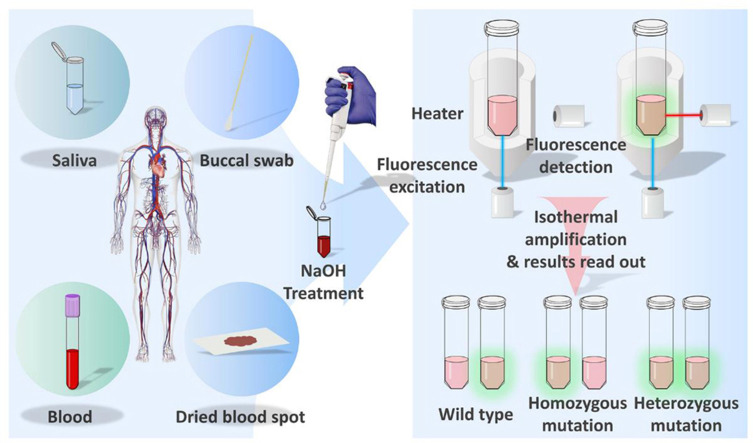
Abstract
Genotyping of single nucleotide polymorphisms (SNPs) in point-of-care (POC) settings could be further improved through simplifying the treatment of samples. In this study, we devised an accurate, rapid and easy-to-use SNP detection system based on direct loop-mediated isothermal amplification (LAMP) without DNA extraction, known as Direct-LAMP. Samples from various sources (including whole blood, dried blood spot, buccal swab and saliva), treated with NaOH, can be used directly in amplification. The turnaround time was about 30 min from sample collection to provision of results. The accuracy was evaluated by assessing the polymorphisms of methylenetetrahydrofolate reductase (MTHFR) C677T and aldehyde dehydrogenase-2 (ALDH2) Glu504Lys, which are better known for their critical role in folate and ethanol metabolism, respectively. Completely consistent genotyping results reveal that Direct-LAMP is generally concordant with sequencing. This system can serve as a very promising platform in the fields of disease predisposition, drug metabolism and personalized medicine.
Keywords: Single nucleotide polymorphism, Isothermal amplification, Multifarious sample types, MTHFR, ALDH2
Graphical abstract
Highlights
-
•
Devised a SNP detection system combining DNA amplification directly with LAMP without DNA purification.
-
•
Biological samples for the Direct-LAMP system are various.
-
•
The genotyping result can be provided within 30 min.
-
•
The Direct-LAMP system can serve as a promising POCT for genotyping.
1. Introduction
The most abundant source of genetic variation (approximately 90%) in the human genome is represented by single nucleotide polymorphisms (SNPs), which can account for heritable inter-individual differences in complex phenotypes (Mhlanga and Malmberg, 2001, Yu et al., 2006). Hence, SNP detection has great potential for direct clinical application by providing highly accurate diagnostic information for facilitating early diagnosis, prevention, and treatment of genetic diseases (Everitt et al., 2012, Moore et al., 2010). Because of its accuracy, DNA sequencing is considered to be the gold standard for SNP detection analysis. However, given the subsequent massive data analysis required by the method, sequencing is more suitable for discovery of novel SNPs rather than detection of specific known SNPs (Grant et al., 2011, Hendre et al., 2012, Muhammad et al., 2013). Other technologies rely on sample amplification, including DNA microarrays (Li et al., 2013, Michal et al., 2010, Zhang et al., 2010), real-time polymerase chain reaction (RT-PCR) (Martinez-Serra et al., 2014, Psifidi et al., 2011), self-sustained sequence replication (3SR) (Ren et al., 2014), strand displacement amplification (SDA) (Toley et al., 2015), high resolution melting (HRM) (Norambuena and Copeland, 2009), Mass ARRAY (Kriegsmann et al., 2015, Suthandiram et al., 2014), and multiplex PCR-RFLP method (Loo et al., 2012) also offer sensitive approaches for SNP detection. However, the tremendous potential of SNP detection in clinical practice has so far often been limited by the lack of efficient screening methods which cannot meet the requirements of point-of-care (POC) testing strategies due to their complex experimental procedures and long testing times (Tost and Gut, 2005). Based on the status quo, development of a new detection platform that shortens assay times and simplifies procedures would be highly desirable. Saving time from DNA purification, a conventional step in SNP detection that not only complicates the procedure, but also enhances the risk of invalid results and cross-contamination, is a promising approach. It has been reported that new SNP detection methods have been developed based on PCR that do not require DNA extraction from whole blood, hair root and saliva (Cascella et al., 2015, Hallie and Reena, 2015, Hayashida et al., 2009). By treating them with specific chemicals, samples can be used directly in PCR without an initial DNA purification step, which would save approximately 30 min compared with conventional SNP detection systems. However, PCR-based detection methods still require long processing times due to thermal cycling. In order to shorten the testing time further, it would be interested to adopt another simple and fast detection method, instead of PCR.
