Abstract
Testing donations for pathogens and deferring selected blood donors have reduced the risk of transmission of known pathogens by transfusion to extremely low levels in most developed countries. Protecting the blood supply from emerging infectious threats remains a serious concern in the transfusion medicine community. Transfusion services can employ indirect measures such as surveillance, hemovigilance, and donor questioning (defense), protein-, or nucleic acid based direct testing (detection), or pathogen inactivation of blood products (destruction) as strategies to mitigate the risk of transmission-transmitted infection. In the North American context, emerging threats currently include dengue, chikungunya, and hepatitis E viruses, and Babesia protozoan parasites. The 2003 SARS and 2014 Ebola outbreaks illustrate the potential of epidemics unlikely to be transmitted by blood transfusion but disruptive to blood systems. Donor-free blood products such as ex vivo generated red blood cells offer a theoretical way to avoid transmission-transmitted infection risk, although biological, engineering, and manufacturing challenges must be overcome before this approach becomes practical. Similarly, next generation sequencing of all nucleic acid in a blood sample is currently possible but impractical for generalized screening. Pathogen inactivation systems are in use in different jurisdictions around the world, and are starting to gain regulatory approval in North America. Cost concerns make it likely that pathogen inactivation will be contemplated by blood operators through the lens of health economics and risk-based decision making, rather than in zero-risk paradigms previously embraced for transfusable products. Defense of the blood supply from infectious disease risk will continue to require innovative combinations of surveillance, detection, and pathogen avoidance or inactivation.
Abbreviations: CHIKV, chikungunya virus; CMV, cytomegalovirus; EID(s), Emerging infectious disease(s); FDA, Food and Drug Administration; HBV, hepatitis B virus; HBsAg, surface antigen of the hepatitis B virus; HCV, hepatitis C virus; hESC(s), human embryonic stem cell(s); HIV, Human Immunodeficiency virus; hPSC(s), human pluripotent stem cell(s); HSC(s), hematopoietic stem cells; HSPC(s), hematopoietic stem and progenitor cells; HTLV, human T-lymphotropic virus; iPSC(s), induced pluripotent stem cell(s); NAT, nucleic acid testing; NGS, next generation sequencing; PCR, polymerase chain reaction; PI, pathogen inactivation; QALY, quality adjusted life years; RBC, red blood cell; SARS, severe acute respiratory syndrome; SURPI, sequence-based ultra-rapid pathogen identification; TT, transfusion transmission; TTI(s), transfusion transmissible infection(s); UCSF, University of California San Francisco; UCSF/AVDDC, UCSF/Abbott Viral Diagnostics and Discovery Center; vCJD, variant Creutzfeldt-Jakob disease; WHO, World Health Organization; WNV, West Nile Virus
Keywords: Blood-borne pathogens, Transfusion, Transfusion-transmitted infection, Next generation sequencing, Pathogen inactivation, Health economics
Highlights
-
•
A symposium on blood-borne pathogens was held September 26, 2015, in Toronto, Canada.
-
•
Transmission-transmitted infections remain a threat to the blood supply.
-
•
The residual risk from established pathogens is small; emerging agents are a concern.
-
•
Next generation sequencing and donor-free blood are not yet practical approaches.
-
•
Pathogen inactivation technology is being increasingly used around the world.
-
•
Health economic concerns will likely guide future advances in this area.
Canadian Blood Services (CBS) manages the blood system for all Canadians except those residing in the province of Québec. Funded primarily by the provincial and territorial ministries of health, CBS is responsible for collecting, testing, and manufacturing blood, blood components, and stem cells, and for distributing these products to hospitals in nine provinces and three territories across Canada. CBS also receives funds through the Canadian federal health agency to conduct research, development, and educational efforts related to the safe and effective provision of blood and blood products. Accordingly, since 2003, CBS has hosted an annual international symposium to address topical areas in transfusion medicine [1], [2], [3], [4], [5], [6], [7], [8]. The 13th iteration in this series was held on September 26, 2015, in Toronto, Ontario, and was entitled “Blood-borne Pathogens: Defend, Detect, And Destroy”.
Protecting the blood supply from established and emerging pathogens has been a top priority for CBS since its inception in 1998, following viral contamination of blood and blood products used in Canada and many other nations with hepatitis C and HIV in the 1980s and 1990s. While great strides in blood and blood product safety have been taken since that time, further reducing the risk of transmitting pathogens by transfusion remains an important concern in the field of transfusion medicine. During 2014 and 2015, a devastating Ebola virus outbreak spread through West Africa and for the first time, chikungunya virus (CHIKV) appeared in the Americas. This period also saw the first North American regulatory approval of a pathogen inactivation technology for use on plasma and single donor apheresis platelets, with the approval of the Intercept system by the United States Food and Drug Administration (FDA). Against this backdrop, the attendees of the 2015 annual symposium were provided insights into the state of the field by the day's line-up of expert speakers.
Blood-Borne Pathogens: Defend
Prevalence and Risk of Blood-Borne Pathogens in the Canadian Blood Supply
Key Messages
-
•
Through improvements in screening, testing, and real time surveillance, the residual risk of transmissible diseases in the Canadian blood supply remains very low.
-
•
We continue to deal with infectious diseases which emerge or re-emerge, such as chikungunya, Babesiosis, and hepatitis E, as well as infectious diseases such as influenza, that threaten the security of the blood supply despite not being transfusion-transmissible.
-
•
New paradigms for transmissible disease prevention must become more cost effective in their scope, using targeted surveillance, donor screening, and risk-based decision making.
Dr Margaret Fearon, CBS Medical Director, Medical Microbiology, and Assistant Professor, University of Toronto, discussed the current prevalence of classical transfusion-transmissible infections (TTIs) in CBS blood donors, new and emerging infectious diseases, how CBS prepares for and manages new risks, and also addressed new paradigms for risk management.
Dr Fearon began by emphasizing that several layers of protection in the Canadian blood supply have likely reduced the risk of the classical TTIs, (hepatitis B virus [HBV], hepatitis C virus [HCV], human immunodeficiency viruses [HIV] 1 and 2, human T-lymphotropic viruses [HTLV] I and II, and syphilis). The success can be largely attributed to intensive donor testing for TTIs, supplemented by donor education and deferral of donors with risk factors. The two latter approaches have reduced the number of donors with window-period infections and contributed to a decrease in confirmed transmissible disease-positive allogeneic donors over the last decade in Canada, most notably for HBV and HCV [9]. Thus, the residual risk of TTIs is low by any standard.
The estimated residual risk in Canada calculated in 2012, using incidence rates from observed donor seroconversions 2006 to 2009, is 1 per 8 million donations for HIV, 1 per 6.7 million donations for HCV, and is 1 per 1.7 million donations for HBV [9]. Dr Fearon noted that updated residual risks are currently being calculated, but that compared to the 2012 report, the risks are not expected to change dramatically.
Dr Fearon next turned her attention to newer and emerging infectious diseases that threaten the blood supply. Some of these diseases have led to the introduction of new TTI testing paradigms at CBS (ie, seasonal and selective testing for West Nile Virus [WNV] and Chagas Disease). Other emerging infections are being monitored (eg, babesiosis, hepatitis E, CHIKV) while others such as influenza feature in contingency planning, in spite of not being transfusion-transmitted, due to their potential to disrupt blood donation and the health care system. She emphasized her opinion that transmissible disease testing must be context-specific, and account for local disease prevalence, environmental factors, and resource allocation.
WNV is a mosquito-borne zoonotic arbovirus that emerged in North America in 1999 and was found to be transfusion-transmissible in 2002 [10]. In humans, febrile illness occurs in 20% to 30% of WNV cases and 1% of patients have serious neurologic symptoms. Since most cases are asymptomatic, TTI testing is the primary means of preventing transmission [11]. Universal donor testing was adopted in 2003 using nucleic acid testing (NAT). However, given the seasonal nature of WNV outbreaks, a more nuanced testing methodology was introduced by CBS in June 2015 that recognizes the lack of local transmission during the winter months. Now, all donors are tested from June 1 to November 30 and only donors who travel outside of Canada are tested during the rest of the calendar year.
