Abstract
Human norovirus (NoV) is the number one cause of foodborne illness. Despite tremendous research efforts, human NoV is still poorly understood and understudied. There is no effective measure to eliminate this virus from food and the environment. Future research efforts should focus on developing: (1) an efficient cell culture system and a robust animal model, (2) rapid and sensitive detection methods, (3) novel sanitizers and control interventions, and (4) vaccines and antiviral drugs. Furthermore, there is an urgent need to build multidisciplinary and multi-institutional teams to combat this important biodefense agent.
Keywords: Human norovirus, Foodborne illness, Acute gastroenteritis, Epidemiology, Detection methods, Prevention and control strategies, Vaccine development
Key points
-
•
Human norovirus (NoV) is the number 1 cause of foodborne disease outbreaks worldwide, accounting for more than 60% of foodborne illness and 95% of nonbacterial acute gastroenteritis.
-
•
Human NoV is highly stable, contagious, and only a few virus particles can cause illness. Symptoms of human NoV infection include diarrhea, vomiting, nausea, abdominal cramping, chills, headache, dehydration, and a high-grade fever.
-
•
Human NoV is difficult to study, because it cannot be grown in cell culture system and lacks a small animal model.
-
•
It has been technically challenging to develop rapid, accurate, and sensitive detection methods for human NoV in foods and environment. Most detection methods focus on genomic RNA-based assays.
-
•
There is no effective measure to control human NoV. Commonly used sanitizers are not effective against human NoV. High-pressure processing is promising to inactivate human NoV in foods.
-
•
No vaccine or antiviral drug for human NoV has been approved by the US Food and Drug Administration. Human NoV VLP–based vaccines and live vectored vaccines have been developed and tested in animal models or human clinical trials.
Introduction
The most recent data from the Centers of Disease Control (CDC) estimate that human norovirus (NoV) is responsible for more than 21 million total cases of illness annually, causing 95% of all nonbacterial gastroenteritis reported each year.1 Human NoV is highly infectious, resistant to common disinfectants, and causes debilitating illness; for these reasons, the virus is considered a category B biodefense agent by the National Institute of Allergies and Infectious Disease. In recent years, the importance of viruses as a cause of foodborne disease has been increasingly appreciated. Of the viruses commonly associated with foodborne disease, human NoV is the most important and is estimated to account for 58% of all foodborne illness reported every year.2 Based on data available in 2011, more than 5 million cases of food-related illness caused by human NoV are estimated to occur each year, leading to 15,000 hospitalizations and nearly 150 deaths.3, 4 The estimated annual cost of human NoV foodborne disease, based on hospitalizations and lost wages, reaches nearly 2 billion US dollars.3, 5 Despite the considerable impact of human NoV on public health, there are no approved antiviral drugs or vaccines to combat the virus.
Human NoV causes severe gastroenteritis, characterized by vomiting, diarrhea, and stomach cramps. Vomiting is seen more commonly in infants and children, whereas adults usually present with diarrhea. The diarrhea associated with the disease is free of blood, mucus, and leukocytes.6 This characteristic differentiates NoV-associated diarrhea from diarrhea caused by bacterial pathogens such as E coli O157:H7, in which blood appears in the stools. The incubation period for the disease is usually 10 to 51 hours and the duration of the disease is 28 to 60 hours.7, 8 NoV affects people of all ages and usually does not require hospitalization. However, severe disease may be observed in infants, children, the elderly, or immunocompromised individuals, all of whom may require supportive care. In immunocompromised patients, chronic NoV infections have been documented, leading to increased morbidity and mortality compared with the general population.9 NoV outbreaks seem to have no clear seasonality, but more cases are reported in the winter months. After infection, individuals may shed virus in the stool for 20 to 40 days at titers as high as 108 to 109 genome copies per gram of stool.10 Human NoV is highly stable in the environment and is resistant to common disinfectants, such as alcohol-based sanitizers and phenolic compounds, so the propagation of disease after a point source outbreak commonly occurs.
Human NoV classification and host susceptibility
The first documented human NoV outbreak occurred in 1968 in the town of Norwalk, Ohio. In 1972, the virus was officially identified using immune electron microscopy (IEM).11 Human NoV is commonly called the stomach flu or winter vomiting disease because of its symptoms and the increase in disease occurrence during the winter months. Human NoV is a member of the genus Norovirus within the family Caliciviridae. The Norovirus genus is subdivided into 5 genogroups: GI to GV, with GI, GII, and GIV causing human disease.12 GIII are bovine NoVs, and GV includes murine NoV. The genogroups are further divided into genotypes, and at least 21 genotypes are assigned to the GII genogroup alone.13 The most prevalent human NoV strains circulating in the human population belong to genogroup II, genotype 4 (GII.4). In the past 10 years, more than 3 global pandemics have occurred, all of which were caused by strains of GII.4.14 The GII.4-2009 New Orleans strain, was identified in the winter of 2009 to 2010 and was the prevalent strain identified in outbreaks in the United States in 2010, displacing the GII.4-2006 Minerva strain.15 More recently a new emerging strain has been identified, the GII.4-2012 Sydney strain, which accounted for more than half of the human NoV outbreaks reported between September and December, 2012.16
It has long been debated whether long-term immunity is acquired after human NoV infection. Data are limited to a few volunteer studies involving just a few human NoV strains. It is believed that the diversity between strains of human NoV plays an integral part in its evasion of the immune system. Even closely related strains of human NoV show major antigenic and receptor binding differences. Host susceptibility also plays an important role in human NoV infections. Early volunteer studies with human NoV strain GI.1 found that some individuals did not show symptoms of disease after exposure to the virus.17 Recent studies have shown that individuals with blood type O are more susceptible to GI.1 strain infections than people with other blood types. Human NoVs use the histoblood group antigens (HBGAs), a family of glycans found on many cell types, as functional receptors.18 HGBAs are found on erythrocytes and on epithelial cells, as well as in some body secretions such as saliva and breast milk. Different strains of human NoV may have different binding affinity to different HBGAs, which include A, B, H, and Lewis antigens.19 The α-1, 3/4 fucosyl transferase (FUT3) and α-1,2-fucosyltransferase (FUT2) genes determine an individual’s status as either a secretor or nonsecretor. Individuals with the FUT3 allele alone are considered nonsecretor, whereas individuals with both the FUT3 and FUT2 alleles are considered secretor. The FUT3 gene encodes the Lewis enzyme, which adds fucose to either the α-1,3 or α-2,4 linkage of the HBGA precursor disaccharide, leading to the synthesis of the trisaccharide required for the Lewis A phenotype. The Lewis A phenotype is also referred to as the nonsecretor phenotype. The FUT2 gene encodes a fucosyltransferase, which adds fucose to α-1,2 linkages of the precursor, creating the H type 1 antigen. Further glycosylation by the Lewis, A, and B enzymes occurs, leading to the expression of other HBGA and the secretor phenotypes.17 An individual’s blood type and secretor/nonsecretor status have been shown to play a role in susceptibility to infection with particular human NoV strains.20
Viral structure, genome organization, and viral proteins
Under electron microscopy (EM), human NoVs look like small round particles ranging from 27 to 38 nm in diameter. It is a nonenveloped virus. The outer shell of the virus particle is a highly stable protein capsid, which carries 32 shallow, cuplike circular indentations and shows icosahedral symmetry. Inside the capsid is the genetic material, which is a single-stranded positive-sense RNA genome. The genome of human NoV is approximately 7.7 kb long and is divided into 3 open reading frames (ORF).21 ORF1 encodes the nonstructural polyprotein, ORF2 encodes the major capsid protein VP1, and ORF3 encodes the minor capsid protein VP2. The polyprotein encoded by ORF1 is further proteolytically cleaved into 6 nonstructural proteins in the order of p48, nucleoside-triphosphatase, p22, VPg, 3CLpro, and RNA dependent RNA polymerase (RdRp).22 The functions of many of these proteins have been deciphered by homologies found in cultivable surrogate viruses such as murine NoV and feline calicivirus.22 During the replication and gene expression, the virus produces subgenomic RNA which only encodes VP1 and VP2. A virally encoded protein, VPg, covalently links to the 5′ end of human NoV genomic and subgenomic RNAs.23 The function of VPg may be involved in the initiation of viral protein translation by recruiting translational machinery.
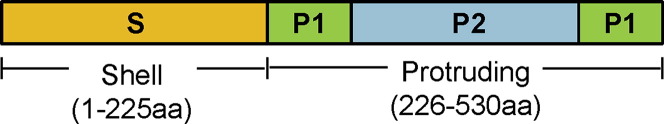
The capsid of human NoV is made up of 90 dimers of the major capsid protein VP1 and 1 or 2 copies of the minor capsid protein VP2. VP1 is composed of ∼530 to 555 amino acids with a molecular weight that ranges from 58 to 60 kDa. Expression of VP1 protein alone can form empty noninfectious viruslike particles (VLPs), which are antigenically and morphologically similar to the native human NoV virions.21 VP1 is vital for the determination of antigenicity, receptor binding activity, immunogenicity, strain specificity, and the classification of NoV genogroups and genotypes.18, 21 VP1 folds to form 2 domains, shell (S) and protrusion (P), linked by a flexible hinge region (Fig. 1 ).24 The S domain is involved in the formation of the continuous shell surface, and the P domain forms the prominent protrusion emanating from the shell.24 The P domain is further divided into 2 subdomains: P1 and P2 (see Fig. 1). The P domain is the primary site of antibody recognition and receptor binding, which plays an important role in human NoV infection and determines the host susceptibility.18
Fig. 1.
Domain organization in NoV capsid (VP1) protein.