Loop-mediated isothermal amplification (LAMP) is a highly specific, sensitive, and rapid gene amplification method that was established by Notomi et al. (Notomi et al., 2000). This method amplifies nucleic acids under isothermal conditions and employs self-recurring strand-displacement synthesis primed by a specially designed set of target-specific primers (Duan et al., 2014, Tomita et al., 2008). LAMP has been used for detection of pathogens via the amplification of gene segments, including Japanese encephalitis virus infection (Mori et al., 2013), rapid genotyping of carcinogenic human papillomavirus and herpesvirus (Mori et al., 2013), and the detection of Middle East Respiratory Syndrome Coronavirus (Bhadra et al., 2015, Mori et al., 2013, Notomi et al., 2015), demonstrating the method's great potential in molecular diagnostics. Based on the advantages of this technology, LAMP has been used for genotyping with buccal swab samples, combining gold nanoparticles functionalized with ssDNA and colorimetric turbidimetry detection (Carlos et al., 2017). Although it no longer requires DNA purification, this method still takes 3 h to perform due to its complex experimental procedure.
Methylenetetrahydrofolate reductase (MTHFR) is the key enzyme in folate metabolism. Mutations in the MTHFR gene would influence the activity of this enzyme and increase the risk of various diseases, such as cancer, neurological disorders, cardiovascular diseases, and pregnancy complications (Nazik et al., 2014). Aldehyde dehydrogenase-2 (ALDH2) is responsible for the oxidation of aldehydes in the liver. Differences in ALDH2 expression contribute to a wide variety of human diseases, including cardiovascular disease, diabetes, and cancer. In addition, genetic polymorphisms of ALDH2 alter susceptibility to ethanol intake, as well as the risk of alcoholism and alcoholic complications, and ALDH2 may possess important therapeutic potential against alcoholism and other forms of myocardial damage (Chen et al., 2014).
In this study, we devised a Direct-LAMP procedure, amplifying nucleic acids with various samples (including whole blood, dried blood spot, buccal swab and saliva) without DNA purification, which is essential for conventional nucleic acid detection methods. The turnaround time was about 30 min from sample collection to provision of results. MTHFR C677T, and ALDH2 Glu504Lys were utilized as models for SNP detection with the Direct-LAMP method. Accuracy was evaluated against clinical samples, and complete consistency between genotyping results reveals that this Direct LAMP procedure generally concords with sequencing.
2. Material and methods
2.1. Direct-LAMP design
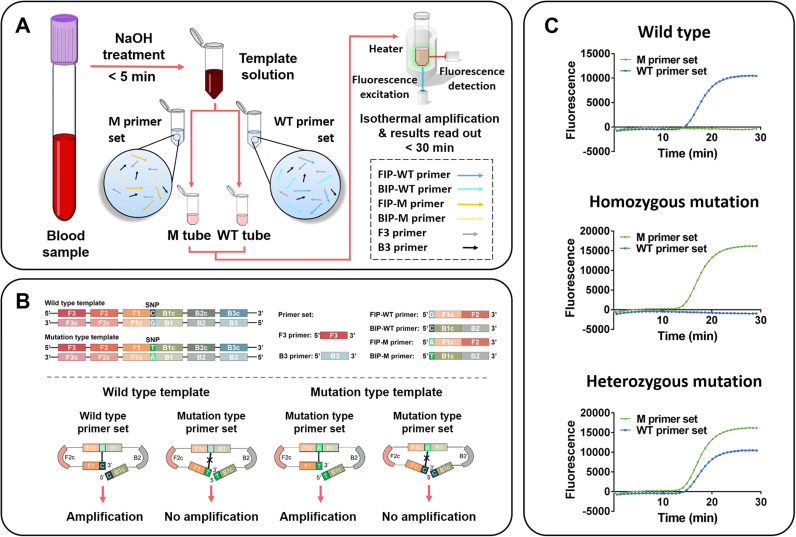
The proof of concept scheme described in this paper uses Direct-LAMP for SNP detection. As exhibited in Fig. 1A, samples are treated with NaOH solution, followed by amplification under isothermal conditions using specifically designed primers. For convenience, we refer to the wild type as “WT” and the mutation type as “M”. For each sample to be detected, two separate reactions (M tube and WT tube) are run simultaneously using the same sample. Two sets of allele-specific primers (WT sequence and M sequence) are added to two different tubes for amplification, respectively. The result can be obtained automatically through a real-time fluorometer. The operating procedures of the Direct-LAMP only take 30 min from sample collection to result.
Fig. 1.
Schematic diagram of Direct-LAMP. (A) Procedure of Direct-LAMP. (B) Schematic diagram of Direct-LAMP reaction process. (C) Genotyping results analyzing based on the signal read-out through a real-time fluorometer automatically.
Based on point mutations in the MTHFR C677T and ALDH2 Glu504Lys genes, four specific primers were designed to discriminate each SNP. As shown in Fig. 1B, the Direct-LAMP procedure relies on the specific primers, with forward inner primer (FIP) and backward inner primer (BIP) that are designed to contain a SNP nucleotide at the 5′ terminus. Each reaction includes two common primers (F3 and B3) and two special primers (FIP-WT and BIP-WT for wild type allele or FIP-M and BIP-M for mutation type allele). The fluorescence signal coming along with target fragment amplification would be read in real-time only when the 5′-end of the specific primer is complementary with the template. The structures of the specific primers used in this study are presented in Fig. 1B. The target SNP was characterized by using four different primer regions specifically designed to recognize six distinct regions on the target gene, which were selected to ensure that the primers would specifically amplify the C677T and Glu504Lys substitutions. The result depicted in Fig. 1C illustrates that the Direct-LAMP could accurately detect all possible homozygotes and heterozygotes of MTHFR C677T with whole blood sample.