Selective testing is also conducted for Chagas disease. Chagas is caused by the protozoan parasite Trypanosoma cruzi and is endemic to Central and South America and Mexico, where it is estimated that 6 – 7 million people have been infected [12]. With increasing northward immigration of people from these regions, it is estimated that > 300,000 people are infected in the United States, and most American blood services have implemented universal donor testing [13], [14]. The rates of immigration from endemic countries are lower in Canada and thus CBS tests donors who are identified to be at risk based on the donor questionnaire. Those considered at high risk include those who were born or lived in an endemic country, or had a mother or maternal grandmother that was born or lived in an endemic country. The safety of this approach was demonstrated in a recent study that identified no evidence of infection amongst donors without risk factors identified on the questionnaire (with the exception of one very unusual transfusion transmission – vertical transmission case) [15]. Interestingly, the selective testing approaches used for WNV and Chagas disease at CBS represent a change from the universal testing approach of the last three decades, which can be summed up as “test everyone for everything”. The newer approach to certain TTIs takes into account geographic location, seasonal effects, and other risk factors in setting the optimal testing strategy.
Dr Fearon called attention to other infectious outbreaks that can impact the security of the blood supply despite not being transfusion-transmissible, and cited severe acute respiratory syndrome (SARS) and pandemic influenza as examples. Outbreaks of these diseases can lead to shortages of staff and donors due to illness and shortages of critical supplies. Contingency planning is necessary to guard against these risks. Staff and donor education, infection control procedures in the clinic, and reassessment of donor deferral criteria are key steps that must be taken to protect the blood supply in this context.
Other transfusion-transmissible diseases are currently being monitored as potential emerging threats to the safety of the blood supply, including babesiosis, hepatitis E, CHIKV, and dengue virus. Babesiosis is caused by the protozoan parasites B microti, B duncani, and others in this genus, and spread by infected ticks. Most infections are asymptomatic or unrecognized, but the spectrum of clinical severity also includes flu-like symptoms, ranging to more severe illness and death in the immunocompromised [16]. Babesia microti is the most frequently transfusion-transmitted microbial pathogen in the United States, especially in the Northeast and Upper Midwest states [17]. There were 160 transfusion-transmitted cases reported from 1979 to 2009 in the United States with one case reported in Canada [18]. Hepatitis E is clinically similar to hepatitis A and it causes water-borne outbreaks in developing countries. In Canada, where it was previously thought to be primarily a disease of travelers, the actual prevalence of endemic hepatitis E is unknown. No cases of transmission by transfusion have been reported in North America, but transfusion transmission has been reported in endemic countries and recently in the United Kingdom [19]. CHIKV and dengue are two viruses common in the tropics that are spread by mosquitos. Both lead to similar acute illnesses with fever, rash, and muscle/joint pain. Transfusion-transmitted cases of dengue have been reported. CHIKV arrived in the Caribbean in 2013 and was thus identified as a threat to North America [20]. However, no transfusion transmitted cases of CHIKV have been reported to date. Current malaria travel deferral provides some protection with respect to many but not all of the affected areas, particularly in the Caribbean.
How can blood operators best prepare for emerging threats? Surveillance is conducted by multiple health agencies, including the World Health Organization (WHO), Centers for Disease Control and Prevention, and the International Society for Infectious Diseases, which operates the ProMED (Program for Monitoring Emerging Diseases). Available to any subscriber, ProMED is an internet-based reporting system dedicated to rapid global dissemination of information on transmissible diseases. The Public Health Agency of Canada is the federal agency responsible for transmissible disease surveillance in Canada. Public Health Agency of Canada encompasses the National Microbiology Laboratory and in collaboration with the provincial public health laboratories, provides diagnostic testing and surveillance data that is useful in guiding CBS decision-making. Testing data provided by the National Microbiology Laboratory on travel- acquired chikungunya was used by CBS to calculate an estimated risk of a case of transfusion transmitted CHIKV in Canada of less than 1 in 11 million.
Collaboration with veterinarians, etymologists, and ornithologists may provide additional information to inform preparative and reactive strategies for emerging agents. For example, active tick surveillance reports provide risk data for Lyme Disease, but are also relevant to other tick-borne, transfusion-transmissible diseases such as babesiosis [21]. A recent Babesia seroprevalence study for B microti in Canadian blood donors demonstrated that donor testing is not warranted in Canada at this time [22]. Dr Fearon presented as-yet-unpublished data on a collaborative CBS and Héma-Quebec (the transfusion service for Quebec province) hepatitis E seroprevalence study that indicated that age is the only significant factor for increasing seroprevalence of hepatitis E. The absence of polymerase chain reaction (PCR)–positive results suggests that the risk of transfusion-transmission of hepatitis E in Canada is extremely low; however, further prevalence data needs to be collected.
A CBS donor travel survey from 2014 also provided data that is used to inform risk assessment. While the United States remains the most popular travel destination for CBS blood donors, nearly 10% of respondents reported travel to the Caribbean. Such donor travel survey data allows CBS to estimate potential donor loss when assessing the risk/benefit of deferring donors who have travelled to countries with outbreaks.
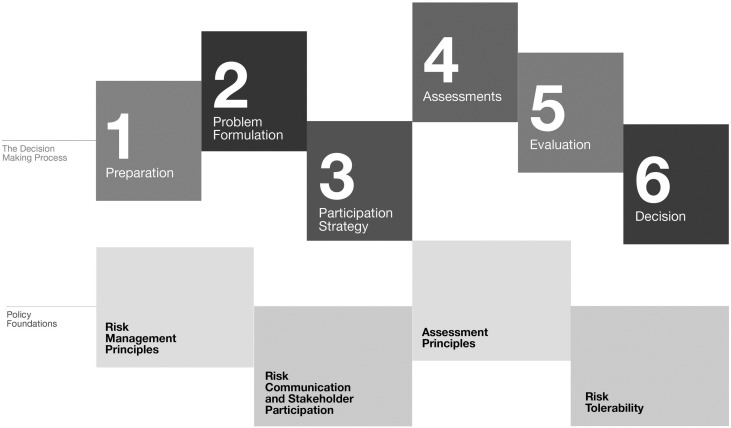
The challenge facing all blood operators is to synthesize all of the available information on existing and emerging threats in order to rapidly make decisions that balance risks, costs, and safety. To meet this challenge, Dr Fearon suggested utilization of the Alliance of Blood Operators' Risk-Based Decision Making Framework for Blood Safety (Fig 1 : Risk-Based Decision Making Framework) [23], [24], [25]. This framework has a health sector focus, can aid evidence-based decisions using risk assessment tools, and accounts for multiple sectors included in the decision making process [23], [26]. The use of this approach also represents a paradigm shift for blood operators, away from “zero-risk” to one that uses a decision-making process that integrates evidence, ethics, social values, economics, public expectations, and historical context with broader health care priorities [26].
Fig 1.
The Risk-Based Decision-Making framework. The Risk-Based Decision-Making framework was designed by the Alliance of Blood Operator to help blood operators identify, assess, act on, and communicate risk in decisions related to blood safety. It is a flexible tool, and its objectives are to optimize the safety of the blood supply while recognizing that elimination of all risk is not possible; allocate resources in proportion to the magnitude and seriousness of the risk and the effectiveness of the interventions to reduce risk; and assess and incorporate the social, economic, and ethical factors that may affect decisions about risk [23], [24], [25].
Current Perspectives on Transfusion-Transmitted Infectious Diseases: Emerging Pathogens Worldwide
Key Messages
-
•
The emergence of infectious diseases is unpredictable.
-
•
Emerging infectious diseases (EIDs) is a global issue that demands international surveillance efforts. Horizon scanning is important.
-
•
The EID tool-kit provides a useful framework for managing infectious threats to the blood supply.
Dr Roger Dodd, Secretary General of the International Society of Blood Transfusion, presented his perspectives on past and current pathogens affecting the safety of the blood supply. The objectives of his presentation were 4-fold: (1) to define what EIDs are and why they occur; (2) to discuss why some EIDs impact blood safety; (3) to review how the impact on the blood supply is managed; and 4) to examine some current examples of emerging infections and how they are being managed.
Dr Dodd began with the Institute of Medicine's definition of an emerging infectious disease as one “whose incidence in humans has increased within the past two decades or threatens to increase in the near future”. The Institute of Medicine further elaborates that “emergence may be due to the spread of a new agent, to the recognition of an infection that has been present in the population but has gone undetected, or to the realization that an established disease has an infectious origin. Emergence may also be used to describe the reappearance (or reemergence) of a known infection after a decline in incidence” [27].
