, in light of the data presented herein, a threshold value of 500 IU/ml is unlikely to be discriminative between CMV pneumonia and pulmonary CMV DNA shedding in our setting. In this sense, we fully support the idea that the magnitude of such a diagnostic cut-off is likely to vary depending upon the patient's characteristics, the BAL procedure and processing, the assay used for CMV DNA quantitation and the severity of CMV pneumonia at the time of sampling.4
Our data should be interpreted with caution as the exclusion of CMV as the probable causative agent can be judged as dubious in some cases provided that no virological methods were used to investigate the presence of CMV in BAL fluid specimens. In particular, there were 4 episodes occurring in 4 patients who died, in whom CMV DNA was detected at high levels in both BAL and plasma specimens (>1000 IU/ml) and no alternative microbial etiology was documented. Nevertheless, the conclusions drawn on the basis of the overall dataset stood when cases in which little or no doubt existed on the lack of involvement of CMV (i.e. bacterial pneumonia, tuberculosis.) were analyzed separately (representative examples are shown in Supplementary Table 4).
CMV is a highly pro-inflammatory and immunosuppressive virus; as such it may act as a synergistic co-pathogen in the absence of documented CMV-induced cytopathogenicity, and may have a relevant impact on patient outcome.3 In this sense, we found that the presence of CMV DNA in BAL fluid specimens at levels >500 IU/ml, in addition to receipt of corticosteroids and low lymphocyte counts at the time of BAL sampling, was associated with increased pneumonia-attributable mortality in Cox multivariate models. The relative scarce number of pneumonia cases in which BAL specimens had CMV DNA loads >500 IU/ml did not allow to investigate whether this apparent effect exhibited a dose-response pattern. Again, this finding must be interpreted cautiously, as in order to avoid overfitting, Cox models were not adjusted to a number of factors that may have had an impact on mortality (i.e., adequacy of antimicrobial treatment, severity of pneumonia, among others). The limited size of the current cohort also precluded any meaningful statistical analysis evaluating the impact of CMV DNA load in BAL at each center, separately. Further studies are urgently needed to validate this observation, since this subset of patients may benefit from short courses of anti-CMV therapy.10
Limitations of the current study, in addition to the ones highlighted above, are its retrospective nature, the potential biased selection of patients requiring bronchoscopy for the etiological diagnosis of pneumonia, the lack of normalization of CMV DNA loads in BAL fluids to cellular DNA content (although this was reported to be expendable in a previous study10) and the use of different DNA extraction platforms and real-time PCR assays for CMV DNA quantitation. Nevertheless, this study has several strengths, including the inclusion of consecutive specimens, the use of fresh BAL fluid specimens and highly-sensitive real-time PCR assays for CMV DNA load quantitation, and the performance of a comprehensive and systematic evaluation of specimens for the presence of RVs pathogens using molecular assays.
In summary, our study highlights the difficulty in establishing universal CMV DNA load thresholds in BAL fluid specimens for distinguishing between CMV pneumonia and pulmonary CMV DNA shedding. Despite this, the potential impact of the presence of CMV in BAL fluid specimens on pneumonia-attributable mortality observed herein merits to be further investigated. To this end, only large multicenter prospective studies using consensus protocols for CMV DNA PCR BAL testing and conventional culture-based virological testing may shed light on this issue.
Conflict of interest
None.
Author's contributions
JLP, AP, JM, MS, MT, JCHB, PA, PM, CC, ABG, AP, CS, JS, GS, CS, attended the patients and collected the clinical data at the participating centers. EG, MDG, VV, EMG and RB, JLP, EG and MDG analyzed the data. DN analyzed the data and wrote the manuscript.
Acknowledgments
This research was supported by a grant (12/1992) from FIS (Fondo de Investigaciones Sanitarias, Ministerio de Sanidad y Consumo, Spain).
Estela Giménez holds a Río Hortega research contract from the Carlos III Health Institute (Ref. CM16/00200).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2019.02.009.