Challenges in human NoV research
The study of human NoV has been hindered by the absence of a cell culture system and the lack of a small animal model. Therefore, many aspects of human NoV such as molecular biology, gene expression, replication, pathogenesis, and immunology are poorly understood. The survival of human NoV and the effectiveness of measures to inactivate human NoV cannot be accurately evaluated. Because human NoV cannot be grown in cell culture, most laboratory efforts to study the virus use cultivable surrogates.25, 26 These surrogates include viruses that are closely related to human NoV in terms of genetic makeup, size, receptor binding, pathogenicity, and environmental stability. Examples of these surrogate viruses include murine NoV, feline calicivirus, porcine sapovirus, and Tulane virus. The major disadvantage of the use of murine NoV and feline calicivirus as surrogates is that both viruses do not cause gastroenteritis. Murine NoV causes systemic infection in mice, whereas feline calicivirus causes respiratory tract infection in cats.25, 26 It has been proposed that porcine sapovirus and Tulane virus may be better surrogates, because they cause symptoms of gastroenteritis in animals.27 Particularly, Tulane virus recognizes the type A and B HBGAs, similar to human NoV. Other surrogates used for the study of human NoV include VLPs and P domain particles (P-particles).28, 29 These particles resemble portions of the human NoV protein capsid, which are important for receptor binding of the virus to the host cell and antigenic recognition of the virus by the immune system. The particles are noninfectious, because they are composed only of protein and lack the viral genome component of the native virus. Although the use of surrogates has aided in the understanding of human NoV, there are limitations in comparing data generated from the use of surrogates with human NoV.
Epidemiology and transmission of human NoV
The mode of transmission of human NoV is typically the fecal-oral route, with direct transmission from person to person; however, indirect transmission can occur through contaminated food, water, surfaces, and fomites. There is also evidence of direct transmission via aerosolized vomitus.30 The infectious dose of human NoV is very low, usually reported as fewer than 10 viral particles. A recent publication31 based on human volunteer studies and mathematical modeling estimated a high risk of infection (49%) caused by exposure to 1 human NoV particle. Human NoV is shed in the stool of infected individuals, and viral shedding peaks 1 to 3 days after infection. Viral shedding typically lasts 20 to 40 days in immunocompetent individuals; however, in immunocompromised individuals, viral shedding has been reported up to 56 days after infection, and in chronic cases, viral shedding can occur for years.9 From 105 to 1011 viral copies per gram of feces can typically be shed by an infected individual.31, 32, 33 Approximately one-third of human NoV–infected individuals are asymptomatic but actively shed the virus, leading to further propagation of disease.13
As mentioned previously, in immunocompromised patients, human NoV infections can be more severe or even chronic. Increased duration of NoV illness has been documented in immunosuppressed patients as a result of congenital immunodeficiency, chemotherapy, immunosuppressive therapy, and human immunodeficiency virus (HIV) infection.34, 35, 36, 37, 38 Complications from human NoV infections in the immunocompromised include dehydration, malnutrition, and dysfunction of the intestinal barrier, which contributes to the higher mortality observed for these individuals.9 Viral shedding is also increased in these patients and can last from weeks to years.39, 40 In addition, in contrast to the general population, who are normally infected by just a few stable variants of human NoV, the clinical samples of immunocompromised patients have tested positive for an array of human NoV variants not normally observed in healthy individuals.40 For these patients, proper hand hygiene should be used to limit human NoV exposure as well as isolation from visitors or staff showing the symptoms of gastroenteritis.
Outbreaks of human NoV have been popularized in the lay press in association with cruise ships, but they can occur in any area where people are in close contact. Human NoV outbreaks have been reported in restaurants, retirement communities, schools, hospitals, nursing homes, hotels, stadia, and military installations.41 Recent outbreak data from the CDC indicate that more than half of the confirmed outbreaks of human NoV in 2010 to 2011 occurred in long-term care facilities.1 Of 1518 confirmed outbreaks in 2010 to 2011, 889 (59%) were attributed to long-term care facilities, 123 (8%) were traced to restaurants, 99 (7%) were sourced to parties or events, 65 (4%) were from hospitals, 64 (4%) from schools, 55 (4%) from cruise ships, and 223 (14%) were from other or unknown sources. The high density of individuals in each of these settings, paired with the fact that food consumed at these locations is normally prepared by others, contributes to the high instance of human NoV outbreaks in these locations.
Human NoV is highly stable in the environment, which makes it difficult to eradicate after primary infections have occurred. It has been estimated that the stool of an individual with an active NoV infection may shed up to 100 billion virus particles per gram of feces.33 This fact, paired with the low infectious dose of human NoV, accounts for the rapid spread of the virus in a closed community as a result of poor hygiene. Because approximately 30% of human NoV infections are asymptomatic,33, 42, 43 consequently, asymptomatic carriers can pass human NoV to other people or to foods that they handle.
Human NoV foodborne disease is commonly associated with foods that undergo little or no processing before consumption, such as fresh produce and raw shellfish, or prepared foods to which a food handler can unknowingly transfer the virus during preparation. Human NoV outbreaks have been associated with many types of food, including fresh cut fruit, lettuce, tomatoes, melons, salads, green onions, strawberries, blueberries, raspberries, salsa, oysters, clams, and other shellfish.44, 45
In the confirmed NoV foodborne outbreaks from 2001 to 2008 that could be traced to a single food commodity, leafy vegetables contributed to 33% of the outbreaks, fruits/nuts were associated with 16% of outbreaks, and mollusks were responsible for 13% of these outbreaks.3 However, complex foods were implicated in 41% of the 2001 to 2008 outbreaks, whereas only 28% was attributed to a simple food.3 Evidence suggests that most of these foods may have become contaminated through the poor hygiene of food handlers, but viral contamination can occur upstream in the food production process. An outbreak of human NoV associated with raspberries was linked to sewage in irrigation water.46, 47, 48 Outbreaks of human NoV have also been associated with oysters that were grown in water contaminated with human waste.49, 50 Hence, prevention measures for production, processing, handling, and preparation should be considered to help minimize human NoV contamination at all steps from farm to fork.
Recent outbreaks of human NoV
Investigations of human NoV outbreaks are complicated. Outbreaks associated with foods, water, fomites, and person-to-person contact are presented in Table 1 to show the many ways in which this virus can be transmitted. Determining human NoV as the cause of outbreaks is often hampered by the limited modes of detection of the virus and the genetic diversity found within the genus Norovirus. The determination of human NoV as the cause of an outbreak is often determined by a combination of symptomatology and the exclusion of other enteric pathogens as the culprit.
Table 1.
Recent NoV outbreaks by various transmission routes
| Dates | Location | Transmission | Description | Genotype(s) | Reference |
|---|---|---|---|---|---|
| November–December, 2010 | United States | Person to person | Players and staff from 13 separate National Basketball Association franchises; direct player-to-player transmission | GII.1 | Desai et al,114 2011 |
| October, 2010 | United States | Fomites | An open-top laminated woven bag; aerosolized vomit | GII.2 | Repp & Keene,115 2012 |
| January–February, 2010 | England | Foodborne | Oysters harvested from category A waters in Europe | Unspecified | Dore et al,116 2010 |
| February, 2009 | Guatemala | Waterborne | Students and chaperones on a school trip at a resort; water | GI.7, GII.12, GII.17 | Arvelo et al,117 2012 |
| January, 2009 | Germany | Foodborne | Outbreak in a military installment; prepared salad | GII.4 | Wadl et al,118 2010 |
| January, 2008 | Korea | Waterborne | Individuals swimming at a water park; groundwater | GI.4 | Koh et al,119 2011 |
| July, 2005 | Spain | Foodborne | Campers at a summer camp; meal, asymptomatic food handler | GII.4 | Barrabeig et al,120 2010 |
| August–September, 2005 | United States | Unknown | Residents of New Orleans displaced after Hurricane Katrina were housed in the Reliant Park Complex in Houston, TX | Multiple strains | Yee et al,121 2007 |
| September, 1998 | United States | Foodborne; person to person | Football players from North Carolina and Florida; a box lunch, person to person | GI.1 | Becker et al,122 2000 |
However, important advances in the surveillance of human NoV outbreaks have been made in recent years. In March 2009, CaliciNet, an outbreak surveillance network for human NoV, was launched by the CDC partnering with state health departments. CaliciNet participants can electronically enter epidemiologic and sequence data for NoV outbreaks, allowing for linking of multistate or common source outbreaks and the identification of emerging virus strains. As of 2011, 20 local and state health departments had been certified to upload laboratory results to CaliciNet.15 The enhanced capacity of health departments to test for human NoV and the database of epidemiologic data will undoubtedly improve the accuracy and efficacy of outbreak investigations.
In the winter of 2009 to 2010, CaliciNet identified the emergence of a prevalent strain of human NoV circulating in the United States, the GII.4-2009 New Orleans strain. In January 2013, the CDC released the CaliciNet surveillance data for September to December 2012, which indicated a new predominant human NoV strain circulating in the United States, the GII.4-2012 Sydney strain, displacing the GII.4-2009 New Orleans strain. This human NoV variant accounted for 141 (53%) of the 266 total outbreaks during the 4-month period. The remaining outbreaks were caused by 10 other GI and GII strains.16 Of the outbreaks associated with GII.4-2012 Sydney in the United States, 72 (51%) were transmitted by direct person-to-person contact, 29 (20%) were foodborne, 1 (1%) was waterborne, and 39 (28%) were transmitted by an unknown route.16 In previous seasons, there has been a peak in human NoV outbreaks in the month of January, so the impact on morbidity and mortality of the GII.2-2012 Sydney strain may not be fully understood until after this threshold.