EIDs often originate from animal-human interactions. A prime example is variant Creutzfeldt-Jakob disease (vCJD) which probably results from human consumption of meat from animals infected with Bovine Spongiform Encephalopathy (also called Mad Cow Disease) [28]. It is estimated that approximately 60% to 70% of current EIDs are zoonoses, and they can be caused by any class of pathogenic agent (viruses, bacteria, parasites, and prions) and spread through several modes of transmission (fecal-oral, sexual contact, etc). Infections emerge for a variety of reasons. Pathogens may undergo a “species jump” as was the case in HIV [29] and SARS [30]. Environmental change, such as global warming, may increase the incidence and range of EIDs such as dengue, malaria, and babesiosis. Drug resistance and mutations may lead to challenges in controlling malaria and HBV and subsequent spread. Human migration and travel contributes to the dissemination of T cruzi (the Chagas disease pathogen) and CHIKV. The migration patterns of birds, reservoirs for WNV, are also associated with the spread of this disease.
Certain parts of the world are considered to be “hot spots” for the emergence of infectious disease for a variety of reasons. For example, China is considered a prime site for the emergence of new strains of viruses (influenza and SARS) due to the close proximity of human-human interactions and human-animal interactions, which can lead to the evolution of animal viral strains into novel strains that can infect humans. In a general sense, this evolution can be potentiated by the consumption of wild meat (meat from non-domesticated mammals, reptiles, amphibians, and birds). Through careful phylogenetic analysis of simian and human viruses, Africa has been recognized as the “hot spot” for HIV emergence, probably due to consumption of primate bush-meat by humans, [29]. Urbanization, poor sanitation, and crowding in the developing world have also been linked to hepatitis E emergence [31]. Although our understanding of the factors contributing to specific EID transmission has improved, these transmissions are likely multifactorial in nature.
Dr Dodd emphasized that EIDs are both a local and a global issue. Some infections may emerge explosively in new areas if appropriate conditions (eg, the vector or environment) are met, as is the case with WNV, dengue, and CHIKV. Other EIDs, such as Chagas disease, may expand slowly as a result of population movements, but can become constrained in their new environment. Tick-borne infections, such as those caused by Babesia in the United States, may be constrained regionally. Infections that are characterized by direct human to human transmission may spread worldwide but at differing rates, depending on the mode of transmission such as the rapid respiratory spread of influenza and SARS and slower sexual transmission of HIV.
Despite our understanding of infectious disease transmission, the emergence of these diseases remains largely unpredictable. Understanding that EIDs may be spread by human travel and through animal contact allows us to understand the probability of acquiring an infection if specific conditions are met, but it does not allow us to predict which infectious disease will emerge and when. Such unpredictability requires regular surveillance and hemovigilance efforts to be in place around the world. Disease surveillance has many challenges, but warning signals may help focus efforts to better monitor disease spread. These warning signals may include disease outbreaks in particularly susceptible populations (such as the immunocompromised), and/or the blood-borne nature of a disease.
Why do some emerging infections impact blood safety? Dr Dodd noted that when an epidemic occurs, only a minute proportion is attributable to transfusion. For example, only 2% of HIV cases were transfusion-transmitted during the HIV epidemic [32]. Similarly, only 23 of 400,000 WNV cases (0.006%) were established as TTIs [33]. In other words, outbreaks are not likely to start from a transfusion, and transfused patients are not necessarily more likely to acquire an infection than the general population. For an infectious disease to be considered a transfusion-transmissible disease, certain pathogen-related and recipient-related characteristics have to be present. First, the pathogen must have an asymptomatic blood-borne phase (such as hepatitis B), which may either be acute or chronic. Second, the pathogen must be able to survive the donated blood processing and storage procedures, including temperature changes, leukoreduction, and centrifugation. Third, the disease must be transmissible by the intravenous route. Fourth, the recipient has to be susceptible to infection. Fifth, the disease must be a recognizable entity in the recipient once symptoms appear.
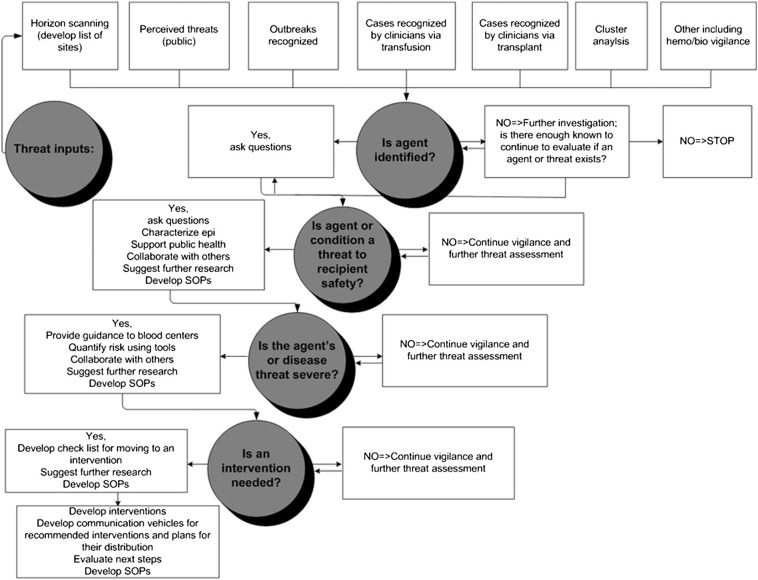
Next, Dr Dodd turned his attention to how the impact of EIDs on the blood supply is managed. In 2009, the AABB published a list of 68 EIDs of interest. Each EID was categorized based on the threat it poses to transfusion safety, the level of regulatory concern, the lack of effective intervention, and the amount of public concern. The top priority was assigned to 3 pathogens towards which intellectual and future resources should be focused: dengue viruses; Babesia species; and prions causing human vCJD [34]. The list also included CHIKV, Plasmodium species, T cruzi, human parvovirus B19, HIV, and hepatitis E. An EID tool-kit (Fig 2 ) was subsequently developed as a framework to guide health professionals and public health officials in triaging and managing infectious threats to the blood supply [35]. The EID tool-kit presents: a variety of methods for EID surveillance; key questions to be asked when a threat is suspected; and potential courses of action based on the situation.
Fig 2.
Emerging Infectious Disease ToolKit. The Emerging Infectious Disease ToolKit is a framework developed by the AABB Transfusion-Transmitted Diseases, Emerging Infectious Diseases subgroup to guide health professionals and public health officials in triaging and managing infectious threats to the blood supply. The EID tool-kit presents: a variety of methods for EID surveillance; key questions to be asked when a threat is suspected; and potential courses of action based on the situation [35].
Included among the 68 pathogens identified in the 2009 AABB report, several agents are being actively monitored both on a regional and global level. Due to increasing reports of transfusion-transmitted cases, their poor prognosis in immunocompromised transfusion recipients, and the lack of effective prevention strategies, Babesia species have been classified as a top priority for future blood supply safety efforts in the United States. Prions such as vCJD are being monitored closely and donor screening has been successful in preventing their spread into the donor pool [28]. Prions are currently being investigated for their potential relationship with other protein-folding diseases such as Alzheimer's disease [36]. Although respiratory infections (eg, Middle East Respiratory Syndrome coronavirus) are not transfusion-transmissible, they do disrupt donor availability and organizational aspects of the blood collection and donation system.
Dr Dodd expanded upon WNV, adding American insights into those earlier provided in the Canadian context by Dr Fearon. West Nile fever is caused by a flavivirus transmitted by culicine mosquitoes. The virus spread from southern Europe, Africa, and the Middle East to India, and arrived in the United States in 1999. By 2004 WNV was endemic in most of the continental US and Canada [33]. The experience of WNV in the US demonstrated that imported infections can be overwhelming and unpredictable. While WNV was considered a stable disease elsewhere in the world, in North America it was experienced as an explosive outbreak in 2002 and 2003 infecting over 400,000 individuals. Public concern was high as WNV, previously unknown in North America, spread rapidly across the continent via infected birds, and then to humans via mosquitoes. Although human to human transmission is not possible, there is potential for transfusion-associated transmission if the donation occurs during periods of pre-symptomatic viremia. NAT of pooled donor samples offered a rapid route to testing. Like other North American blood operators, CBS tests donors in pools. If a pool tests positive, individual donor testing is initiated. After NAT was initiated in the US in 2003, cases of transfusion-transmitted WNV have only rarely been encountered [33].