Appendix. Supplementary materials
References
- 1.Erard V., Guthrie K.A., Seo S., Smith J., Huang M., Chien J., et al. Reduced mortality of cytomegalovirus pneumonia after hematopoietic cell transplantation due to antiviral therapy and changes in transplantation practices. Clin Infect Dis. 2015;61(1):31–39. doi: 10.1093/cid/civ215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Travi G., Pergam S.A. Cytomegalovirus pneumonia in hematopoietic stem cell recipients. J Intensive Care Med. 2014;29:200–212. doi: 10.1177/0885066613476454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez Romero P., Blanco P., Giménez E., Solano C., Navarro D. An update on the management and prevention of cytomegalovirus infection following allogeneic hematopoietic stem cell transplantation. Futur Virol. 2015;10:113–134. [Google Scholar]
- 4.Ljungman P., Boeckh M., Hirsch H.H., Josephson F., Lundgren J., Nichols G., et al. Definitions of cytomegalovirus infection and disease in transplant patients for use in clinical trials. Clin Infect Dis. 2017;64:87–91. doi: 10.1093/cid/ciw668. [DOI] [PubMed] [Google Scholar]
- 5.Crawford S.W., Hackman R.C., Clark J.G. Open lung biopsy diagnosis of diffuse pulmonary infiltrates after marrow transplantation. Chest. 1988;94:949–953. doi: 10.1378/chest.94.5.949. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt G.M., Horak D.A., Niland J.C., Duncan S.R., Forman S.J., Zaia J.A. A randomized, controlled trial of prophylactic ganciclovir for cytomegalovirus pulmonary infection in recipients of allogeneic bone marrow transplants; The City of Hope-Stanford-Syntex CMV Study Group. N Engl J Med. 1991;324:1005–1011. doi: 10.1056/NEJM199104113241501. [DOI] [PubMed] [Google Scholar]
- 7.Zaia J.A., Schmidt G.M., Chao N.J., Rizk N.W., Nademanee A.P., Niland J.C., et al. Preemptive ganciclovir administration based solely on asymptomatic pulmonary cytomegalovirus infection in allogeneic bone marrow transplant recipients: long-term follow-up. Biol Blood Marrow Transpl. 1995;1:88–93. [PubMed] [Google Scholar]
- 8.Tan S.K., Burgener E.B., Waggoner J.J., Gajurel K., Gonzalez S., Chen S.F., et al. Molecular and culture-based bronchoalveolar lavage fluid testing for the diagnosis of cytomegalovirus pneumonitis. Open Forum Infect Dis. 2016;10:3. doi: 10.1093/ofid/ofv212. ofv212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iglesias L., Perera M.M., Torres-Miñana L., Pena-López M.J. CMV viral load in bronchoalveolar lavage for diagnosis of pneumonia in allogeneic hematopoietic stem cell transplantation. Bone Marrow Transpl. 2017;52:895–897. doi: 10.1038/bmt.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeckh M., Stevens-Ayers T., Travi G., Huang M.L., Cheng G.S., Xie H., et al. Cytomegalovirus (CMV) DNA quantitation in bronchoalveolar lavage fluid from hematopoietic stem cell transplant recipients with CMV pneumonia. J Infect Dis. 2017;215:1514–1522. doi: 10.1093/infdis/jix048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beam E., Germer J.J., Lahr B., Yao J.D., Limper A.H., Binnicker M.J., et al. Cytomegalovirus (CMV) DNA quantification in bronchoalveolar lavage fluid of immunocompromised patients with CMV pneumonia. Clin Transpl. 2017 doi: 10.1111/ctr.13149. [DOI] [PubMed] [Google Scholar]
- 12.Piñana J.L., Martino R., Barba P., Margall N., Roig M.C., Valcárcel D. Cytomegalovirus infection and disease after reduced intensity conditioning allogeneic stem cell transplantation: single-centre experience. Bone Marrow Transpl. 2010;45:534–542. doi: 10.1038/bmt.2009.180. [DOI] [PubMed] [Google Scholar]
- 13.Gleaves C.A., Reed E.C., Hackman R.C., Meyers J.D. Rapid diagnosis of invasive cytomegalovirus infection by examination of tissue specimens in centrifugation culture. Am J Clin Pathol. 1987;88:354–358. doi: 10.1093/ajcp/88.3.354. [DOI] [PubMed] [Google Scholar]