The GII.4-2012 Sydney strain was first identified in Australia in March 2012 and has been correlated with increased outbreaks in Europe and Japan compared with previous seasons.16, 51 New GII.4 variants have emerged every 2 to 3 years since 1995, which is believed to be caused by population immunity and genetic drift.16, 51 Gene and protein sequence analysis identified GII.4-2012 Sydney as phylogenetically distinct from the GII.4-2009 New Orleans and the GII.4-2007 Apeldoorn strains. GII.4-2012 Sydney had amino acid changes in the P2 domain of VP1 in the major epitopes involved in cell receptor binding.51 These changes to the P2 domain could explain the high incidence of outbreaks associated with the new variant.
Detection methods for human NoV
Clinically, diagnosis of human NoV infection is usually based on the symptoms, such as acute onset of vomiting; watery, nonbloody diarrhea with abdominal cramps; nausea; low-grade fever; and headaches. However, to confirm the cause, we must rely on laboratory diagnostic tools, particularly because many human NoV infections are asymptomatic. Because human NoVs cannot be grown in cell culture, viral RNA, viral proteins, or viral particles are targets for detection. Limitations for NoV detection are low concentration of viruses in a sample and extreme genetic and antigenic diversity seen within the genus Norovirus. There are no cross-reactive antibodies that can detect all circulating strains using enzyme immunoassays (EIAs). Likewise, nucleic acid detection assays are also hampered by low sequence homology because of genetic diversity. Thus, a single primer pair is insufficient for detecting all NoV strains and yet be free of false-positive reactions. For viral particle detection, EM, IEM, and solid-phase IEM (SPIEM) are expensive, require a highly trained observer to distinguish NoVs from other enteric viruses, and a large number of outbreak specimens cannot be rapidly examined.
Detection of human NoV in implicated foods is complicated by the complexity of the food matrix and low levels of viruses.52 In general, determination of foodborne outbreaks associated with human NoV relies on epidemiologic investigations or laboratory testing. The virus must be isolated from people who have become ill after consumption of the same food items. Sometimes, an outbreak may be traced to a food handler who also harbors human NoV. The recent trend in food microbiology to focus on viruses will certainly lead to improved molecular detection methods for human NoV in foods.
A summary of detection methods can be found in Table 2 . Initially, RNA detection methods for NoVs were reverse-transcriptase polymerase chain reaction (RT-PCR) assays.53, 54 RT quantitative PCR (RT-qPCR) assays are considered to be the gold standard for NoV detection and are used in many public health, clinical, food, environmental, and research laboratories.55, 56, 57 In addition to RT-PCR and RT-qPCR, other amplification variations, such as RT multiplex PCR,55, 58, 59 RT-nested PCR,60, 61 direct RT-PCR,62 RT-nested, real-time PCR,63 RT-booster PCR,64 and nucleic acid sequence-based amplifications50, 51 have been used for the detection of NoVs in various specimens. Recently, a reverse transcription loop-mediated isothermal amplification approach has also been used for the rapid detection of NoVs.65, 66, 67 To have a 90% probability for detecting an NoV as a cause for an outbreak, at least 3 samples from the same patient need to be tested using a standard RT-PCR assay.68
Table 2.
Detection methods for human NoV
| Detection Methods | Comments/Issues |
|---|---|
| Reverse-transcriptase polymerase chain reaction (RT-PCR) | Early amplification method for NoV detection; amplicons useful for confirming NoVs by sequencing or probes; risk for carryover contamination resulting in false-positive results; enzyme inhibitors result in false-negative results; primers determine specificity but can lead to false-negative results |
| RT quantitative PCR | Gold standard for NoV detection; faster detection than RT-PCR; less chance for carryover contamination (single closed vessel format); generally more sensitive; quantitative assay; more expensive equipment and reagents |
| RT multiplex PCR | Detects >1 target (eg, genogroup); similar annealing temperatures suggested for primer sets; potential false-negative results for targets with low initial sample copy number |
| RT-nested PCR | Risk for carryover contamination; enhanced sensitivity (compared with RT-PCR), up to 10,000 increase in sensitivity |
| Direct RT-PCR | Eliminates RNA extraction and purification; more rapid throughput; potential for less operator carryover contamination |
| RT-nested, real-time PCR | Risk for carryover contamination |
| RT-booster PCR | Double-round PCR; enhanced sensitivity; greater contamination risk |
| Nucleic acid sequence-based amplification | Isothermal amplification; excellent sensitivity; rapid assay; can be multiplexed |
| Reverse transcription loop-mediated isothermal amplification | Simple to use NoV genogroup assay; excellent sensitivity and specificity; reduced assay time; no carryover contamination (single-step format) |
| EIA | Low cost; fairly rapid assay (4 h); excellent sensitivity and specificity when homologous NoVs or antigens are used, lower sensitivity and specificity with heterologous sporadic and outbreak specimens; not recommended for diagnosing sporadic cases; false-positive results in neonates; RIDASCREEN third-generation FDA-approved test has higher sensitivity and specificity |
| Immunochromatographic | Useful for screening and point-of-care testing (POCT); easy to use; simple sample preparation; extremely rapid test (15–30 min); reduced sensitivity; applicable for outbreak cases; negative results should be confirmed |
| EM, IEM, and SPIEM | Useful for detecting new viruses when primers repeatedly fail (ie, outbreaks or cases negative by molecular approaches should be screened by EM); pooling and concentrating samples may enhance detection when other methods are negative; direct EM has limited sensitivity for NoV detection; specific antisera needed for immune aggregation with IEM and SPIEM; useful for determining NoV antigenic types; reduced throughput rate for specimen examination; excellent for detecting new viruses; used to detect or confirm NoV outbreaks |
Because NoV molecular detection methods, like RT-qPCR, are not always cost-effective or adaptable to some health care settings (eg, physician offices, local health departments, small laboratories, off-site clinics, nursing homes, field sites) commercial EIAs have been developed for testing human specimens.69, 70, 71, 72, 73 Immunologic detection of NoVs has shown limited application of early-generation EIAs/enzyme-linked immunosorbent assays (ELISAs). A review of 10 EIA/ELISA studies indicated that the sensitivity for NoV detection ranged from 31% to 90% and specificity ranged from 65% to 100%.74 The evaluation of third-generation EIAs has shown vast improvement in both sensitivity and specificity.59, 60 Consequently, some commercial EIA kits offer an improvement for rapid diagnosis of sporadic infections and also are more applicable for outbreak screenings. To have a 90% probability for detecting an NoV as a cause for an outbreak, a minimum of 6 specimens from the same patient have to be tested when using earlier-generation ELISAs.68
Immunochromatographic (ICGs) assays have been developed and could be helpful, especially for screening specimens from sporadic and outbreak cases.75, 76, 77, 78, 79 In addition, simple, sensitive, specific, rapid, and inexpensive point-of-care tests (POCTs) would be a helpful medical and public health asset. The best POCTs for NoVs are ICG assays. POCT kits for human NoVs have the potential to be improved in sensitivity and specificity as a result of recent developments in fluorescence immunochromatography.80
Human NoVs have been detected by EM procedures.11, 69, 74 Although direct EM has limited sensitivity, in any outbreak in which human NoV is the suspected cause and molecular detection results are repeatedly negative for human NoV, patient specimens should be examined by direct EM for a potential viral pathogens. Because of the low specificity of many of the human NoV molecular assays, a correct diagnosis may be missed by these techniques and direct EM would elucidate human NoV as the cause. Also, other nonspecified viral agents may be identified using direct EM and may not be detected by clinical laboratory assays. Because many diagnostic laboratories may not have the capacity for direct EM analysis of viruses causing gastroenteritis, a partnering with the public health system would be required for this type of identification. In addition, pooling outbreak specimens and concentrating specimen pools would speed the detection of any cause. Once a potential cause has been detected in a pool, then more time can be taken to examine individual specimens for an agent that matches any agent found in a concentrated pool.
It is difficult to predict future trends for the detection of human NoVs. However, there is a high probability that the current RT-qPCR approach for detecting NoVs in clinical specimens will be modified by using nanoparticle probes. A nucleic acid, multiplexed test, based on nanosphere and microarray technology, is already available for the detection of respiratory viruses.81 The complete process, from sample to final results, takes approximately 2.5 hours.81 With this technology, it should be feasible to detect in clinical specimens a wide variety of NoV genogroups, genotypes, and new genetic variants all in the same specimen on a real-time basis. The future is bright for the rapid and accurate point-of-care detection of NoVs.
Prevention methods for human NoV contamination and infection
Human NoV has high environmental stability and a low infectious dose, which makes controlling the transmission of the virus challenging. The CDC has published guidelines for disinfection procedures after a human NoV outbreak, and the recommended disinfectant for surface disinfection is 1000 to 5000 ppm of household bleach (sodium hypochlorite).13 However, because human NoV cannot be cultivated, the efficacy of this treatment and other disinfectants approved by the US Environmental Protection Agency for human NoV has been established using surrogate viruses.82, 83, 84 These surrogate viruses may not accurately represent the disinfection kinetics of human NoV, but they remain the most suitable representation. The CDC recommends increasing cleaning wards to twice daily and contact surfaces to 3 times daily with 1000 to 5000 ppm chlorine or an EPA-approved disinfectant during a human NoV outbreak, to increase the efficacy of decontamination. A summary of current data on the efficacy of sanitizers against human NoV clinical isolates can be found in Table 3 .85 Most of these data have relied solely on RT-qPCR assessment of genomic RNA copies; however, a method coupling genomic RNA detection with human NoV binding ability to HBGAs has recently been used to more accurately determine viral inactivation.86
Table 3.