Horizon scanning efforts are actively monitoring other potential EIDs. Dr Dodd described several emerging agents for which there is some evidence that that have or an assumption that they may be transfusion-transmitted including Severe Fever with Thrombocytopenia Syndrome Virus (no reported transfusion transmission [TT]), Q Fever (one report of TT, but no definitive evidence), Hepatitis E Virus (good evidence of occasional TT), vCJD, and other prions (evidence for TT) [35]. Dr Dodd focused on two viruses that are current causes for concern. Dengue virus is an important arbovirus. Like WNV, it is a flavivirus and is spread by mosquitos (Aedes genus). Humans are the amplifying host, and while a vaccine is under investigation there is currently no vaccine or specific treatment. Vector control is the only effective intervention. There are an estimated 390 million infections per annum worldwide [37]. In 50% to 80% of cases infection is asymptomatic. Dengue viruses have been found to be transfusion-transmitted in 7 separate geographic clusters in Hong Kong, Singapore, Puerto Rico, and Brazil. Currently, there is no FDA-licensed test for dengue RNA [37], [38]. CHIKV, which has caused recent massive outbreaks in the Caribbean and co-exists with dengue virus, is a potential threat to the North American blood supply due to its geographic proximity and donor travel patterns [39]. CHIKV recently appeared in the Caribbean. Should it arrive in North America, donors could be deferred for exposure or symptoms, NAT testing for CHIKV RNA could be initiated, and red cell and plasma collections could even be stopped. Dr Dodd stated that pathogen inactivation will likely become increasingly important in preventing transfusion-transmission of emerging agents. The American Red Cross in Puerto Rico is currently involved in a trial to investigate and monitor the safety of INTERCEPT pathogen inactivation technology (Cerus Corporation). INTERCEPT was recently approved by the FDA for use with apheresis platelets. Use of INTERCEPT to make available pathogen-reduced apheresis platelets could prevent interruptions in the local platelet supply in areas where viruses like CHIKV emerge [40].
Preventing the Spread of Pathogens Around the Globe—A Clinical and Public Health Perspective
Key Messages
-
•
The 2014 Ebola outbreak in West Africa was the largest in history.
-
•
The difficulties in identifying the virus in West Africa and in containing its spread were related to poverty, growing populations and deforestation, lack of healthcare infrastructure and resources. These fundamental issues must be addressed in order to prevent and/or contain future outbreaks.
Dr Allison McGeer, Director of Infection Control at Toronto's Mount Sinai Hospital, used the recent Ebola outbreak in West Africa to provide a thought-provoking global perspective regarding pathogens and their spread. As a consultant on a WHO-initiated mission to Liberia, Dr McGeer obtained first-hand information and impressions of the situation on the ground in West Africa, where she navigated issues related to policy and healthcare set-up.
Ebola virus is difficult to study because of its high mortality rates, and because it often occurs in areas that are difficult to access due to poor infrastructure or because of conflict or political turmoil. Ebola infections seem to emerge due to the interconnection of enzootic and epizootic cycles [41]. The first sequence involves bats as the most likely reservoir host for the virus, which is spread by enzootic transmission within the bat population. From this pool of virus carriers, transmission to other non-human species is thought to happen in an epizootic cycle. Initial transmission to humans involves contact with infected bats or other species through hunting or accidental contact with ill wild animals. Human-to-human spread via direct close contact is then very efficient.
Once humans are infected, the Ebola virus first appears to target the immune system and subsequently destroys the vascular system, leading to blood leakage. Initial attacks on dendritic cells lead to decreased interferon production and macrophage and endothelial cell degradation [42]. This pattern results in clinical presentations of hemorrhage, hypotension, drop in blood pressure, followed by shock and death [43].
The 2014 Ebola outbreak, which primarily affected Guinea, Sierra Leone, and Liberia in West Africa, is the most recent in the history of Ebola epidemics [44]. Since 1976, more than 20 outbreaks have been recorded in Sub-Saharan Africa leading to hundreds of cases and deaths [45]. Historically, infection has been controlled by local communities with the isolation of any patient showing symptoms. However, in 2014 in West Africa, the outbreak was on an unprecedented scale. Dr McGeer provided an eye-opening overview of health infrastructure in the affected West African countries. Guinea, Liberia, and Sierra Leone have populations of 11.5, 4.2, and 6 million, respectively (vs 35 million in Canada). Spending on health (total expenditure per capita) is US$59, US$88, US$228, and US$4759 in Guinea, Liberia, Sierra Leone, and Canada, respectively [46]. This imbalance is mirrored in the number of doctors per 100,000 population: 10, 1.4, 2.2 for Guinea, Liberia, Sierra Leone, respectively, compared to 245.2 in the United States of America [47]. Under-resourcing of public health and healthcare delivery was an important contributor to the unprecedented scale of the epidemic.
The first step in this epidemic was the spread of the virus from a reservoir in West Africa, but the outbreak was aided by many economic, political, and geographic factors. Being now relatively stable after periods of civil war, these countries have increasing birth rates and increasing populations. The median age in Guinea, Liberia, and Sierra Leone is 18.5, 18.4, and 20, respectively (vs 40 in Canada). The ever-growing population and deforestation is believed to have accelerated the frequency of the epizootic cycle. Once rich in forests, West Africa has been intensively logged over the last decade. Guinea's rainforests have been reduced by 80%, while Liberia has sold logging rights to over half its forests. Some analysts have predicted the complete deforestation of Sierra Leone within the next few years [48]. The forests are the habitat for fruit bats, Ebola's probable reservoir host. With the loss of their habitat, the bats escape to urban environments to hunt for food, coming in contact with humans and triggering more frequent transmissions of the virus.
Dr McGeer personally witnessed the huge barriers to effective control of the epidemic in hospitals in Liberia. She pointed to the rudimentary nature of facilities and equipment, poor hygiene, and a lack of infrastructure for disposing of hospital waste. Dr McGeer commented that due to the lack not only of resources but also of any form of emergency plan, the base from which to fight the epidemic was completely lacking. The lack of resources for infection control and personal protective equipment are the main reasons for nosocomial transmission [44], and affected healthcare workers can act as amplifiers spreading the virus into the community. In the 2014 epidemic, 1 in 20 healthcare workers were infected, and of those, 1 in 10 are believed to have died [43]. This devastated the ability of front-line healthcare workers to control the epidemic, and led to hospital closures.
Poor general infrastructure hindered transportation of medical supplies and expertise, isolating rural areas and limiting access. Dr McGeer highlighted the absence of public health and health care infrastructure as a fundamental issue in being able to fight this disease, not just on the ground in West Africa, but globally. This issue is illustrated by a fragmented global health system is which the institutions, laws, and strategies are not interconnected. Experience of this outbreak has led to calls to reform the worldwide health systems architecture and the WHO [49]. Some media reports led to widespread misunderstanding of the Ebola outbreak as an “African problem,” and unhelpfully perpetuated prejudicial colonial-era stereotypes.
Dr McGeer took the audience through the evolution of the 2014 epidemic in Liberia which peaked in August/September 2014. During July and August 2014 the number of confirmed cases per day increased from about 10 to about 50, and Ebola treatment units and burial systems were overwhelmed. In Monrovia, the Liberian capital city, about 80% of patients with Ebola virus disease were being managed at home. Despite this, hospitals were overwhelmed with Ebola patients, filling to 300% of its capacity. Many health care workers were infected, which ultimately led to the decision to close hospitals. At the peak of the outbreak, the President of Liberia quarantined West Point, a township particularly badly affected. This decision led to riots in that area. Similar actions took place in other parts of Liberia [50].
As a consequence of the deteriorating situation in West Africa, international responses were initiated [49]. By September 2014, more than 100 non-governmental organizations were on the ground in Liberia to contribute to the fight against Ebola. Their work spanned a wide range of activities including: setting up medical care; contact tracing; opening orphanages; providing food for people in quarantine; building roads for improved access to cemeteries; identifying how to de-sludge septic tanks from Ebola treatment units; and sourcing and supplying personal protection equipment for Ebola treatment units.