2.2. Primer design and synthesis
For MTHFR C677T and ALDH2 Glu504Lys polymorphism genotyping, respectively, two sets of primers for Direct-LAMP were designed using the Primer 5.0 software program (Primer-E Ltd., Plymouth, UK), including two outer primers (F3 and B3) and four inner primers (FIP-WT, FIP-M, BIP-WT and BIP-M) that recognize six distinct regions of each target gene. Two primers were also designed for sequencing. The sequences of the primers are shown in Supplementary Table S1. All primers were synthesized by Invitrogen Biotechnology Ltd. (Shanghai, China).
2.3. Sample preparation
Matched fresh human whole blood and dried blood spot samples were collected from 50 unrelated Chinese volunteers at the Shaanxi Provincial People's Hospital (Xi’an, China) with informed consent. Whole blood sample was collected using EDTA-coated tubes. Dried blood spot sample was prepared by dropping the peripheral blood on the filter paper and drying in the air. Matched saliva and buccal swab samples were obtained from 50 Chinese volunteers at Northwest University (Xi’an, China) with informed consent. Saliva samples were collected in Eppendorf tubes. Buccal swab sample was collected by swabbing the insides of cheeks 40 times. The study was approved by the Ethics Committee of the College of Life Sciences, Northwest University (Xi’an, China).
Peripheral blood and saliva samples were collected in EDTA-coated tubes and Eppendorf tubes, respectively, followed by mixing of the samples with 100 mM NaOH in a 1:2 ratio, with a final volume of 30 μL, and incubation at room temperature for 3 min. In total, 1 μL of the mixture was taken for subsequent amplification. The buccal swab head was cut off (about 5 mm under the head) and placed into 100 μL of 100 mM NaOH, followed by incubation at room temperature for 3 min, and 1 μL of the mixture was taken for subsequent amplification.
Dried blood spot of 5 mm diameter was put into an Eppendorf tube and mixed with 100 μL of 100 mM NaOH, followed by incubation at room temperature for 3 min, and 1 μL of the mixture was taken for subsequent amplification. The mixture must be used immediately after preparation. Samples treated with NaOH were observed using an optical microscope (IX71, Olympus Optical Co., Ltd., Tokyo, Japan) with 100 W halogen light source (U-LH100L-3, Olympus); cells to be observed were stained with rapid Wright-Giemsa Staining Solution Kit (Sangon Biotech Co., Ltd., Shanghai, China).
2.4. Optimization of the Direct-LAMP
The initial conditions of the Direct-LAMP procedure were adopted as described by Zhang et al. (2016). The Direct-LAMP was performed with different combinations of primer concentration (0.2 μM FIP/BIP with 0.05 μM F3/B3, 0.4 μM FIP/BIP with 0.1 μM F3/B3, 0.8 μM FIP/BIP with 0.2 μM F3/B3 and 1.2 μM FIP/BIP with 0.3 μM F3/B3) to determine the best primer concentration. The best reaction temperature was also investigated by running the Direct-LAMP at 58, 59, 60, 61, or 62 °C for 30 min.
The four sample types were used to establish the Direct-LAMP, and the genotypes of the used samples had been sequenced. The reaction mixture contained 15 μL of Isothermal Master Mix (OptiGene Ltd., UK) (including Geobacillus DNA polymerase, thermostable inorganic pyrophosphatase, optimized buffer including MgCl2, dNTPs and ds-DNA dye), 5 μL of primer mix consisting of four primers each for F3 and B3 primers at 0.2 μM, FIP and BIP primers at 0.8 μM, 1 μL sample treated solution, and ddH2O up to 25 μL. The reaction was performed in 8-well 0.2-mL tubes, with incubation at 60 °C for 30 min. The fluorescence intensity of the ds-DNA dye was simultaneously monitored in a real-time fluorometer (Genie II from OptiGene Ltd. UK).
2.5. Evaluation of the Direct-LAMP
The detection limit of optimized Direct-LAMP was evaluated by a serial dilution of the target concentrations (100%, 50%, 25%, 12.5%, 6.25%, 3.125%, 1.5625%) of whole blood sample and saliva sample with two different genotypes (wild type and homozygous mutation, confirmed by sequencing) of MTHFR C677T and ALDH2 Glu504Lys. The four sample types with two different genotypes (wild type and homozygous mutation which confirmed by sequencing) of MTHFR C677T and ALDH2 Glu504Lys, respectively, were used to evaluate the specificity of the detection system.
2.6. Clinical application of the Direct-LAMP
The accuracy of the optimized Direct-LAMP was further verified using clinical samples, which had been obtained with informed consent. The study was approved by the Ethics Committee of the College of Life Sciences, Northwest University (Xi’an, China). The genotype of each sample was analyzed by Direct-LAMP and compared with the results obtained via sequencing by BGI (Beijing Genomic Institute, Beijing, China). Based on the statistical data, the coincidence rate and total agreements of three genotypes of MTHFR C677T and ALDH2 Glu504Gly were calculated to evaluate the accuracy of our method.