Methods for the inactivation of human NoVs
| Treatment | Effectiveness | References |
|---|---|---|
| Sanitizers | ||
| Chlorine (1000 ppm) | Surface wiping; 1 log reduction in GI.4 RNA and 1.5 log reduction in GII.4 RNA | Tuladhar et al,123 2012 |
| Sodium hypochlorite (160 ppm) | Surface treatment for 30 s; 5 log reduction in GI.1 RNA | Liu et al,124 2010 |
| Alcohol or isopropanol (50%–75%) | Not efficient for GII.4 RNA | Nowak et al,125 2011 |
| Alcohol or isopropanol (90%) | <2 log reduction in GII.4 RNA | Park et al,126 2010 |
| Alcohol (95%) | Ineffective in reducing GI.1 RNA | Liu et al,124 2010 |
| Quaternary ammonium compounds | Not efficient for GII.4 RNA | Nowak et al,125 2011 |
| Chlorine dioxide (200 ppm) | Not efficient for GII.4 RNA | Nowak et al,125 2011 |
| Hydrogen peroxide (2.1%) | Treatment for 5 min; 2 log reduction in GI.8 RNA and 1 log reduction in GII.4 RNA | Li et al,127 2011 |
| Thermal Processing | ||
| 64°C | 64°C for 1 min; 0.9 logs reduction of GI.1 in binding to gastric mucin–coated beads | Dancho et al,95 2012 |
| 73°C | 73°C for 2 min; 3.1 logs reduction of GI.1 in binding to gastric mucin–coated beads | Dancho et al,95 2012 |
| 70°C | 70°C for 3 min; 1 log reduction in GI.8 RNA, but no reduction in GII.4 RNA | Li et al,127 2011 |
| Nonthermal Processing | ||
| HPP (600 MPa at 6°C for 5 min) | Oysters seeded with GI.1 strain treated by HPP; no infection (0/10) in human volunteers consuming oysters; complete inactivation | Leon et al,94 2011 |
| HPP (400 MPa at 25° for 5 min) | 60% (3/5) infection in human volunteers consuming HPP-treated oysters; incomplete inactivation | Leon et al,94 2011 |
| HPP (400 MPa at 6° for 5 min) | 21% (3/14) infection in human volunteers consuming HPP-treated oysters; incomplete inactivation | Leon et al,94 2011 |
| HPP (600 MPa at 6°C for 5 min) | GI.1 and GII.4 strains reduced binding to gastric mucin–coated beads to 0.3% and 4.0%; 4.7-log RNA reduction | Dancho et al,95 2012 |
| Ultraviolet light | 2.0 J/cm2 treatment; 3.8 log reduction in GI.1 RNA | Dancho et al,95 2012 |
| Gaseous ozone | 1 log reduction for NoV RNA on surfaces | Hudson et al,128 2007 |
Although sanitizers can be used on human NoV–contaminated surfaces, most are not approved for food use. According to the US Food and Drug Administration (FDA), sodium hypochlorite at the concentration of less than 200 ppm may be used for food sanitization purposes (FDA CFR 178.1010, 2011). This concentration of chlorine is not effective (1–2 log virus reduction) in removing viral contaminants.87, 88, 89, 90, 91 The food matrix and organic material also affect the ability of the sanitizers to inactive viruses. Thermal treatment is an effective means for inactivation of most pathogens; however, appropriate D values (the temperature and time required to eliminate 1 log of a pathogen) have not been established for human NoV. Recent data on the thermal inactivation of human NoV are presented in Table 3. However, the highest-risk foods for human NoV contamination (fresh produce and shellfish) are normally minimally processed, eaten raw, or mildly heated.
Several nonthermal processing options exist for the treatment of fresh produce and shellfish, including: high-pressure processing (HPP), γ irradiation, ultraviolet irradiation, ozone, and pulsed electric field. Many of these technologies have been evaluated for efficacy against human NoV using surrogates (such as murine NoV and feline calicivirus).29, 92, 93 Research in nonthermal processing on human NoV clinical isolates is summarized in Table 3. The most promising human NoV inactivation technology seems to be HPP. Human NoV–inoculated oysters treated with 600 MPa for 5 minutes were subsequently fed to human volunteers and these oysters did not cause infection in humans, indicating virus inactivation.94 Similarly, pressures of 700 MPa for 45 minutes could inhibit the binding of human NoV VLPs to their HBGA receptors.28 Another study using high-pressure treatment of 600 MPa for 5 minutes to treat GI.1 and GII.4 NoV isolates significantly decreased the ability of the virus to bind to HBGA receptors.95 These studies of HPP are promising; however, further research using human NoV isolates is required to substantiate these findings, as well as to more appropriately evaluate other nonthermal processes for viral inactivation.
Potential vaccine candidates against human NoV
The Need to Develop a Vaccine for Human NoV
Vaccination is the most effective strategy to protect humans from infectious diseases. There is no FDA-approved vaccine for human NoV. Although human NoV causes self-limiting illness, it causes significant health, economical, and emotional burdens. Recent epidemiologic studies found that severe clinical outcomes including death are often associated with high-risk populations such as the elderly, children, and immunocompromised individuals. The CDC estimates that 900,000 clinic visits by children in the developed world occur annually as a result of NoV infections, leading to an estimated 64,000 hospitalizations.96 From 1999 to 2007, human NoV caused, on average, 797 deaths per year in the United States; however, this estimate has been reduced in recent years.96 Mortality of NoV-associated infection increases during the epidemic seasons, and the burden of human NoV is greater in the developing world. The CDC estimates that NoV causes the death of 200,000 children younger than 5 years every year in developing countries.97 An effective vaccine would be highly beneficial. The increasing clinical significance of human NoV infections suggests that there is an urgent need for an efficacious vaccine against human NoV, particularly for the populations at high risk, such as food handlers, military personnel, elderly, infants, children, and immunocompromised individuals. An effective vaccine would not only prevent acute gastroenteritis caused by this virus but also block transmission routes and thus improve food safety, public health, and biodefense.
Protein-Based Subunit Vaccine Candidates
Because human NoV is not cultivable, most vaccine studies have been focused on a subunit vaccine using VP1 as the antigen. The VP1 protein has been expressed in many expression systems, including yeast, Escherichia coli, insect cells, mammalian cell lines, tobacco, and potatoes.21, 98, 99 In most expression systems, VP1 can self-assemble into VLPs that are structurally and antigenically similar to native virions. These VLPs contain optimal epitopes that can trigger human NoV–specific immune responses in hosts. A baculovirus-insect cell expression system has been shown to be the most efficient expression system for VLPs.100 Mice immunized with VLP-based vaccine candidates stimulated a variable level of antibody, T-cell, and intestinal and vaginal mucosal immunities, which were dependent on vaccination dosage, route, and type of adjuvants.99, 100 However, it is not known whether these immunities are protective, because mice are not susceptible to human NoV infection. Recently, it was found that gnotobiotic pigs inoculated with human NoV developed symptoms of gastroenteritis, including mild diarrhea, viral shedding in feces, and pathologic changes in the small intestine.101 Subsequently, it was found that gnotobiotic pigs vaccinated with VLPs and mucosal adjuvants (immunostimulating complexes [ISCOM] or mutant E coli LT toxin [mLT, R192G]) triggered NoV-specific antibody responses, thyroxine 1 (Th1)/Th2 serum cytokines and cytokine-secreting cells, and mucosal immune responses.102 Both vaccine candidates induced increased protection rates against viral shedding and diarrhea compared with unvaccinated controls.102 These data suggest that the VLP-based vaccine is protective in gnotobiotic pigs.
The VLP-based vaccine candidate has been tested in human clinical trials (Table 4 ). In 1999, Ball and colleagues103 performed the first clinical study to show that baculovirus-expressed human NoV VLPs were safe and immunogenic in humans when administered orally. El-Kamary and colleagues104 and Tacket and colleagues105 (2003) performed a human volunteer study using Norwalk VLPs as antigens. Thirty-six healthy adult volunteers received 250 μg (n = 10), 500 μg (n = 10), or 2000 μg (n = 10) of orally administered VLPs (without adjuvant) or placebo (n = 6). All vaccinees developed significant increases in IgA anti-VLP antibody-secreting cells. Ninety percent who received 250 μg developed increases in serum anti-VLP IgG. However, neither the rates of seroconversion nor antibody titers increased at the higher vaccination doses. Later, the effects of VLP (containing chitosan) vaccination dose on immune responses were further compared in human volunteers. Only 20% of individuals developed serum IgG and 40% of individuals developed serum IgA when receiving 15 μg of VLPs. The rate of IgG and IgA was increased to 56% and 72%, respectively, when the vaccination dose increased to 50 μg. Although these studies showed that VLP-based vaccine candidates are safe and immunogenic, it was not determined whether they can protect humans from human NoV–induced gastroenteritis. Recently, a human study was conducted in healthy adults to assess the protection efficacy of a VLP vaccine candidate (with chitosan and monophosphoryl lipid A [MPL] as adjuvants) to prevent acute viral gastroenteritis after challenge with a homologous viral strain, Norwalk virus (genotype GI.1).96 Within 98 human subjects, 50 participants received the VLP vaccine, 48 participants received placebo, and 90 received both doses (47 participants in the vaccine group and 43 in the placebo group). Norwalk virus-specific IgA antibody was detected in 70% of vaccine recipients. After challenge with Norwalk virus, vaccination significantly reduced the frequency of Norwalk virus gastroenteritis. Sixty-nine percent of placebo recipients developed gastroenteritis, whereas only 37% of vaccine recipients had symptoms. In addition, 82% of placebo recipients had Norwalk virus infection, whereas only 61% of vaccine recipients had infection. It was concluded that the VLP vaccine candidate provided protection against illness and infection after challenge with a homologous virus (see Table 4).96
Table 4.