- 14.Crawford S.W., Hackman R.C., Clark J.G. Open lung biopsy diagnosis of diffuse pulmonary infiltrates after marrow transplantation. Chest. 1988;94:949–953. doi: 10.1378/chest.94.5.949. [DOI] [PubMed] [Google Scholar]
- 15.Cathomas G., Morris P., Pekle K., Cunningham I., Emanuel D. Rapid diagnosis of cytomegalovirus pneumonia in marrow transplant recipients by bronchoalveolar lavage using the polymerase chain reaction, virus culture, and the direct immunostaining of alveolar cells. Blood. 1993;81:1909–1914. [PubMed] [Google Scholar]
- 16.Tamm M., Traenkle P., Grilli B., Solèr M., Bolliger C.T., Dalquen P., et al. Pulmonary cytomegalovirus infection in immunocompromised patients. Chest. 2001;119:838–843. doi: 10.1378/chest.119.3.838. [DOI] [PubMed] [Google Scholar]
- 17.Haslam P.L., Baughman R.P. Report of ERS Task Force: guidelines for measurement of acellular components and standardization of BAL. Eur Respir J. 1999;14:245–248. doi: 10.1034/j.1399-3003.1999.14b01.x. [DOI] [PubMed] [Google Scholar]
- 18.Solano C., Muñoz-Cobo B., Giménez E., Remigia M.J., Amat P., Clari M.A., et al. Re-emptive antiviral therapy for active CMV infection in adult allo-SCT patients guided by plasma CMV DNAemia quantitation using a real-time PCR assay: clinical experience at a single center. Bone Marrow Transpl. 2013;48:1010–1012. doi: 10.1038/bmt.2012.286. [DOI] [PubMed] [Google Scholar]
- 19.Clari M.Á., Bravo D., Costa E., Muñoz-Cobo B., Solano C., Remigia M.J., et al. Comparison of the new Abbott Real Time CMV assay and the Abbott CMV PCR Kit for the quantitation of plasma cytomegalovirus DNAemia. Diagn Microbiol Infect Dis. 2013;75:207–209. doi: 10.1016/j.diagmicrobio.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Solano C., Giménez E., Piñana J.L., Vinuesa V., Poujois S., Zaragoza S., et al. Preemptive antiviral therapy for CMV infection in allogeneic stem cell transplant recipients guided by the viral doubling time in the blood. Bone Marrow Transpl. 2016;51:718–721. doi: 10.1038/bmt.2015.303. [DOI] [PubMed] [Google Scholar]
- 21.Solano C., Giménez E., Piñana J.L., Albert E., Vinuesa V., Hernández-Boluda J.C., et al. Impact of cytomegalovirus DNAemia on overall and non-relapse mortality in allogeneic stem cell transplant recipients. Transpl Infect Dis. 2017;19(4) doi: 10.1111/tid.12717. [DOI] [PubMed] [Google Scholar]
- 22.Montesinos P., Sanz J., Cantero S., Lorenzo I., Martín G., Saavedra S., et al. Incidence, risk factors, and outcome of cytomegalovirus infection and disease in patients receiving prophylaxis with oral valganciclovir or intravenous ganciclovir after umbilical cord blood transplantation. Biol Blood Marrow Transpl. 2009;15:730–740. doi: 10.1016/j.bbmt.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Pillet S., Bourlet T., Pozzetto B. Comparative evaluation of the QIAsymphony RGQ system with the easyMAG/R-gene combination for the quantitation of cytomegalovirus DNA load in whole blood. Virol J. 2012;9:231. doi: 10.1186/1743-422X-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fryer J.F., Heath A.B., Anderson R., Minor P.D., . the collaborative study group. World Health Organization; 2010. Collaborative Study to Evaluate the Proposed 1st WHO International Standard for Human Cytomegalovirus (HCMV) for Nucleic Acid Amplification (NAT)-based aSsays. WHO ECBS Report; p. 2138. WHO/BS/10. [Google Scholar]