3. Results
3.1. Effect of NaOH on the Direct-LAMP
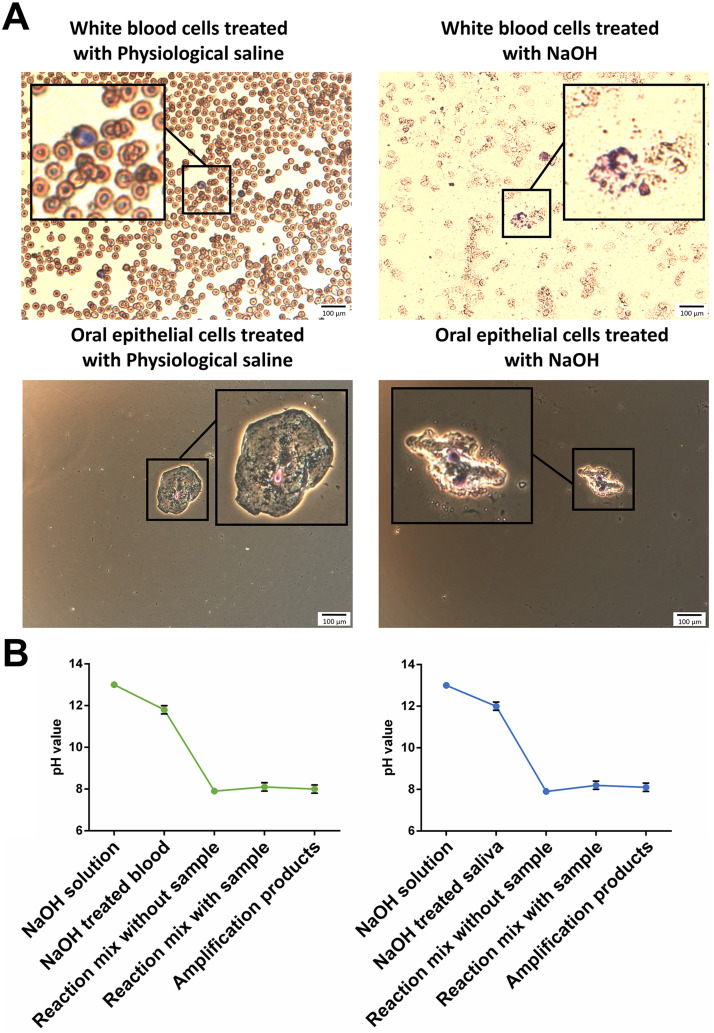
In the current study, samples were treated with NaOH to induce cells lysis and DNA release, saving time ordinarily spent on extraction of DNA from sample. To evaluate the effect of NaOH on the performance of Direct-LAMP, the morphologies of leukocytes and oral epithelial cells in whole blood and saliva were observed, and the pH value at each step of Direct-LAMP process was assessed. In Fig. 2A, a great number of cellular debris were found with the whole blood treated with NaOH, while integrated cells were observed when blood was incubated with physiological saline. Similar results were found for saliva treated with NaOH and physiological saline, respectively. The results indicated that NaOH treatment lead to cell lysis, followed by release of DNA for use in subsequent amplification.
Fig. 2.
(A) Micrographs of whole blood and saliva treated with physiological saline and NaOH, respectively. (B) pH value at each stage of Direct-LAMP.
In Fig. 2B, it is shown that the pH value of NaOH solution was 13.0, and by adding the NaOH solution to blood sample and saliva sample, the pH value of the mixture was 11.8–12.0. However, the pH value drops dramatically to 8.1–8.2 by mixing the NaOH treated sample with reaction buffer, and no significant difference in pH value was observed between the reaction buffer with and without NaOH treated sample. The reason can be explained as the buffering effect of Tris-HCl buffer, which provided a benign working environment for DNA polymerase, according to Verma et al. (2014).
3.2. Optimization of the Direct-LAMP
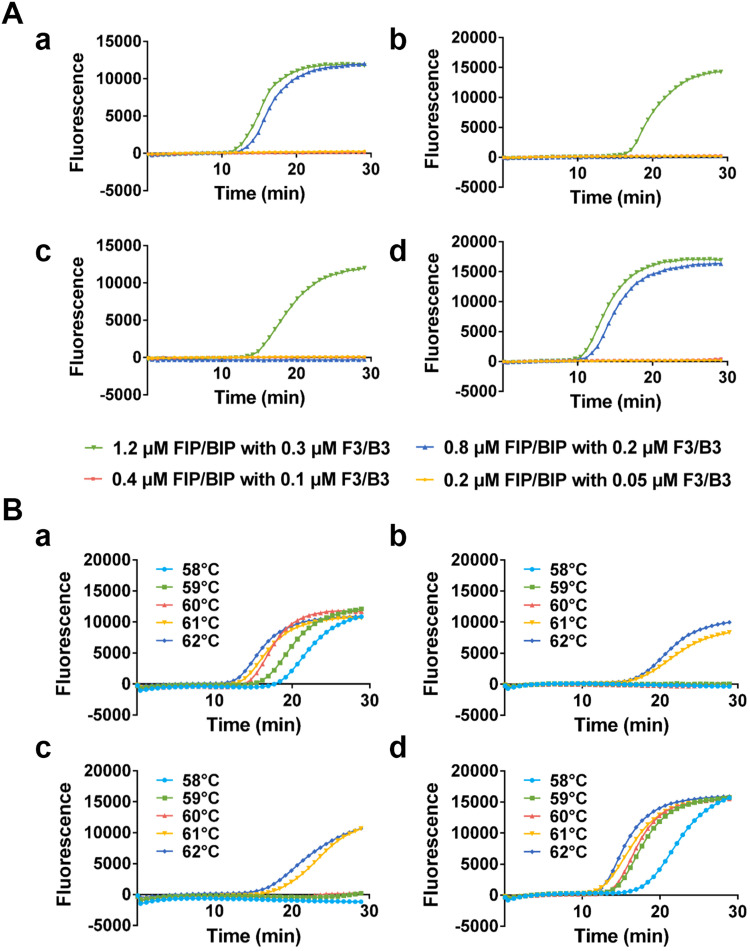
To determine the best working condition, the performance of the Direct-LAMP was evaluated under different concentrations of primers and reaction temperatures by using whole blood sample with two different genotypes of MTHFR C677T (wild type and homozygous mutation, which were confirmed by sequencing). Firstly, the best combination of primer concentrations was found to be 0.8 μM of the FIP/BIP and 0.2 μM of the F3/B3 ( Fig. 3A). The reaction temperature of Direct-LAMP was also optimized, as shown in Fig. 3B; the best amplification efficiency and specificity was obtained when the reaction temperature was 60 °C.