Vaccine candidates against human NoV
| Vaccine Candidates | Dosage (μg) | Adjuvants | Vaccination Routes and Numbers of Dose | Animal Model or Human Subject | Immune Response and Protection Efficacy | References |
|---|---|---|---|---|---|---|
| Baculovirus-derived VLPs | 100 | Liquid water, no adjuvant | Two doses, orally (days 1 and 21) | 5 human subjects | 60% subjects developed serum IgG, 80% serum IgA, no fecal IgA | Ball et al,103 1999 |
| Baculovirus-derived VLPs | 250 | Liquid water, no adjuvant | Two doses, orally (days 1 and 21) | 15 human subjects | 100% subjects developed serum IgG, 80% serum IgA, 10% fecal IgA | Ball et al,103 1999 |
| Baculovirus-derived VLPs | 250 | Liquid water, no adjuvant | Two doses, orally (days 0 and 21) | 10 human subjects | 90% subjects developed serum IgG, 90% serum IgA, 40% salivary IgA, 28.5% fecal IgA, 80% vaginal IgA | Tacket et al,129 2003 |
| Baculovirus-derived VLPs | 500 | Liquid water, no adjuvant | Two doses, orally (days 0 and 21) | 10 human subjects | 70% subjects developed serum IgG, 60% serum IgA, 30% salivary IgA, 42.9% fecal IgA, 66.7% vaginal IgA | Tacket et al,129 2003 |
| Baculovirus-derived VLPs | 2000 | Liquid water, no adjuvant | Two doses, orally (days 0 and 21) | 10 human subjects | 80% subjects developed serum IgG, 100% serum IgA, 50% salivary IgA, 30% fecal IgA | Tacket et al,129 2003 |
| Baculovirus-derived VLPs | 250 | Liquid containing ISCOM or mutant E coli LT toxin | Three doses (1 oral and 2 intranasal) (days 0, 10, 21) | 8 gnotobiotic piglets | 100 seroconversion, Th1/Th2 serum cytokines and cytokine-secreting cells, increased IgM, IgA, and IgG antibody-secreting cells; protection against viral shedding and diarrhea (75%–100%) | Souza et al,102 2007 |
| Baculovirus-derived VLPs | 15 | Dry powder containing chitosan (MPL at 25 μg) | Two doses, intranasally (days 0 and 21) | 5 human subjects | 20% subjects developed serum IgG; and 40% developed serum IgA | El-Kamary et al,104 2007 Tacket et al,105 2009 |
| Baculovirus-derived VLPs | 50 | Dry powder containing chitosan (MPL at 25 μg) | Two doses, intranasally (days 0 and 21) | 20 human subjects | 56% subjects developed serum IgG; and 72% developed serum IgA | Tacket et al,105 2009 |
| Baculovirus-derived VLPs | 100 | Dry powder containing chitosan (MPL at 25 μg) | Two doses, intranasally (days 0 and 21) | 20 human subjects | 63% subjects developed serum IgG; and 79% developed serum IgA | Tacket et al,105 2009 |
| Baculovirus-derived VLPs | 100 | Lyophilized, containing MPL and chitosan | Two doses, intranasally, (3 wk apart) | 50 human subjects | 70% of vaccine recipients developed IgA seroresponse; significantly reduced gastroenteritis (69% of placebo recipients vs 37% of vaccine recipients) and NoV infection (82% of placebo recipients vs 61% of vaccine recipients) | Atmar et al,96 2011 |
| Baculovirus-derived VLPs (VP1 + VP2) | 50 | Liquid containing alhydrogel | Two doses, intramuscularly (days 0 and 30) | 2 chimpanzees | No cross-protection. Chimpanzees vaccinated with GI VLPs, but not GII VLPs vaccine, were protected from Norwalk virus (GI.1) infection | Bok et al,106 2011 |
| VSV vectored Vaccine | 106 PFU | Liquid Dulbecco’s modified Eagle’s medium, no adjuvant | One dose, intranasally (day 3) | 5 gnotobiotic piglets | 100% serum IgG, fecal, nasal, and vaginal IgA; protection against intestinal pathologic changes | Ma et al,130 2011 |
The advantage of a VLP-based vaccine candidate is that it is safe and immunogenic in humans. However, the duration of the immune response may be limited because VLPs are nonreplicating proteins. It is unknown whether it can provide cross-protection against heterogeneous strains of human NoV; however, as discussed earlier, no long-term immunity is acquired after human NoV infection because of strain diversity, so cross-protection against heterogeneous strains is unlikely. For example, chimpanzees vaccinated with VLPs derived from GII.4 strains failed to protect Norwalk virus (GI.1 strain), providing evidence that VLPs may not provide cross-protection against different genotype of NoV.106 In addition, production of VLPs in vitro is time consuming and expensive. Immunization usually requires a high dosage of VLPs and multiple booster immunizations. The efficacy of VLP-based vaccines relies on the addition of mucosal adjuvants such as cholera toxin, E coli toxin, ISCOM, chitosan, and MPL.
Live Vectored Vaccine Candidates
The first live-virus vector vaccine was reported by Smith and colleagues in 1983.107 A recombinant vaccinia virus expressing hepatitis B surface antigen–induced hepatitis B–specific antibodies in rabbits. This discovery has inspired the development of many other live-virus vectors, DNA viruses (adenoviruses and herpesviruses); positive-strand RNA viruses (alphaviruses and flaviviruses); negative-sense RNA viruses (vesicular stomatitis virus [VSV], and Newcastle disease virus). In general, a live vectored vaccine may be suitable for the following 3 conditions: viruses that cause persistent infections, such as HIV and hepatitis C virus (HCV); viruses that are highly lethal such as severe acute respiratory syndrome, Ebola, and Marburg viruses; and viruses that cannot be grown in cell culture, such as human NoV.
Three live vectored vaccine candidates have been developed for human NoV. Harrington and colleagues108 first developed a Venezuelan equine encephalitis (VEE) vectored human NoV vaccine candidate. VEE replicons expressing Norwalk VLPs induced systemic, mucosal, and heterotypic immunities against NoV. Recently, adenovirus expressing capsid protein of human NoV has been constructed.109 Mice vaccinated by the adenovirus-vectored human NoV vaccine produced systemic, mucosal, and cellular Th1/Th2 immune responses. A combination of an adenovirus-vectored vaccine and a VLP-based subunit vaccine can enhance human NoV–specific immunity.110 Recently, Ma and Li111 generated a recombinant VSV vectored human NoV vaccine candidate (rVSV-VP1). Mice inoculated with a single dose (106 PFU) of rVSV-VP1 through intranasal and oral routes stimulated a significantly stronger humoral and cellular immune response than baculovirus-expressed VLP vaccination. Furthermore, recombinant rVSV-VP1 triggered strong human NoV–specific immunity in gnotobiotic piglets and protected pigs from the challenge of a human NoV GII.4 strain, showing that live vectored human NoV vaccine is protective in an animal model.112
Although live vectored vaccine candidates are promising, it may be challenging to implement their use in human clinical trials. For example, the biosafety of VEE may be an issue, because VEE is a biodefense pathogen and the use of functional VEE genes is restricted. Delivery of the adenovirus-vectored vaccine may be hampered because a large portion of the global population has preexisting immunities against the adenovirus vector.113 Although VSV is not a human pathogen, there is little experience with VSV administration in humans. At least 3 independent phase I human clinical trials are being performed to test the safety, immune response, and effectiveness of the VSV-based HIV vaccines and oncolytic therapy in humans. It seems clear that detailed information on safety and efficacy of VSV-based vaccines in humans will be forthcoming. The outcomes of these studies will facilitate future clinical trials of VSV vectored NoV vaccine candidates in humans.
Summary
Human NoV is the number 1 cause of foodborne illness. Despite the research efforts, human NoV is still poorly understood and understudied. There is no effective measure to eliminate this virus from food and the environment. Future research efforts should focus on developing: (1) an efficient cell culture system and a small animal model, (2) rapid and sensitive detection methods, (3) novel sanitizers and control interventions, and (4) vaccines and antiviral drugs. Furthermore, there is an urgent need to build multidisciplinary and multi-institutional teams to combat this important biodefense agent.
Footnotes
Disclosure: This study was supported by a special emphasis grant (2010-01498) from the National Integrated Food Safety Initiative (NIFSI) of the USDA and a food safety challenge grant (2011-68003-30005) from the Agriculture and Food Research Initiative (AFRI) of the USDA National Institute of Food and Agriculture.
References
- 1.Norovirus: Trends and outbreaks. Centers for Disease Control and Prevention. Available at: http://www.cdc.gov/norovirus/trends-outbreaks.html. Accessed March 19, 2012.