The crisis response also involved a coordinated international response of unprecedented scale to accelerate vaccine development. No vaccine had ever been tested in humans prior to 2014, but several were fast-tracked through phases I and II, and in June of 2015, an international group of scientists published the interim results of an open-label, cluster-randomized ring vaccination trial of an engineered Vesicular Stomatitis Virus–based Ebola vaccine developed at Canada's National Microbiology Laboratory [51]. Ring vaccination seeks to create a buffer of protection around each case, so that the virus cannot continue to spread. This initial trial found that 100% of recipients were protected from the virus; further study of this vaccine and trials of other vaccines are on-going.
With improvements in the coordination of the overall emergency response, more than a year later, the outbreak is under control. Occasional cases were still appearing in Liberia in November 2015, but on November 7, 2015, WHO declared the end of Ebola virus transmission in Sierra Leone, as 42 days had passed since the second negative test of the last confirmed patient with Ebola in the country [52]. Control of the outbreak was only achieved through the institution of effective control and quarantine measures and an understanding of local practices and challenges that impacted the spread of the virus. Dr McGeer added her voice to the chorus of experts recommending coordinated national and international efforts to prevent future outbreaks. Early warning systems should be developed in connection with local communities in high-risk areas, and provision of clearly defined response recommendations specific to the needs of each community [53]. Recent advances in diagnostics, risk mapping, mathematical modeling, and pathogen genome sequencing have the potential to improve substantially the quantity and quality of information available to guide the public health response to outbreaks [54].
However, prevention remains extremely difficult [55]. The World Bank estimated that an ~ $26 M investment in public health in West Africa would have prevented more than 90% of cases in West Africa. This missed opportunity eventually led to a costly emergency response estimated by the UN mission for Ebola emergency response at about $1.6 billion. Future epidemic control measures in poverty-stricken areas, including worldwide response teams and pre-approved emergency funds may improve outbreak response, but addressing the fundamental issues of poverty, infrastructure, education, healthcare, workforce development, and communications will be needed if outbreaks are to be prevented [56].
An Alternative Method of Maintaining a Clean Blood Supply: Ex Vivo Generated red Blood Cells
Key Messages
-
•
Several approaches for the production of red blood cells (RBCs) ex vivo exist and have demonstrated feasibility; however, none can yet be conducted on a scale that would allow replacement of donor-derived RBCs
-
•
To improve the chances of success in bringing ex vivo RBCs to the clinic, a multidisciplinary approach and an integrated plan are required to navigate the long path from concept to commercial product.
An alternative means to keep the blood system safe from pathogens is to move away from the current paradigm of donor-derived products. This topic was addressed by Dr Marc Turner, Medical Director at the Scottish National Blood Transfusion Service and Professor of Cellular Therapy at the University of Edinburgh. Dr Turner began by mentioning some milestones in the history of blood transfusion, highlighting a number of events that took place in his adopted home city of Edinburgh, Scotland. For example, in 1816 the first systematic experiments in intra-species transfusion were carried out by John Henry Leacock, who was studying in Edinburgh at the time. James Blundell, a University of Edinburgh medical school graduate, is credited with conducting the first successful human transfusions several years later. Since early in the last century, transfusion has become a mainstay of clinical practice with around 92 million RBC transfusions conducted annually world-wide [57]. For the most part, and particularly in developed countries, blood transfusion is safe. However, limitations remain, including sufficiency of supply, immunological compatibility of the donor and recipient, the risk of TTIs, and the risk of other complications such as iron overload. Many of these limitations could potentially be overcome by the production of RBCs for transfusion in the laboratory. Dr Turner's talk focused on human stem cells and their potential use for the ex vivo generation of RBCs for transfusion. Dr Turner discussed the biological and engineering limitations that must be overcome in order for ex vivo generated cells to become viable alternatives to donor-derived RBCs.
The first conclusive observation of stem cells was in 1960 by McCulloch and Till in the host city of this symposium – Toronto [58]. Stem cells have the capability for self-renewal and can differentiate into multiple lineages, ultimately resulting in the generation of differentiated cells or tissues. There are various types of stem cells, including those derived from embryos (eg, human embryonic stem cells [hESCs]) and from adults. Not all stem cells are the same. For example, hESCs are pluripotent and have unlimited ability to replicate, whereas adult-derived hematopoietic stem and progenitor cells (HSPCs) can differentiate into cells of the hematopoietic and immune systems and have limited ability to replicate.
Found in the bone marrow, HSPCs are morphologically indistinct from other bone marrow cells, and are distinguished as CD34 + cells using flow cytometry. In vivo, HSPCs are found in a hypoxic environment at the edges of the bone marrow. As they differentiate, they move more centrally in the marrow into an environment with a higher O2 tension. It has long been known that when placed into agar/semi-solid medium and provided with the right cytokine support, HSPCs can differentiate along different hematopoietic lineages, forming granulocyte/monocyte, and erythroid colonies with 7 to 14 days [59]. Culturing HSPCs in suspension using a two-phase culture system has several advantages over solid-phase culture [60], the in vitro environment can be more precisely controlled and can be separated more easily [61], [62]. Dr Turner provided an overview of a two stage culture system which uses combinations of cytokines to control cell differentiation and proliferation and can be used to produce erythroid cells from CD34 + adult peripheral blood cells. During the enucleation process, which moves the cells into the early reticulocyte stage, the nucleus is extruded from the cell, and engulfed by macrophages. By days 10 to 14 of the two-stage culture, the population consists of approximately 50% normoblasts and 50% early (R1) reticulocytes. Other 3-stage systems have had demonstrated success expanding CD34 + HSPC from peripheral blood, bone marrow, or cord blood into functional RBCs [63]. These proofs-of-principle demonstrated that RBC generation ex vivo is possible, but processes described to date have limited scalability and do not yet constitute a workable approach to generate RBCs for transfusion.
Dr Turner spent the remainder of his talk discussing the two major types of challenges facing the ex vivo generation of RBCs for transfusion: biological challenges and engineering/logistical challenges. From the biological point of view, one major challenge is to determine the best source from which to derive the cells. Adult hematopoietic stem cells (HSCs), one potential source of ex vivo RBCs, have a limited replication capacity; they could generate a small number of units of RBC but the reliance on donors remains. Unlike HSCs, hESCs have indefinite expansion capabilities and can self-renew, and were a source of much excitement when their derivation was first described [64]. hESCs are pluripotent – they can be cultured indefinitely as cell lines and are able to differentiate into all the cells of the body including hematopoietic cells [65]. In 2006, a considerably less ethically-controversial source of pluripotent stem cells was discovered– induced pluripotent stem cells (iPSCs) [66], [67] earning the discoverer the Nobel Prize for Medicine in 2012. Initially generated by Takahashi and Yamanaka from human somatic skin fibroblasts by the use of four genes (Oct4/Sox2/Klf4/Myc), iPSCs are very similar to hESCs, can be differentiated into all three germ layers and have huge potential for regenerative medicine applications and disease modeling. Like hESCs, human iPSCs can be differentiated into hematopoietic cells in vitro [68], [69], [70], [71], and this is an area of intensive research [72]. Dr Turner provided an overview of a system by which RBCs can be derived from human pluripotent stem cells (hPSC) in vitro using a feeder/serum free approach and cytokine mixes that drive the hPSC to HSPCs, then erythroblasts, normoblasts, and eventually reticulocytes over an approximately 30-day time frame. The differentiation process is complex and uses mixes of multiple cytokines and small molecules. Several issues remain including the long differentiation time, and the fact that while hPSC-derived RBCs can enucleate, the resulting cells are fragile and difficult to maintain in culture. The hPSC-derived RBCs express α/γ globin chains, the same combination of hemoglobin polypeptides expressed in fetal RBCs (as opposed to α/β globin chains in adult RBCs); however, the oxygen-delivering characteristics of the hPSC-derived RBCs are acceptable and therefore this would not preclude their use. Efforts are underway to optimize erythrocyte differentiation and enucleation and modify the hemoglobin to a more adult form [73], [74].
Another approach that holds promise is to immortalize multipotent stem cells using well-understood and established methods. Conditional immortalization from hPSCs or adult HSPC can be achieved using human papilloma virus E6/E7 or combinations of transcription factors. This approach can establish immortalized human erythroid progenitor cell lines [75]. Switching off the immortalization allows the production of enucleate RBCs ex vivo. This approach has several benefits over PSC-as a starting material: the process is less complex and the differentiation time is shorter; there are reduced costs related to cytokines, growth factors, and media; and the final RBC phenotype is closer to an adult phenotype. The feasibility of this approach to produce enucleated RBCs has been shown [75]. Potential drawbacks of this approach include concerns regarding: the introduction of oncogenes into the cells; cell line stability; and the likelihood of being able to meet good manufacturing practice standards and produce clinical grade products.