Fig. 3.
Fluorescence curve of Direct-LAMP performed with (A) different reaction temperatures ((a) WT primer set with WT template (b) WT primer set with M template (c) M primer set with WT template (d) M primer set with M template) and (B) different primer concentrations ((a) WT primer set with WT template (b) WT primer set with M template (c) M primer set with WT template (d) M primer set with M template).
3.3. Performance of the Direct-LAMP
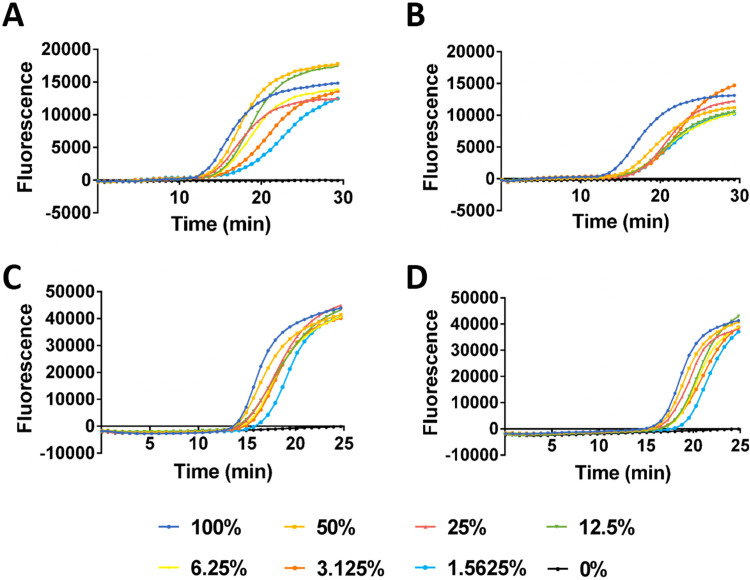
To evaluate the performance of the Direct-LAMP, a serial dilution of the target concentrations of whole blood sample and saliva sample with two different genotypes (wild type and homozygous mutation which confirmed by sequencing) of MTHFR C677T and ALDH2 Glu504Lys, respectively, were used to determine the detection limit. The amplification efficiency reflected by the threshold time is inversely proportional to the dilution ratio of biological sample ( Fig. 4). Accurate genotyping results were still obtained with the Direct-LAMP when biological sample was diluted to 64 times. The specificity of the detection system for every target and sample type was evaluated, accurate genotyping results were still obtained under interference of background in all cases (Supplementary Fig. S1 and Fig. S2), demonstrating great potential for direct clinical application.
Fig. 4.
Fluorescence curve of Direct-LAMP performed with different percentages of whole blood which contain (A) homozygous mutation gene and (B) wild type gene for MTHFR C677T genotyping. Fluorescence curve of Direct-LAMP performed with different percentages of saliva which contain (C) homozygous mutation gene and (D) wild type gene for ALDH2 Glu504Lys genotyping.