- 25.Costa E., Rodríguez-Domínguez M., Clari M.Á., Giménez E., Galán J.C., Navarro D. Comparison of the performance of 2 commercial multiplex PCR platforms for detection of respiratory viruses in upper and lower tract respiratory specimens. Diagn Microbiol Infect Dis. 2015;82:40–43. doi: 10.1016/j.diagmicrobio.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baron E.J., Miller J.M., Weinstein M.P., Richter S.S., Gilligan P.H., Thomson R.B., Jr, et al. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM) Clin Infect Dis. 2013;57:e22–e121. doi: 10.1093/cid/cit278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris B., Geyer A.I. Diagnostic evaluation of pulmonary abnormalities in patients with hematologic malignancies and hematopoietic cell transplantation. Clin Chest Med. 2017;38:317–331. doi: 10.1016/j.ccm.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Pauw B., Walsh T.J., Donnelly J.P., Stevens D.A., Edwards J.E., Calandra T. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alanio A., Hauser P.M., Lagrou K., Melchers W.J., Helweg-Larsen J., Matos O., et al. ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J Antimicrob Chemother. 2016;71:2386–2396. doi: 10.1093/jac/dkw156. [DOI] [PubMed] [Google Scholar]
- 30.Glucksberg H., Storb R., Fefer A. Clinical manifestations of graft-versus-hostmdisease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18:295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 31.Preiksaitis J.K., Hayden R.T., Tong Y., Pang X.L., Fryer J.F., Heath A.B., et al. Are we there yet? Impact of the first international standard for cytomegalovirus DNA on the Harmonization of results reported on plasma samples. Clin Infect Dis. 2016;63:583–589. doi: 10.1093/cid/ciw370. [DOI] [PubMed] [Google Scholar]
- 32.Vinuesa V., Giménez E., Solano C., Gimeno C., Navarro D. Spanish society for infectious diseases and clinical microbiology quality control study group would kinetic analyses of plasma cytomegalovirus DNA load help to reach consensus criteria for triggering the initiation of preemptive antiviral therapy in transplant recipients? Clin Infect Dis. 2016;63:1533–1535. doi: 10.1093/cid/ciw608. [DOI] [PubMed] [Google Scholar]
- 33.Hayden R.T., Sun Y., Tang L., Procop G.W., Hillyard D.R., Pinsky B.A., et al. Progress in quantitative viral load testing: variability and impact of the who quantitative international standards. J Clin Microbiol. 2017;55:423–430. doi: 10.1128/JCM.02044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westall G.P., Michaelides A., Williams T.J., Snell G.I., Kotsimbos T.C. Human cytomegalovirus load in plasma and bronchoalveolar lavage fluid: a longitudinal study of lung transplant recipients. J Infect Dis. 2004;190:1076–1083. doi: 10.1086/422327. [DOI] [PubMed] [Google Scholar]
- 35.Chemaly R.F., Yen-Lieberman B., Chapman J., Reilly A., Bekele B.N., Gordon S.M., et al. Clinical utility of cytomegalovirus viral load in bronchoalveolar lavage in lung transplant recipients. Am J Transpl. 2005;5:544–548. doi: 10.1111/j.1600-6143.2005.00747.x. [DOI] [PubMed] [Google Scholar]
- 36.Bauer C.C., Jaksch P., Aberle S.W., Haber H., Lang G., Klepetko W., et al. Relationship between cytomegalovirus DNA load in epithelial lining fluid and plasma of lung transplant recipients and analysis of coinfection with Epstein-Barr virus and human herpesvirus 6 in the lung compartment. J Clin Microbiol. 2007;45:324–328. doi: 10.1128/JCM.01173-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerna G., Lilleri D., Rognoni V., Agozzino M., Meloni F., Oggionni T., et al. Preemptive therapy for systemic and pulmonary human cytomegalovirus infection in lung transplant recipients. Am J Transpl. 2009;9:1142–1150. doi: 10.1111/j.1600-6143.2009.02616.x. [DOI] [PubMed] [Google Scholar]
- 38.Lodding I.P., Schultz H.H., Jensen J.U., Kirkby N., Perch M., Andersen C., et al. Cytomegalovirus viral load in bronchoalveolar lavage to diagnose lung transplant associated CMV pneumonia. Transplantation. 2017 doi: 10.1097/TP.0000000000001927. [DOI] [PubMed] [Google Scholar]
- 39.Shah D.P., Ghantoji S.S., Ariza-Heredia E.J., Shah J.N., El Taoum K.K., Shah P.K., et al. Immunodeficiency scoring index to predict poor outcomes in hematopoietic cell transplant recipients with RSV infections. Blood. 2014;123:3263–3268. doi: 10.1182/blood-2013-12-541359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.