- 2.Scallan E., Hoekstra R.M., Angulo F.J. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall A.J., Eisenbart V.G., Etingue A.L. Epidemiology of foodborne norovirus outbreaks, United States, 2001-2008. Emerg Infect Dis. 2012;18(10):1566–1573. doi: 10.3201/eid1810.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batz M.B., Hoffmann S., Morris J.G., Jr. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J Food Prot. 2012;75(7):1278–1291. doi: 10.4315/0362-028X.JFP-11-418. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann S., Batz M.B., Morris J.G., Jr. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J Food Prot. 2012;75(7):1292–1302. doi: 10.4315/0362-028X.JFP-11-417. [DOI] [PubMed] [Google Scholar]
- 6.Glass P.J., White L.J., Ball J.M. Norwalk virus open reading frame 3 encodes a minor structural protein. J Virol. 2000;74(14):6581. doi: 10.1128/jvi.74.14.6581-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopman B.A., Reacher M.H., Vipond I.B. Clinical manifestation of norovirus gastroenteritis in health care settings. Clin Infect Dis. 2004;39(3):318–324. doi: 10.1086/421948. [DOI] [PubMed] [Google Scholar]
- 8.Rockx B., De Wit M., Vennema H. Natural history of human calicivirus infection: a prospective cohort study. Clin Infect Dis. 2002;35(3):246–253. doi: 10.1086/341408. [DOI] [PubMed] [Google Scholar]
- 9.Bok K., Green K.Y. Norovirus gastroenteritis in immunocompromised patients. N Engl J Med. 2012;367(22):2126–2132. doi: 10.1056/NEJMra1207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass R.I., Parashar U.D., Estes M.K. Norovirus gastroenteritis. N Engl J Med. 2009;361(18):1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapikian A.Z., Wyatt R.G., Dolin R. Visualization by immune electron-microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. 1972;10(5):1075–1081. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng D.P., Ando T., Fankhauser R.L. Norovirus classification and proposed strain nomenclature. Virology. 2006;346(2):312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention Updated norovirus outbreak management and disease prevention guidelines. MMWR Recomm Rep. 2011;60(RR-3):1–18. [PubMed] [Google Scholar]
- 14.Siebenga J.J., Vennema H., Zheng D.P. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J Infect Dis. 2009;200(5):802–812. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- 15.Vega E., Barclay L., Gregoricus N. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg Infect Dis. 2011;17(8):1389–1395. doi: 10.3201/eid1708.101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention (CDC) Notes from the field: emergence of new norovirus strain GII.4 Sydney–United States, 2012. MMWR Morb Mortal Wkly Rep. 2013;62:55. [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson E.F., Lindesmith L.C., Lobue A.D. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol Rev. 2008;225(1):190. doi: 10.1111/j.1600-065X.2008.00680.x. [DOI] [PubMed] [Google Scholar]
- 18.Tan M., Jiang X. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 2005;13(6):285–293. doi: 10.1016/j.tim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 19.de Rougemont A., Ruvoen-Clouet N., Simon B. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J Virol. 2011;85(9):4057–4070. doi: 10.1128/JVI.02077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Pendu J., Ruvoen-Clouet N., Kindberg E. Mendelian resistance to human norovirus infections. Semin Immunol. 2006;18(6):375–386. doi: 10.1016/j.smim.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X., Wang M., Graham D.Y. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66(11):6527. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy M.E. Norovirus protein structure and function. FEMS Microbiol Lett. 2005;253(1):1–8. doi: 10.1016/j.femsle.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 23.Daughenbaugh K.F., Fraser C.S., Hershey J.W. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22(11):2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad B.V., Hardy M.E., Dokland T. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286(5438):287. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 25.Wobus C.E., Thackray L.B., Virgin H.W., 4th Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol. 2006;80(11):5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doultree J.C., Druce J.D., Birch C.J. Inactivation of feline calicivirus, a Norwalk virus surrogate. J Hosp Infect. 1999;41(1):51–57. doi: 10.1016/s0195-6701(99)90037-3. [DOI] [PubMed] [Google Scholar]
- 27.Farkas T., Sestak K., Wei C. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J Virol. 2008;82(11):5408–5416. doi: 10.1128/JVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lou F., Huang P., Neetoo H. High-pressure inactivation of human norovirus virus-like particles provides evidence that the capsid of human norovirus is highly pressure resistant. Appl Environ Microbiol. 2012;78(15):5320–5327. doi: 10.1128/AEM.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng K., Divers E., Ma Y. Inactivation of a human norovirus surrogate, human norovirus virus-like particles, and vesicular stomatitis virus by gamma irradiation. Appl Environ Microbiol. 2011;77(10):3507–3517. doi: 10.1128/AEM.00081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marks P.J., Vipond I.B., Regan F.M. A school outbreak of Norwalk-like virus: evidence for airborne transmission. Epidemiol Infect. 2003;131(1):727–736. doi: 10.1017/s0950268803008689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teunis P.F., Moe C.L., Liu P. Norwalk virus: how infectious is it? J Med Virol. 2008;80(8):1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 32.Aoki Y., Suto A., Mizuta K. Duration of norovirus excretion and the longitudinal course of viral load in norovirus-infected elderly patients. J Hosp Infect. 2010;75(1):42–46. doi: 10.1016/j.jhin.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Atmar R.L., Opekun A.R., Gilger M.A. Norwalk virus shedding after experimental human infection. Emerg Infect Dis. 2008;14(10):1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armbrust S., Kramer A., Olbertz D. Norovirus infections in preterm infants: wide variety of clinical courses. BMC Res Notes. 2009;2:96. doi: 10.1186/1756-0500-2-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludwig A., Adams O., Laws H.J. Quantitative detection of norovirus excretion in pediatric patients with cancer and prolonged gastroenteritis and shedding of norovirus. J Med Virol. 2008;80(8):1461–1467. doi: 10.1002/jmv.21217. [DOI] [PubMed] [Google Scholar]
- 36.Fishman J.A. Infections in immunocompromised hosts and organ transplant recipients: essentials. Liver Transpl. 2011;17(Suppl 3):S34–S37. doi: 10.1002/lt.22378. [DOI] [PubMed] [Google Scholar]
- 37.Koo H.L., DuPont H.L. Noroviruses as a potential cause of protracted and lethal disease in immunocompromised patients. Clin Infect Dis. 2009;49(7):1069–1071. doi: 10.1086/605558. [DOI] [PubMed] [Google Scholar]
- 38.Wingfield T., Gallimore C.I., Xerry J. Chronic norovirus infection in an HIV-positive patient with persistent diarrhoea: a novel cause. J Clin Virol. 2010;49(3):219–222. doi: 10.1016/j.jcv.2010.07.025. [DOI] [PubMed] [Google Scholar]
- 39.Sukhrie F.H., Siebenga J.J., Beersma M.F. Chronic shedders as reservoir for nosocomial transmission of norovirus. J Clin Microbiol. 2010;48(11):4303–4305. doi: 10.1128/JCM.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siebenga J.J., Beersma M.F., Vennema H. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J Infect Dis. 2008;198(7):994–1001. doi: 10.1086/591627. [DOI] [PubMed] [Google Scholar]
- 41.Seymour I.J., Appleton H. Foodborne viruses and fresh produce. J Appl Microbiol. 2001;91(5):759–773. doi: 10.1046/j.1365-2672.2001.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham D.Y., Jiang X., Tanaka T. Norwalk virus infection of volunteers: new insights based on improved assays. J Infect Dis. 1994;170(1):34–43. doi: 10.1093/infdis/170.1.34. [DOI] [PubMed] [Google Scholar]
- 43.Phillips G., Lopman B., Tam C.C. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infect Dis. 2009;9:63. doi: 10.1186/1471-2334-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herwaldt B.L., Lew J.F., Moe C.L. Characterization of a variant strain of Norwalk virus from a food-borne outbreak of gastroenteritis on a cruise ship in Hawaii. J Clin Microbiol. 1994;32(4):861–866. doi: 10.1128/jcm.32.4.861-866.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hjertqvist M., Johansson A., Svensson N. Four outbreaks of norovirus gastroenteritis after consuming raspberries, Sweden, June-August 2006. Euro Surveill. 2006;11(9):E060907.1. doi: 10.2807/esw.11.36.03038-en. [DOI] [PubMed] [Google Scholar]
- 46.Falkenhorst G., Krusell L., Lisby M. Imported frozen raspberries cause a series of norovirus outbreaks in Denmark, 2005. Euro Surveill. 2005;10(9):E050922.2. doi: 10.2807/esw.10.38.02795-en. [DOI] [PubMed] [Google Scholar]
- 47.Gaulin C.D., Ramsay D., Cardinal P. Epidemic of gastroenteritis of viral origin associated with eating imported raspberries. Can J Public Health. 1999;90(1):37–40. doi: 10.1007/BF03404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Guyader F.S., Mittelholzer C., Haugarreau L. Detection of noroviruses in raspberries associated with a gastroenteritis outbreak. Int J Food Microbiol. 2004;97(2):179–186. doi: 10.1016/j.ijfoodmicro.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 49.Dowell S.F., Groves C., Kirkland K.B. A multistate outbreak of oyster-associated gastroenteritis: implications for interstate tracing of contaminated shellfish. J Infect Dis. 1995;171(6):1497–1503. doi: 10.1093/infdis/171.6.1497. [DOI] [PubMed] [Google Scholar]