3.4. Evaluation of the Direct-LAMP with clinical samples
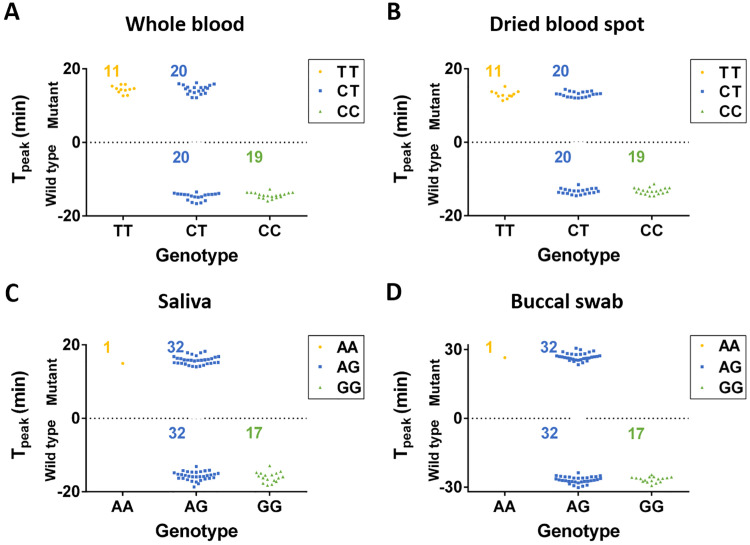
The accuracy of the Direct-LAMP was further verified with clinical samples (matched 50 whole blood and 50 dried blood spot samples, matched 50 buccal swab and 50 saliva samples) with informed consent. The study was approved by the Ethics Committee of the College of Life Sciences, Northwest University (Xi’an, China). MTHFR C677T of whole blood samples and ALDH2 Glu504Lys of saliva sample were also sequenced by BGI. In Fig. 5, the threshold time of wild type were defined as a negative value, the threshold time of mutation type were defined as a positive value. For MTHFR C677T genotyping, 11 samples of homozygous mutation, 19 samples of wild type and 20 samples of heterozygous mutation were observed with whole blood in Fig. 5A, which was same as those found with dried blood spot in Fig. 5B. For ALDH2 Glu504Lys genotyping, 1 sample of homozygous mutation, 32 samples of wild type and 17 samples of heterozygous mutation were observed with saliva in Fig. 5C, which was same as those found with buccal swab in Fig. 5D. The genotyping results provided by the Direct-LAMP showed no discrepancies with those found with sequencing. As shown in Supplementary Table S2, the derived allele frequencies of MTHFR C677T based on the Direct-LAMP detection were 58% and 42% for C and T, respectively (calculated from: C: F1 +F2/2, T: F2/2 +F3), which is in agreement with those reported by Hui et al. (Hui et al., 2016). As shown in Supplementary Table S3, the observed allele frequencies of ALDH2 Glu504Lys were 66% and 34% for G and A respectively, which is also in agreement with those reported by Eng et al. (2007) in the Chinese population.
Fig. 5.
Genotyping result with multiple sample types. (A) Genotyping result of MTHFR C677T with 50 whole blood samples. (B) Genotyping result of MTHFR C677T with 50 dried blood spot samples. (C) Genotyping result of ALDH2 Glu504Lys with 50 saliva samples. (D) Genotyping result of ALDH2 Glu504Lys with 50 buccal swab samples. (the threshold time of wild type were defined as negative).
4. Discussion
Loop-mediated isothermal amplification (LAMP) was established by Notomi et al. and this method has been used for detection of pathogens via the amplification of gene segments, demonstrating the method's great potential in molecular diagnostics. Based on the advantages of this technology, some researchers are working on this technology can be applied for SNPs detection. A few reports indicated that this method has been applied for SNPs detection successfully (Carlos et al., 2017, Iwasaki et al., 2003, Kwong et al., 2017, Kwong et al., 2014, Nakamura et al., 2007, Nakamura and Ito, 2009, Zhang et al., 2016), which shorten the testing time further than PCR. However, DNA purification or complex clinical sample pre-treatment are required for SNPs detection in these methods. The sample pre-treatment procedures including repeating incubation, elution and centrifugation consume a lot of time and energy during the detection process. Although commercial kits simplify the procedure of DNA purification and clinical sample pre-treatment, there are restrictions on the clinical sample types, such as colorless clinical samples are required (FTA™ Indicated Micro Card, Whatman, UK).
The need for rapid SNPs detection to provide highly accurate diagnostic information is realized by all major health organizations, including the Centers for Disease Control and Prevention and the World Health Organization. However, no method combining LAMP exists for SNPs detection that can be performed within a single patient visit (30 min) directly from various clinical samples. Here, we have established a rapid, easy-to-use and accurate SNP detection platform using Direct-LAMP, which enables us to use whole blood, dried blood spot, buccal swab or saliva as samples for genotyping without DNA purification. The whole testing procedure required only 30 min from sample collection to result, which is much faster than with conventional SNP detection systems. This is the first report to introduce the pretreatment of various kinds of samples with a simple NaOH solution before LAMP for genotyping. Designed as a universal genotyping platform, this Direct-LAMP protocol has been successfully applied not only to the four sample types previously mentioned, but also verified with clinical sample, indicating that this genotyping platform could be applied to more sample types in further study.
The treatment of sample with NaOH plays an important role in this Direct-LAMP, making the whole testing procedure convenience and useable. First of all, compared with other SNP detection methods, the DNA purification step is eliminated in our assay with the help of NaOH treatment, reducing the sample preparation time. Secondly, using NaOH-treated sample simplifies the complex procedures of conventional genotyping methods and eliminates the problem of cross-contamination, which happened in traditional DNA purification processes. Furthermore, various natural DNA polymerase inhibitors in whole blood, such as hemoglobin, IgG, lactoferrin, and proteases (Adams, 2005, Connelly et al., 2014, Monroe et al., 2013), can be inactivated by treatment with NaOH solution.