Notwithstanding the difficulties involved, the potential now clearly exists to allow culturing of RBC with rare phenotypes or modification of the RBCs [76] to help meet the needs of hard-to-match transfusion recipients. The first proof-of-principle human transfusion with ex vivo generated RBCs cultured from peripheral CD34 + HSC was conducted in 2011, and showed that post-transfusion survival of ex vivo generated RBCs in a single, healthy subject was comparable to that of donor-derived RBCs [77].
Dr Turner then turned his attention to the engineering technology needed to make ex vivo generated RBCs a viable alternative to donor-derived products. The key issues here are scale-up, process control, and intensification. Dr Turner described the use of the Ambr bioreactor technology (from TAP Biosystems, part of the Sartorius Stedim Biotech Group) as an approach to optimising the in vitro culture environment. Stirred tank bioreactors are a mature technology that is scalable and well-established for production of biotherapeutics. They allow for precise control of several physico-chemical parameters and economic/rapid development screening at a variety of scales (10 mL, 250 mL, multi-liter). There are two gross limits to system efficiency. The first is the absolute density limit, which is calculated from specific oxygen uptake rate of cells and the mass transfer coefficient of the system. In this regard, the bioreactor system performs well; high density can be achieved, and the potential is certainly there to succeed in scaling it up to the needs of producing erythroid cells. The second limit is media volumetric productivity (liters of media/units of blood). Volumetric productivity is precisely determined by cell growth rate and specific support capacity of the media—consumption and supplementation. Specific rates are unstable and supplementation with glucose and glutamine does not improve growth, suggesting these are not limiting factors. In order for potential scale-up of this system, the limiting factors need to be identified.
Currently, Dr Turner estimates that the costs of ex vivo-derived RBCs stand at many times the cost of donor-derived RBCs. At this stage this technology is therefore only likely to be considered for “boutique” applications in patients for whom a donor-derived matched product is difficult or impossible to source. For more generalized application, many challenges remain, including control over the genetic and epigenetic stability of cell lines; optimization of the differentiation pathway to allow stable enucleation; efficiency of the differentiation pathway; process control over multiple physical and biochemical factors; scale up and intensification to control the cost of goods; detailed characterization of the product; demonstration of preclinical safety and efficacy; quality control and regulatory compliance; and the design and execution of pivotal clinical trials [78].
Many of the challenges Dr Turner mentioned are common to all cell-based therapeutics. Regarding quality control and product characterization, one advantage in this field is that there is more than 50 years of RBC product characteristic information, knowledge, and experience on which to draw. Dr Turner ended by noting that a sea change in the level of process control would be required to make the production of the ex vivo RBCs a reality. Dr Turner outlined the environment, resources, and approaches he believes are necessary in order for this type of innovation to take place and achieve commercial success [78]. This includes multidisciplinarity, integrated planning, and lengthy time lines and in these regards Dr Turner acknowledged the commitment of the research teams and funders working in this space.
Blood-Borne Pathogens: Detect
The Technical Aspects of Pathogen Testing in Canada
Key Messages
-
•
Pathogen testing in Canada is centralized, automated, and closely regulated by the federal government.
Ms. Nancy Angus, Director of Testing at CBS, provided an overview of the current state of testing at this organization, with an emphasis on donor testing. Focusing on tests in use in Canada, Ms. Angus described laboratory tests for the detection of blood-borne pathogens and summarized the differences in sensitivity among the tests currently in use.
CBS testing sites include donor testing, diagnostic services, the national testing laboratory, national reference laboratories, quality control product laboratories, and the human leukocyte antigen laboratory. Donor testing, which includes transmissible disease testing, is performed at two sites: Calgary, Alberta, where all blood collected west of Ontario is tested; and Toronto, where all blood collected in Ontario and east of Ontario is tested. Each site is a mirror-image of the other in terms of equipment, and can act as a back-up for the other site for business continuity reasons should the need arise.
All blood collected by CBS is tested for a number of blood-borne pathogens: Syphilis; HIV-1 and − 2; HBV; HCV; and HTLV I and II (Table 1 ). Since implementation in 2003, WNV testing has been performed on all collected blood; however, as of 2015, that testing is now performed seasonally. In addition to mandatory testing performed on all collected blood, there are tests that are performed based on risk. Approximately 40% of collected blood is tested for cytomegalovirus (CMV) in order to supply hospital demand and based on an algorithm identifying which donors are most likely to be CMV-free. Testing for Chagas is performed selectively, based on risk; donations from donors indicating on their questionnaires that they or their mothers or maternal grandmothers originate in a Chagas-endemic country or that they have had extended stays in endemic areas for greater than six months.
Table 1.
Transmissible disease testing performed at Canadian Blood Services
| Transmissible disease | Assay(s) | Sensitivity | Implementation year |
|---|---|---|---|
| HIV | Serological:
NAT
|
1985 2001 |
|
| HBV | Serological:
NAT
|
|
1972 2005 2011 |
| HCV | Serological
NAT
|
|
1990 1999 |
| HTLV I/II | Serological
|
100% (99.48%-100%) | 1990 (HTLV I); 1998 (HTLV I/II) |
| WNV | NAT
|
40.3 copies/mL§ | 2003 (Seasonal, 2015) |
| Syphilis | Serological | 100% (0.555-0.97) | 1949 |
| CMV⁎ | Serological | 99.4% (99.4%-100%) | 1984 |
| Chagas⁎ | Serological | 98.47% (94.59-99.81%) | 2010 |
anti-HIV 1 and 2, antibodies to HIV-1 groups M and O and antibodies to HIV-2; NAT, nucleic acid testing; anti-HBc, total antibodies to hepatitis B core antigen; anti-HCV, antibodies to hepatitis C virus; anti-HTLV I/II, antibodies to human T-lymphotropic virus I and human T-lymphotropic virus type II.
Only selected units are tested.
Single unit limit of detection; theoretical sensitivity is calculated by multiplying single unit limit of detection by 6.
Chemiluminescent assays are performed using the Abbott PRISM platform to detect the surface antigen of the hepatitis B virus (HBsAg), total antibody to hepatitis B core antigen, antibodies to hepatitis C virus, antibodies to HIV-1 groups M and O and/or antibodies to HIV-2, antibodies to human T-lymphotropic virus type I and/or human T-lymphotropic virus type II, and antibodies to T cruzi. Agglutination assays are performed on the Beckman Coulter PK7300 platform: syphilis infection is identified using a micro-hemagglutination assay to detect Treponema pallidum antibodies, and CMV infection is detected using a passive particle agglutination assay to detect total cytomegalovirus antibodies. NAT is performed using the Roche Cobas 201 platform to detect HIV-1 RNA (groups M and O), HIV-2 RNA, HCV RNA, HBV DNA, and WNV RNA. Samples are screened in pools of 6, and single unit testing is performed on selected donations from the same geographic region when a positive donation for WNV is identified.
In Canada, all testing platforms require approval by Health Canada. Other platforms currently available include the Immucor NEO platform, which is a solid phase system that allows for the serological detection of syphilis and CMV, and the Grifols Tigris platform, in use at Hema-Quebec, which performs NAT to detect HIV RNA, HCV RNA, HBV DNA, and WNV RNA.
If a blood donation is found to be serologically reactive, confirmatory testing is performed to determine if the donor is a “true” positive. Confirmatory testing is either performed at CBS or at an external agency. Within CBS, confirmatory tests include HBsAg neutralization assay, immunoblots for HIV-1, HTLV I, HTLV II, and HCV, and enzyme immunoassay to detect HIV-2. Syphilis confirmatory testing is performed at either the Alberta Public Health Laboratory or the Ontario Public Health Laboratory and Chagas confirmatory testing is performed at the National Reference Centre for Parasitology in Montreal.
The Medical Services and Innovation division at CBS performs surveillance to identify blood-borne pathogens that may pose a threat to the blood supply. If it is decided that a new test needs to be implemented to identify a new pathogen or new equipment is required to replace equipment at the end of its life, Health Canada licensure is required, unless there is an emerging threat. Currently on the horizon, CBS is considering the possibility of introducing hepatitis E virus NAT, and testing for Babesia
Pathogen Detection
Next Generation Sequencing: The Future of Pathogen Testing?