- 50.Morse D.L., Guzewich J.J., Hanrahan J.P. Widespread outbreaks of clam- and oyster-associated gastroenteritis. Role of Norwalk virus. N Engl J Med. 1986;314(11):678–681. doi: 10.1056/NEJM198603133141103. [DOI] [PubMed] [Google Scholar]
- 51.van Beek J., Ambert-Balay K., Botteldoorn N. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill. 2013;18(1):8–9. [PubMed] [Google Scholar]
- 52.Le Guyader F.S., Neill F.H., Dubois E. A semiquantitative approach to estimate Norwalk-like virus contamination of oysters implicated in an outbreak. Int J Food Microbiol. 2003;87(1–2):107–112. doi: 10.1016/s0168-1605(03)00058-8. [DOI] [PubMed] [Google Scholar]
- 53.Atmar R.L., Estes M.K. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin Microbiol Rev. 2001;14(1):15–37. doi: 10.1128/CMR.14.1.15-37.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green J., Norcott J.P., Lewis D. Norwalk-like viruses: demonstration of genomic diversity by polymerase chain reaction. J Clin Microbiol. 1993;31(11):3007–3012. doi: 10.1128/jcm.31.11.3007-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antonishyn N.A., Crozier N.A., McDonald R.R. Rapid detection of norovirus based on an automated extraction protocol and a real-time multiplexed single-step RT-PCR. J Clin Virol. 2006;37(3):156–161. doi: 10.1016/j.jcv.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 56.Kageyama T., Kojima S., Shinohara M. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41(4):1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richards G.P., Watson M.A., Fankhauser R.L. Genogroup I and II noroviruses detected in stool samples by real-time reverse transcription-PCR using highly degenerate universal primers. Appl Environ Microbiol. 2004;70(12):7179–7184. doi: 10.1128/AEM.70.12.7179-7184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoehne M., Schreier E. Detection of norovirus genogroup I and II by multiplex real-time RT-PCR using a 3'-minor groove binder-DNA probe. BMC Infect Dis. 2006;6:69. doi: 10.1186/1471-2334-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pang X.L., Preiksaitis J.K., Lee B. Multiplex real time RT-PCR for the detection and quantitation of norovirus genogroups I and II in patients with acute gastroenteritis. J Clin Virol. 2005;33(2):168–171. doi: 10.1016/j.jcv.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 60.Green J., Henshilwood K., Gallimore C.I. A nested reverse transcriptase PCR assay for detection of small round-structured viruses in environmentally contaminated molluscan shellfish. Appl Environ Microbiol. 1998;64(3):858–863. doi: 10.1128/aem.64.3.858-863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Neill H.J., McCaughey C., Wyatt D.E. Gastroenteritis outbreaks associated with Norwalk-like viruses and their investigation by nested RT-PCR. BMC Microbiol. 2001;1:14. doi: 10.1186/1471-2180-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishimura N., Nakayama H., Yoshizumi S. Detection of noroviruses in fecal specimens by direct RT-PCR without RNA purification. J Virol Methods. 2010;163(2):282–286. doi: 10.1016/j.jviromet.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 63.Boxman I.L., Verhoef L., Dijkman R. Year-round prevalence of norovirus in the environment of catering companies without a recently reported outbreak of gastroenteritis. Appl Environ Microbiol. 2011;77(9):2968–2974. doi: 10.1128/AEM.02354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Medici D., Suffredini E., Crudeli S. Effectiveness of an RT-booster-PCR method for detection of noroviruses in stools collected after an outbreak of gastroenteritis. J Virol Methods. 2007;144(1–2):161–164. doi: 10.1016/j.jviromet.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 65.Fukuda S., Takao S., Kuwayama M. Rapid detection of norovirus from fecal specimens by real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol. 2006;44(4):1376–1381. doi: 10.1128/JCM.44.4.1376-1381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iturriza-Gomara M., Xerry J., Gallimore C.I. Evaluation of the Loopamp (loop-mediated isothermal amplification) kit for detecting norovirus RNA in faecal samples. J Clin Virol. 2008;42(4):389–393. doi: 10.1016/j.jcv.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 67.Yoda T., Suzuki Y., Yamazaki K. Application of a modified loop-mediated isothermal amplification kit for detecting norovirus genogroups I and II. J Med Virol. 2009;81(12):2072–2078. doi: 10.1002/jmv.21626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duizer E., Pielaat A., Vennema H. Probabilities in norovirus outbreak diagnosis. J Clin Virol. 2007;40(1):38–42. doi: 10.1016/j.jcv.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 69.Castriciano S., Luinstra K., Petrich A. Comparison of the RIDASCREEN norovirus enzyme immunoassay to IDEIA NLV GI/GII by testing stools also assayed by RT-PCR and electron microscopy. J Virol Methods. 2007;141(2):216–219. doi: 10.1016/j.jviromet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 70.Costantini V., Grenz L., Fritzinger A. Diagnostic accuracy and analytical sensitivity of IDEIA norovirus assay for routine screening of human norovirus. J Clin Microbiol. 2010;48(8):2770–2778. doi: 10.1128/JCM.00654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gray J.J., Kohli E., Ruggeri F.M. European multicenter evaluation of commercial enzyme immunoassays for detecting norovirus antigen in fecal samples. Clin Vaccine Immunol. 2007;14(10):1349–1355. doi: 10.1128/CVI.00214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morillo S.G., Luchs A., Cilli A. Norovirus 3rd generation kit: an improvement for rapid diagnosis of sporadic gastroenteritis cases and valuable for outbreak detection. J Virol Methods. 2011;173(1):13–16. doi: 10.1016/j.jviromet.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 73.Siqueira J.A., Linhares Ada C., Oliveira Dde S. Evaluation of third-generation RIDASCREEN enzyme immunoassay for the detection of norovirus antigens in stool samples of hospitalized children in Belem, Para, Brazil. Diagn Microbiol Infect Dis. 2011;71(4):391–395. doi: 10.1016/j.diagmicrobio.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 74.MacCannell T., Umscheid C.A., Agarwal R.K. Guideline for the prevention and control of norovirus gastroenteritis outbreaks in healthcare settings. Infect Control Hosp Epidemiol. 2011;32(10):939–969. doi: 10.1086/662025. [DOI] [PubMed] [Google Scholar]
- 75.Bruggink L.D., Witlox K.J., Sameer R. Evaluation of the RIDA(®)QUICK immunochromatographic norovirus detection assay using specimens from Australian gastroenteritis incidents. J Virol Methods. 2011;173(1):121–126. doi: 10.1016/j.jviromet.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 76.Bruins M.J., Wolfhagen M.J., Schirm J. Evaluation of a rapid immunochromatographic test for the detection of norovirus in stool samples. Eur J Clin Microbiol Infect Dis. 2010;29(6):741–743. doi: 10.1007/s10096-010-0911-5. [DOI] [PubMed] [Google Scholar]
- 77.Kirby A., Gurgel R.Q., Dove W. An evaluation of the RIDASCREEN and IDEIA enzyme immunoassays and the RIDAQUICK immunochromatographic test for the detection of norovirus in faecal specimens. J Clin Virol. 2010;49(4):254–257. doi: 10.1016/j.jcv.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 78.Park K.S., Baek K.A., Kim D.U. Evaluation of a new immunochromatographic assay kit for the rapid detection of norovirus in fecal specimens. Ann Lab Med. 2012;32(1):79–81. doi: 10.3343/alm.2012.32.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thongprachum A., Khamrin P., Tran D.N. Evaluation and comparison of the efficiency of immunochromatography methods for norovirus detection. Clin Lab. 2012;58(5–6):489–493. [PubMed] [Google Scholar]
- 80.Pyo D., Yoo J. New trends in fluorescence immunochromatography. J Immunoassay Immunochem. 2012;33(2):203–222. doi: 10.1080/15321819.2011.618863. [DOI] [PubMed] [Google Scholar]
- 81.Buchan B, Anderson N, Jannetto P, et al. Simultaneous detection of influenza A and its subtypes (H1, H3, 2009 H1N1), influenza B, and RSV A and B in respiratory specimens on an automated, random access, molecular platform. 21st European Congress of Clinical Microbiology and Infectious Diseases (ECCMID). Milan, May 7, 2011.
- 82.Predmore A., Li J. Enhanced removal of a human norovirus surrogate from fresh vegetables and fruits by a combination of surfactants and sanitizers. Appl Environ Microbiol. 2011;77(14):4829–4838. doi: 10.1128/AEM.00174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feliciano L., Li J., Lee J. Efficacies of sodium hypochlorite and quaternary ammonium sanitizers for reduction of norovirus and selected bacteria during ware-washing operations. PLoS One. 2012;7(12):e50273. doi: 10.1371/journal.pone.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J., Predmore A., Divers E. New interventions against human norovirus: progress, opportunities, and challenges. Annu Rev Food Sci Technol. 2012;3:331–352. doi: 10.1146/annurev-food-022811-101234. [DOI] [PubMed] [Google Scholar]
- 85.Norovirus in healthcare facilities fact sheet. Centers for Disease Control and Prevention. Available at: http://www.cdc.gov/hai/pdfs/norovirus/229110-ANoroCaseFactSheet508.pdf. Accessed March 19, 2013.