By using Direct-LAMP, qualitative analysis can be carried out by reading the fluorescence signal automatically in real time. The major advantage of this assay system is rapid qualitative answer in ‘‘Yes’’ or ‘‘No’’ according to the fluorescence signal. Compared with conventional SNP detection methods based on PCR or DNA sequencing, Direct-LAMP shows significant advantages (Jung et al., 2015, Ngo et al., 2016). For conventional genotyping methods that require purified DNA as template, such as PCR-microarray, qPCR and DNA sequencing, the process of DNA purification required a fair amount of time during the whole testing, whereas the present assay system requires less than 5 min for sample preparation. Traditional genotyping techniques based on PCR always require more than 1.5 h for DNA amplification, whereas the present methods based on LAMP shorten the testing time further. The process of nucleic acid amplification under isothermal conditions, employing self-recurring strand-displacement synthesis primed by a specially designed set of target-specific primers, enable this special identification system, which is faster than PCR-based methods, which use only two primers to recognize two regions (Duan et al., 2014, Tomita et al., 2008). Thus, compared with conventional SNP detection techniques usually requiring a complex operation procedure that limits their application, the present methods provide an easy-to-operate on-site technique for genotyping with high efficiency. Therefore, the Direct-LAMP will greatly benefit molecular diagnostics by dramatically reducing turnaround time, which makes this SNP detection method very promising for POC testing in clinical diagnostics. With further development for additional SNPs and sample types, Direct-LAMP could enable rapid clinical decision-making, improve management of disease prevention and facilitate personalized medicine in each level of medical institutions.
5. Conclusions
In this study, we presented a Direct-LAMP for SNP detection by using whole blood, dried blood spot, buccal swab or saliva as samples without DNA purification. This is the first report that a SNPs detection platform combined the direct amplification and LAMP for genotyping with various clinical sample validation. Our data have demonstrated that this system is highly applicable for detection of SNPs in clinical samples due to its unparalleled advantages, such as short processing time, simple procedure and accuracy. Since detection of SNPs has significant clinical value, in our laboratory this method has already been extended to detection of SNPs in other genes related to folic acid metabolism. It is envisaged that our newly established Direct-LAMP could be quickly adapted for the detection of other SNPs of genes that are associated with disease risk, drug metabolism, or drug reaction.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81772289), National Natural Science Foundation of China (31771083), Shaanxi Provincial Nano-Biomedical Detection Innovation Team Founds (2017KCT-25) and Northwest University Graduate Innovation and Creativity Funds (YZZ15008).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bios.2018.05.021.
Contributor Information
Kai Hua, Email: kaihua@nwu.edu.cn.
Yali Cui, Email: yalicui@nwu.edu.cn.
Appendix A. Supplementary material
Supplementary material
References
- Adams D.N. J. Clin. Microbiol. 2005;43:2932–2933. doi: 10.1128/JCM.43.6.2932-2933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. PLoS One. 2015;10:e0123126. doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos F.F., Veigas B., Matias A.S., Doria G., Flores O., Baptista P.V. Biotechnol. Rep. 2017;16:21–25. doi: 10.1016/j.btre.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella R., Strafella C., Ragazzo M., Zampatti S., Borgiani P., Gambardella S., Pirazzoli A., Novelli G., Giardina E. Pharm. J. 2015;15:196–200. doi: 10.1038/tpj.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H., Ferreira J.C., Gross E.R., Mochlyrosen D. Physiol. Rev. 2014;94:1. doi: 10.1152/physrev.00017.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C.M., Porter L.R., Termaat J.R. BMC Med. Genet. 2014;15:1–9. doi: 10.1186/s12881-014-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Zhang X., Ge C., Wang Y., Cao J., Jia X., Wang J., Zhou M. Sci. Rep. 2014;4:7094. doi: 10.1038/srep07094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng M.Y., Luczak S.E., Wall T.L. Alcohol Res. Health J. Natl. Inst. Alcohol Abus. Alcohol. 2007;30:22–27. [PMC free article] [PubMed] [Google Scholar]
- Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J. Int. J. Infect. Dis. 2012;484:519–523. [Google Scholar]
- Grant J.R., Arantes A.S., Liao X., Stothard P. Bioinformatics. 2011;27:2300–2301. doi: 10.1093/bioinformatics/btr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallie A.M.P.S., Reena R.P.D. J. Forensic Sci. 2015;60:1542–1552. [Google Scholar]