Key Messages
-
•
Next generation sequencing (NGS) is an advanced technology approach that involves sequencing all DNA found in a clinical sample.
-
•
Innovative bioinformatic approaches are required to minimize the computational time required to find pathogen DNA in the sample and to maximize accuracy.
-
•
NGS is being increasingly used for otherwise indeterminate clinical diagnoses, for tracking of infectious disease outbreaks, and to detect novel pathogens.
-
•
NGS is unlikely to play a role in screening the blood supply for infectious disease markers in the near to medium term.
Dr Samia Naccache, Associate Specialist, Department of Laboratory Medicine, University of California San Francisco (UCSF) School of Medicine, introduced attendees to the use of next generation sequencing, metagenomics, and bioinformatics for the detection and identification of infectious agents. Dr Naccache first pointed out that she was a member of the laboratory of Dr Charles Chiu, which is home to the UCSF/Abbott Viral Diagnostics and Discovery Center (UCSF/AVDDC), and that the Center works closely with the UCSF Clinical Microbiology Laboratory to provide advanced technology assistance with the most challenging problems in infectious disease diagnosis and tracking.
Dr Naccache commenced her presentation by contrasting “classical” DNA sequencing methods with next generation sequencing (NGS). Frederick Sanger and colleagues invented a method of DNA sequence determination in the 1970s [79] that was intensively used by scientists for the next 25 years [80]. Sanger sequencing was based on the selective and partial incorporation of chain-terminating dideoxyribonucleotides into copies of the DNA strand being sequenced, and their separation by denaturing electrophoresis into a readable sequence “ladder”. Dr Naccache stressed that Sanger sequencing, even in later, high throughput versions, was employed on one limited piece of DNA (typically a small DNA sector amplified using PCR) at a time to yield a single output sequence. Although this method was sufficiently advanced to be used to sequence first the human mitochondrial genome [81] and then the entire human genome [82], [83], its robustness pales in comparison to NGS approaches. NGS yields huge amounts of DNA sequence in parallel; in other words, many sequences in a sample can be analyzed simultaneously [84]. For this reason, NGS is also called deep or massively parallel or high throughput sequencing. Its information output is such that thousands or millions of sequences can be provided concurrently, in a matter of hours. NGS has made possible metagenomic approaches, in which all DNA sequences from all genomes present in a clinical sample can be identified. This approach can now be used to determine if a blood sample contains only human DNA, or human DNA plus the DNA of an infectious pathogen, as well as to identify that pathogen if its sequence is known.
A brief technical overview of NGS methodology was provided by Dr Naccache. All DNA/RNA present in a sample is first rapidly extracted (or copied using PCR) and fragmented into pieces of uniform length, providing a library. Short artificial DNA sequences are then bonded onto either end of all library DNA fragments. These adaptors allow hybridization of one strand of the modified DNA to complementary, tethered pieces of DNA in a flow cell. They are then “sequenced by synthesis” using fluorescent deoxyribonucleotide triphosphate building blocks. As each base is added to the growing chain, its position is noted via imaging and the positional information is captured in parallel. NGS can therefore provide 10 to 100 Gigabytes of data—much of it “redundant” in that the same sequence has been detected and read multiple times—to ensure sufficient coverage of all DNA sequence present in a sample. This huge amount of data is first processed using algorithms that detect and remove low quality reads, and then the “host” or human genome sequence information is subtracted. The remaining DNA is assembled into contiguous arrays by alignment of overlapping sequences, and compared to reference genomes of known pathogens. If an identified pathogen differs slightly from reference genomes, taxonomic classification can then be done to determine how recently the variant has diverged from known sequences. Dr Naccache stressed that this approach will work on bacterial, viral, fungal, and parasitic pathogens, all encoded by RNA- or DNA-based genomes, but not on prions, which are protein-based pathogens that infect by causing host proteins to take on pathological conformations.
NGS is an advanced technology approach fully dependent on the characteristics of the instrumentation employed. Dr Naccache surveyed the rapid development of commercial deep sequencing machines [85]. First to market in 2005 was Roche, with its 454 Sequencer, capable of 500 000 reads of 350 to 550 bp. Illumina and Ion Torent produced instruments that generated millions to billions of reads of slightly shorter sizes of 50 to 300 bp, with similar overall run times, between 2009 and 2014. These products all worked on the paradigm of sequencing by synthesis. The most recent entries into this instrumentation field work on a different principle, called nanopore sequencing: Pac Bio's RS apparatus (2011); and Oxford Nanopore Technology's MinION (2014). Both instruments are capable of a smaller number of reads than earlier machines (up to 100,000) but the reads are much longer, up to 40,000 bp. The technology works on the principle of detecting a growing DNA chain electrically when the chain is extruded through an engineered protein pore of nanometer diameter. The MinION instrument is amazingly small to essentially the size of a large memory stick [86].
Effective exploitation of the “mountain” of DNA information produced by NGS from a clinical or blood bank sample requires minimizing computation time and maximizing the accuracy of the diagnostic output. Dr Naccache and co-workers developed a bioinformatics platform for these tasks called SURPI, Sequence-based Ultra-Rapid Pathogen Identification [87]. Such platforms are necessary given the size of the NGS output DNA sequence, the size of pathogen reference sequence databases, and the fact that pathogen sequences typically constitute no more than 0.001% to 1% of reads in the NGS output. This presents a needle in a haystack-type problem. To achieve rapid and robust detection, SURPI employs two analyzer programs that work on both DNA and translated protein alignments, using bacterial and viral databases. The platform can be employed in either fast or comprehensive mode; in fast mode pathogenic sequences can be identified in a clinical sample in minutes to hours, while comprehensive mode is more appropriate for detection of a novel pathogen's entire genome or to rule out infection in a clinically complex case. Dr Naccache noted that 20% to 30% of clinically significant respiratory infections are currently of unknown etiology.
Dr Naccache presented data comparing the time of completion of analysis of NGS data using either SURPI mode on a variety of clinical sample types. Serum took less time to analyze than biological materials open to the environment (eg, stool samples); HIV spiked into plasma could be detected in minutes in either mode, down to 100 viral copies per milliliter, with successful identification of strain specificity. SURPI was extensively tested and optimized using such clinical samples prior to its employment in a prospective clinical case series carried out at the UCSF between April 2013 and December 2014. This was a single-site study with respect to analysis, but included cases referred from across the United States and also from Europe. The study included acutely or chronically ill, hospitalized patients with clinical features suggestive of an infectious disease but who tested negative for all candidate agents.
Dr Naccache highlighted two of the diagnoses achieved by NGS in the clinical series. The first involved an adolescent boy whose fever and headaches evolved over the course of four months to hydroencephalopathy that forced his physicians to induce a coma to stabilize him [88]. Following over a hundred inconclusive laboratory tests, NGS revealed the presence of Leptospira santarosai, a pathogenic bacterium, in cerebrospinal fluid samples. In view of the patient's poor status and the safety of the specific treatment for Leptospira had the NGS-based diagnosis been incorrect, intravenous penicillin treatment was commenced before confirmation of Leptospira infection was received from the Center for Disease Control. Following 2 weeks' treatment followed by rehabilitation, the patient made a full recovery. In the second case highlighted by the speaker, a 42-year-old man underwent a bone marrow transplant, with immunosuppression, for treatment of chronic lymphocytic leukemia [89]. A month later he developed tinnitus and partial deafness, which progressed rapidly and was accompanied by increasing mental deterioration. Brain biopsy tissue was assessed by NGS, which detected neuroinvasive astrovirus infection, for which there is no known efficacious therapy; the patient died 4 months post-NGS diagnosis and 7.5 months after the onset of symptoms. NGS sample to answer turnaround times were reported to be 48 hours in the first case [88] and 96 hours in the second [89].