- 86.Tian P., Yang D., Jiang X. Specificity and kinetics of norovirus binding to magnetic bead-conjugated histo-blood group antigens. J Appl Microbiol. 2010;109(5):1753–1762. doi: 10.1111/j.1365-2672.2010.04812.x. [DOI] [PubMed] [Google Scholar]
- 87.Bae J., Schwab K.J. Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl Environ Microbiol. 2008;74(2):477–484. doi: 10.1128/AEM.02095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baert L., Vandekinderen I., Devlieghere F. Efficacy of sodium hypochlorite and peroxyacetic acid to reduce murine norovirus 1, b40-8, Listeria monocytogenes, and Escherichia coli o157:H7 on shredded iceberg lettuce and in residual wash water. J Food Prot. 2009;72(5):1047–1054. doi: 10.4315/0362-028x-72.5.1047. [DOI] [PubMed] [Google Scholar]
- 89.Baert L., Uyttendaele M., Vermeersch M. Survival and transfer of murine norovirus 1, a surrogate for human noroviruses, during the production process of deep-frozen onions and spinach. J Food Prot. 2008;71(8):1590–1597. doi: 10.4315/0362-028x-71.8.1590. [DOI] [PubMed] [Google Scholar]
- 90.Gulati B.R., Allwood P.B., Hedberg C.W. Efficacy of commonly used disinfectants for the inactivation of calicivirus on strawberry, lettuce, and a food-contact surface. J Food Prot. 2001;64(9):1430–1434. doi: 10.4315/0362-028x-64.9.1430. [DOI] [PubMed] [Google Scholar]
- 91.Dawson D.J., Paish A., Staffell L.M. Survival of viruses on fresh produce, using MS2 as a surrogate for norovirus. J Appl Microbiol. 2005;98(1):203–209. doi: 10.1111/j.1365-2672.2004.02439.x. [DOI] [PubMed] [Google Scholar]
- 92.Lou F., Neetoo H., Chen H. Inactivation of a human norovirus surrogate by high-pressure processing: effectiveness, mechanism, and potential application in the fresh produce industry. Appl Environ Microbiol. 2011;77(5):1862–1871. doi: 10.1128/AEM.01918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lou F., Neetoo H., Li J. Lack of correlation between virus barosensitivity and the presence of a viral envelope during inactivation of human rotavirus, vesicular stomatitis virus, and avian metapneumovirus by high-pressure processing. Appl Environ Microbiol. 2011;77(24):8538–8547. doi: 10.1128/AEM.06711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leon J.S., Kingsley D.H., Montes J.S. Randomized, double-blinded clinical trial for human norovirus inactivation in oysters by high hydrostatic pressure processing. Appl Environ Microbiol. 2011;77(15):5476–5482. doi: 10.1128/AEM.02801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dancho B.A., Chen H., Kingsley D.H. Discrimination between infectious and non-infectious human norovirus using porcine gastric mucin. Int J Food Microbiol. 2012;155(3):222–226. doi: 10.1016/j.ijfoodmicro.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 96.Atmar R.L., Bernstein D.I., Harro C.D. Norovirus vaccine against experimental human Norwalk virus illness. N Engl J Med. 2011;365(23):2178–2187. doi: 10.1056/NEJMoa1101245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patel M.M., Widdowson M.A., Glass R.I. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14(8):1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mason H.S., Ball J.M., Shi J.J. Expression of Norwalk virus capsid protein in transgenic tobacco and potato and its oral immunogenicity in mice. Proc Natl Acad Sci U S A. 1996;93(11):5335–5340. doi: 10.1073/pnas.93.11.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang X.R., Buehner N.A., Hutson A.M. Tomato is a highly effective vehicle for expression and oral immunization with Norwalk virus capsid protein. Plant Biotechnol J. 2006;4(4):419–432. doi: 10.1111/j.1467-7652.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 100.Ball J.M., Hardy M.E., Atmar R.L. Oral immunization with recombinant Norwalk virus-like particles induces a systemic and mucosal immune response in mice. J Virol. 1998;72(2):1345. doi: 10.1128/jvi.72.2.1345-1353.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Souza M., Cheetham S.M., Azevedo M.S. Cytokine and antibody responses in gnotobiotic pigs after infection with human norovirus genogroup II.4 (HS66 strain) J Virol. 2007;81(17):9183–9192. doi: 10.1128/JVI.00558-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Souza M., Costantini V., Azevedo M.S. A human norovirus-like particle vaccine adjuvanted with ISCOM or MLT induces cytokine and antibody responses and protection to the homologous GII.4 human norovirus in a gnotobiotic pig disease model. Vaccine. 2007;25(50):8448. doi: 10.1016/j.vaccine.2007.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ball J.M., Graham D.Y., Opekun A.R. Recombinant Norwalk virus-like particles given orally to volunteers: phase I study. Gastroenterology. 1999;117(1):40–48. doi: 10.1016/s0016-5085(99)70548-2. [DOI] [PubMed] [Google Scholar]
- 104.El-Kamary S, Pasetti M, Tacket C, et al. Phase 1 dose escalation, safety and immunogenicity of intranasal dry powder norovirus vaccine. 3rd International Calicivirus Conference. Cancun, November 10, 2007.
- 105.Tacket C, Frey S, Bernstein D, et al. Phase 1 dose-comparison, safety and immunogenicity of intranasal dry powder Norwalk VLP vaccine. 5th International Conference on Vaccines for Enteric Diseases. Malaga, September 9, 2009.
- 106.Bok K., Parra G.I., Mitra T. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc Natl Acad Sci U S A. 2011;108(1):325–330. doi: 10.1073/pnas.1014577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smith G.L., Mackett M., Moss B. Infectious vaccinia virus recombinants that express hepatitis B virus surface antigen. Nature. 1983;302(5908):490–495. doi: 10.1038/302490a0. [DOI] [PubMed] [Google Scholar]
- 108.Harrington P.R., Yount B., Johnston R.E. Systemic, mucosal, and heterotypic immune induction in mice inoculated with Venezuelan equine encephalitis replicons expressing Norwalk virus-like particles. J Virol. 2002;76(2):730–742. doi: 10.1128/JVI.76.2.730-742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo L., Wang J., Zhou H. Intranasal administration of a recombinant adenovirus expressing the norovirus capsid protein stimulates specific humoral, mucosal, and cellular immune responses in mice. Vaccine. 2008;26(4):460–468. doi: 10.1016/j.vaccine.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 110.Guo L., Zhou H., Wang M. A recombinant adenovirus prime-virus-like particle boost regimen elicits effective and specific immunities against norovirus in mice. Vaccine. 2009;27(38):5233–5238. doi: 10.1016/j.vaccine.2009.06.065. [DOI] [PubMed] [Google Scholar]
- 111.Ma Y., Li J. Vesicular stomatitis virus as a vector to deliver virus-like particles of human norovirus: a new vaccine candidate against an important noncultivable virus. J Virol. 2011;85(6):2942. doi: 10.1128/JVI.02332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Duan Y., Ma Y., Hughes J. American Society for Virology; Madison (WI): 2012. A vesicular stomatitis virus (VSV)-based human norovirus vaccine provides protection against norovirus challenge in a gnotobiotic pig model. [Google Scholar]
- 113.Sekaly R.P. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J Exp Med. 2008;205(1):7–12. doi: 10.1084/jem.20072681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Desai R., Yen C., Wikswo M. Transmission of norovirus among NBA players and staff, winter 2010–2011. Clin Infect Dis. 2011;53(11):1115–1117. doi: 10.1093/cid/cir682. [DOI] [PubMed] [Google Scholar]
- 115.Repp K.K., Keene W.E. A point-source norovirus outbreak caused by exposure to fomites. J Infect Dis. 2012;205(11):1639–1641. doi: 10.1093/infdis/jis250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dore B., Keaveney S., Flannery J. Management of health risks associated with oysters harvested from a norovirus contaminated area, Ireland, February-March 2010. Euro Surveill. 2010;15(19):pii/19567. [PubMed] [Google Scholar]
- 117.Arvelo W., Sosa S.M., Juliao P. Norovirus outbreak of probable waterborne transmission with high attack rate in a Guatemalan resort. J Clin Virol. 2012 Sep;55(1):8–11. doi: 10.1016/j.jcv.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 118.Wadl M., Scherer K., Nielsen S. Food-borne norovirus-outbreak at a military base, Germany, 2009. BMC Infect Dis. 2010;10(30) doi: 10.1186/1471-2334-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koh S.J., Cho H.G., Kim B.H. An outbreak of gastroenteritis caused by norovirus-contaminated groundwater at a waterpark in Korea. J Korean Med Sci. 2011;26(1):28–32. doi: 10.3346/jkms.2011.26.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barrabeig I., Rovira A., Buesa J. Foodborne norovirus outbreak: The role of an asymptomatic food handler. BMC Infect Dis. 2010;10(269) doi: 10.1186/1471-2334-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yee E.L., Palacio H., Atmar R.L. Widespread outbreak of norovirus gastroenteritis among evacuees of hurricane katrina residing in a large “Megashelter” In Houston, Texas: Lessons learned for prevention. Clin Infect Dis. 2007;44(8):1032–1039. doi: 10.1086/512195. [DOI] [PubMed] [Google Scholar]
- 122.Becker K.M., Moe C.L., Southwick K.L. Transmission of norwalk virus during football game. N Engl J Med. 2000;343(17):1223–1227. doi: 10.1056/NEJM200010263431704. [DOI] [PubMed] [Google Scholar]
- 123.Tuladhar E., Hazeleger W.C., Koopmans M. Residual viral and bacterial contamination of surfaces after cleaning and disinfection. Appl Environ Microbiol. 2012;78(21):7769–7775. doi: 10.1128/AEM.02144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu P., Yuen Y., Hsiao H.M. Effectiveness of liquid soap and hand sanitizer against norwalk virus on contaminated hands. Appl Environ Microbiol. 2009;76(2):394–399. doi: 10.1128/AEM.01729-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nowak P., Topping J.R., Fotheringham V. Measurement of the virolysis of human gii.4 norovirus in response to disinfectants and sanitisers. J Virol Methods. 2011;174(1-2):7–11. doi: 10.1016/j.jviromet.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 126.Park G.W., Barclay L., Macinga D. Comparative efficacy of seven hand sanitizers against murine norovirus, feline calicivirus, and gii.4 norovirus. J Food Prot. 2011;73(12):2232–2238. doi: 10.4315/0362-028x-73.12.2232. [DOI] [PubMed] [Google Scholar]
- 127.Li D., Baert L., Van Coillie E., Uyttendaele M. Critical studies on binding-based rt-pcr detection of infectious noroviruses. J Virol Methods. 2011;177(2):153–159. doi: 10.1016/j.jviromet.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 128.Hudson J.B., Sharma M., Petric M. Inactivation of norovirus by ozone gas in conditions relevant to healthcare. J Hosp Infect. 2007;66(1):40–45. doi: 10.1016/j.jhin.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 129.Tacket C.O., Sztein M.B., Losonsky G.A. Humoral, mucosal, and cellular immune responses to oral Norwalk virus-like particles in volunteers. Clin Immunol. 2003;108(3):241–247. doi: 10.1016/s1521-6616(03)00120-7. [DOI] [PubMed] [Google Scholar]
- 130.Ma Y., Li J. Vesicular stomatitis virus as a vector to deliver virus like particles of human norovirus: a new vaccine candidate against an important noncultivable virus. J Virol. 2011;85(6):2942–2952. doi: 10.1128/JVI.02332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]