- Hayashida M., Iwao-Koizumi K., Murata S., Kinoshita K. Analytical Sciences the International. J. Jpn. Soc. Anal. Chem. 2009;25:1487. doi: 10.2116/analsci.25.1487. [DOI] [PubMed] [Google Scholar]
- Hendre P.S., Kamalakannan R., Varghese M. Plant Biotechnol. J. 2012;10:646–656. doi: 10.1111/j.1467-7652.2012.00699.x. [DOI] [PubMed] [Google Scholar]
- Hui W., Zhang S., Zhang C., Wan Y., Zhu J., Zhao G., Wu S., Xi D., Zhang Q., Li N. Nanoscale. 2016;8:3579. doi: 10.1039/c5nr07547e. [DOI] [PubMed] [Google Scholar]
- Iwasaki M., Yonekawa T., Otsuka K., Suzuki W., Nagamine K., Hase T., Tatsumi K.I., Horigome T., Notomi T., Kanda H. Genome Lett. 2003;2(3):119–126. [Google Scholar]
- Jung Y.K., Kim J., Mathies R.A. Anal. Chem. 2015;87:3165. doi: 10.1021/ac5048696. [DOI] [PubMed] [Google Scholar]
- Kriegsmann M., Arens N., Endris V., Weichert W., Kriegsmann J. Diagn. Pathol. 2015;10:132. doi: 10.1186/s13000-015-0364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong K.M., Tam C.C., Chan R., Lee W.L., Lau R., Tung E.K.K., Kwok J. Hum. Immunol. 2014;75(6) (482-482) [Google Scholar]
- Kwong K.M., Tam C.C., Chan R., Lee S., Ip P., Kwok J. Clin. Chim. Acta. 2017:478. doi: 10.1016/j.cca.2017.12.013. [DOI] [PubMed] [Google Scholar]
- Li J., Huang Y., Wang D., Song B., Li Z., Song S., Wang L., Jiang B., Zhao X., Yan J. Chem. Commun. 2013;49:3125–3127. doi: 10.1039/c3cc40680f. [DOI] [PubMed] [Google Scholar]
- Loo K.W., Griffiths L.R., Gan S.H. BMC Med. Genet. 2012;13:1–9. doi: 10.1186/1471-2350-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Serra J., Robles J., Nicolàs A., Gutierrez A., Ros T., Amat J.C., Alemany R., Vögler O., Abelló A., Noguera A. J. Blood Med. 2014;5:99. doi: 10.2147/JBM.S64976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhlanga M.M., Malmberg L. Methods. 2001;25:463–471. doi: 10.1006/meth.2001.1269. [DOI] [PubMed] [Google Scholar]
- Michal M., Harma F., Nijman I.J., Ewart D.B., Van dZ.P.J., Victor G., Edwin C. Nucleic Acids Res. 2010;38 (e116-e116) [Google Scholar]
- Monroe C., Grier C., Kemp B.M. Forensic Sci. Int. 2013;228:142. doi: 10.1016/j.forsciint.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Moore J., Mcknight A.J., Simmonds M.J., Courtney A.E., Hanvesakul R., Brand O.J., Briggs D., Ball S., Cockwell P., Patterson C.C. Jama. 2010;303:1282. doi: 10.1001/jama.2010.356. [DOI] [PubMed] [Google Scholar]
- Mori Y., Kanda H., Notomi T. J. Infect. Chemother. 2013;19:404–411. doi: 10.1007/s10156-013-0590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad A., Li X., Lundberg A.E., Marcin K., Siegel P.B., Örjan C., Stefan M. Front. Genet. 2013;4:226. doi: 10.3389/fgene.2013.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Ito K.M. Clin. Biochem. 2009;42(10–11):1158. doi: 10.1016/j.clinbiochem.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Ito K., Takahashi M., Hashimoto K., Kawamoto M., Yamanaka M., Taniguchi A., Kamatani N., Gemma N. Anal. Chem. 2007;79(24):9484–9493. doi: 10.1021/ac0715468. [DOI] [PubMed] [Google Scholar]
- Nazik F.H., Sameer A.S., Ganaie B.A. Gene. 2014;533:11. doi: 10.1016/j.gene.2013.09.063. [DOI] [PubMed] [Google Scholar]
- Ngo H.T., Gandra N., Fales A.M., Taylor S.M., Vodinh T. Biosens. Bioelectron. 2016;81:8–14. doi: 10.1016/j.bios.2016.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norambuena P.A., Copeland J.P. Clin. Biochem. 2009;42:1308–1316. doi: 10.1016/j.clinbiochem.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Mori Y., Tomita N., Kanda H. J. Microbiol. 2015;53:1–5. doi: 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- Psifidi A., Dovas C., Banos G. PLoS One. 2011;6:e14560. doi: 10.1371/journal.pone.0014560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R., Wang L.L., Ding T.R., Li X.M. Biosens. Bioelectron. 2014;54:122–127. doi: 10.1016/j.bios.2013.10.063. [DOI] [PubMed] [Google Scholar]
- Suthandiram S., Gan G.G., Zain S.M., Haerian B.S., Ping C.B., Lian L.H., Chang K.M., Ong T.C., Mohamed Z. J. Hum. Genet. 2014;59:280–287. doi: 10.1038/jhg.2014.19. [DOI] [PubMed] [Google Scholar]
- Toley B.J., Covelli I., Belousov Y., Ramachandran S., Kline E., Scarr N., Vermeulen N., Mahoney W., Lutz B.R., Yager P. Analyst. 2015;140:7540–7549. doi: 10.1039/c5an01632k. [DOI] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Nat. Protoc. 2008;3:877. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Tost J., Gut I.G. Clin. Biochem. 2005;38:335. doi: 10.1016/j.clinbiochem.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Verma K., Dalal J., Sharma S. Int. J. Pharm. Sci. Res. 2014;5:3086–3095. [Google Scholar]
- Yu A., Geng H., Zhou X. BMC Genom. 2006;7:143. doi: 10.1186/1471-2164-7-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Yao Y., Zhu J.L., Zhang S.N., Zhang S.S., Wei H., Hui W.L., Cui Y.L. Sci. Rep. 2016;6:26533. doi: 10.1038/srep26533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zeng D., Ma H., Feng G., Hu J., He L., Li C., Fan C. Small. 2010;6:1854–1858. doi: 10.1002/smll.201000908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material