A substantial list of the different pathogens detected by the UCSF/AVDDC group using NGS in different biological fluids such as CSF, respiratory secretions, and blood, was next presented by the speaker. In the latter category, the agents included pathogens familiar to the transfusion medicine-oriented audience such as the viruses Epstein-Barr virus, cytomegalovirus, HIV-1 and -2, West Nile Virus, hepatitis viruses A through E, and CHIKV virus (CHIKV), the bacterium Pseudomonas aeruginosa (which can be transferred from donor skin into blood products by venipuncture) and the parasite Plasmodium falciparum (one of the causative agents of malaria). Also included on the NGS detection list were less familiar agents, such as RNA viruses enterovirus D68, hantavirus, pegivirus, and rhinovirus C, double-stranded DNA viruses such as four variants of human herpesviruses, and the BK and JC viruses, and various single-stranded DNA viruses of the Cycloviridae and Anelloviridae families, the bacterium Salmonella typhi, and the parasite Leishmania infantum. The sensitivity of NGS is underlined by the detection of viruses that are not usually associated with disease (Anelloviridae) or are only associated with disease in immunosuppressed individuals (the BK and JC viruses) that may be part of the “background flora and fauna” in humans that must be discounted in arriving at a bona fide NGS diagnosis.
Dr Naccache continued with a consideration of recent results from the UCSF/AVDDC group employing Nanopore Sequencing. The UCSF/AVDDC participated in a research program sponsored by Oxford Nanopore Technologies designed to probe and optimize the capabilities of the MinION instrument. Dr Naccache reported that, using MinION and a SURPI-like bioinformatics platform called MetaPORE, plasma samples from individuals separately infected with CHIKV, Ebola virus, and hepatitis C were rapidly identified in real time [90]. For CHIKV and Ebola virus, samples contained 107 to 108 copies/ml and were detected within 4 to 10 minutes of data acquisition; lower-titer hepatitis C virus (105 copies/ml) was detected within 40 minutes. The analyzer algorithms successfully identified these viruses despite the relatively error-prone nature of the sequence data. The total sample to answer time was less than 6 hours, a feat apparently unprecedented in the NGS area.
Dr Naccache then provided a cautionary tale illustrating the extreme sensitivity of NGS which involved parvovirus-like hybrid virus, a previously undescribed novel virus related to both Circoviridae and Parvoviridae families, initially detected in Chinese patients with chronic seronegative hepatitis of unknown etiology [91]. The UCSF/AVDDC group also found parvovirus-like hybrid virus by NGS in their hepatitis cohorts, but eventually realized that the virus was present in commercial silica-type spin columns used for DNA purification/concentration, but not in any original patient sample [92]. The silicates are typically sourced from cell walls of diatoms and a 100% concordant PVH sequence was found in environmental metagenomics databases of samples taken from North American coastal waters. This instance of laboratory contamination illustrates the extreme sensitivity of NGS and the methodological stringency that must be brought to its application.
In wrapping up her presentation, Dr Naccache discussed the likelihood that NGS would contribute directly to blood donation screening. She assessed this outcome as somewhat improbable over the near term, in part because blood donation screening is currently highly effective. NGS is currently quite expensive in its most accurate form, leading Dr Naccache to estimate the cost of a well-covered NGS study of around 10 samples using a total of 300 million sequences on the HiSeq platform at US$3000 , and a more shallow study, of the kind described above for rapid detection of Ebola and CHIKV, of around 6 samples using a total of 30 million sequences on the MiSeq platform, at US$1500. However, these estimates do not truly take into account the substantial infrastructure in place at UCSF/AVVDC, which comprises specialized equipment for sample extraction, library generation, and validation, NGS sequencing, and extensive computational analysis, not to mention the skills of medical technologists, researchers, and bioinformatics specialists. The relevance of NGS with respect to keeping the blood system safe from emerging pathogens will more likely lie in determining patterns of disease transmission and providing the confidence necessary to re-qualify a previously deferred blood donor. Suspected transmission by transfusion can be identified rapidly with great sensitivity and specificity using NGS of all implicated donors and recipients. It can also be used in a general, epidemiological sense, to trace patterns of infection and adaptation and speciation of infectious agents. While the contributions of NGS to clinical diagnosis of intractable cases and to public health, with respect to tracking and understanding disease outbreaks are already substantial, and will likely continue to accumulate exponentially, the prospects of using NGS for routine blood donation screening remain remote due to cost and throughput considerations.
Blood-Borne Pathogens: Destroy
The Biological Impact of Pathogen Inactivation on Blood Product Quality
Key Messages
-
•
Pathogen inactivation (PI) of blood products may be advantageous as they overcome some of the limitations of current strategies (eg, assay sensitivity and threats of emerging pathogens).
-
•
PI techniques bring two sides of a coin into blood banking: improved safety versus a negative effect on blood component quality (damage to ~ 20%-25% of platelets).
-
•
In order to improve the quality of pathogen-inactivated blood products, molecular mechanisms triggered by these technologies need to be identified.
-
•
PI-treatment of whole blood might be the emerging method of choice in this field.
Dr Peter Schubert, Research Associate at the Canadian Blood Services' Centre for Innovation and a Clinical Associate Professor in the Department of Pathology and Laboratory Medicine at the University of British Columbia, focused his talk on the need for PI to ensure the safety of the blood supply. He compared the currently available PI technologies and their mechanisms of action, and discussed the potential impact of this technology on product quality.
Dr Schubert noted that blood safety has historically been achieved by mitigating known risks with interventions such as donor screening and universal donor testing for specific pathogens. However, risks remain, as all tests have a detection limit and current testing does not account for unknown or unexpected pathogens. Thus, PI has the potential to improve the safety of blood products by preventing TTIs, especially in platelet products, where bacterial contamination is a particular risk [93].
In the United States, the INTERCEPT blood system from Cerus Corporation is currently licensed for platelet products, and both INTERCEPT and the Octaplas product from Octapharma are licensed for plasma (Table 2 , [94]). In Canada, the only currently licensed PI product is Octaplas plasma. Three different PI systems for platelet concentrates are currently on the market (Table 2). These exploit the fact that pathogen proliferation occurs by replicating DNA or RNA, a mandatory step for all pathogens except prions. All use UV light, with or without a photosensitizer, to damage nucleic acids and subsequently prevent proliferation of pathogens. Many of these systems are in routine use, mostly in Europe and the Middle East [94], and in many other jurisdictions regulatory approval is initiated and under investigation. Worldwide, the Cerus INTERCEPT blood system is the most adopted system, and has been in routine use for over 10 years. Hemovigilance data from jurisdictions that use these products are highly favorable and support their safety and efficacy [95].
Table 2.
Pathogen reduction technology systems and products currently approved and in use
| System/product | Manufacturer | Mechanism | Product(s) for which use is approved | Used in⁎ |
|---|---|---|---|---|
| INTERCEPT Blood System [131] (technology) |
Cerus | Photosensitizer (amotosalen) + UV A (320-400 nm) illumination | Plasma:
|
Plasma: 13 countries |
Platelets:
|
Platelets: 22 countries | |||
| Mirasol Pathogen Reduction System [132] (technology) |
TerumoBCT | Photosensitizer (riboflavin) + UV B (280-360 nm) illumination | Plasma:
|
Plasma: 11 countries |
Platelets:
|
Platelets: 18 countries | |||
| The THERAFLEX MB-Plasma System§[133], [134] (technology) |
Macopharma | Filtration (0.65 μm), methylene blue + visible light (~ 400-700 nm) illumination | Single unit fresh frozen plasma
|
15 countries |
| Octaplas [135] (product) |
Octapharma | Solvent/detergent treatment (1% trinitrobutyl phosphate/1% Triton X-100) | Pooled plasma
|
32 countries |
Several clinical trials of PI technologies have taken place or are underway, including euroSPRITE (looking at INTERCEPT-treated platelet concentrates [96]), SPRINT (looking at INTERCEPT-treated apheresis platelets [97]), and MIRACLE (looking at Mirasol-treated apheresis platelets [98]). Although safety and levels of adverse transfusion reactions were favorable with PI-treated platelets, one observation was that approximately 20% to 25% of the platelets appear to be damaged by the PI treatment. The SPRINT and MIRACLE trials demonstrated a lower mean 24-hour post-transfusion count increment, increased number of platelet transfusions, and lower 1-hour corrected count increment, respectively, for patients treated with PI platelets [98], [99].
The PREPAReS (Pathogen Reduction Evaluation and Predictive Analytical Rating Score) trial is a recently completed prospective, randomized, single-blinded, multicenter non-inferiority trial comparing Mirasol-treated and standard of care pooled platelet products in hemato-oncological patients [100]. Initiated in the Netherlands in November 2010, PREPAReS is sponsored by the Sanquin Blood Supply Foundation, the national blood operator in the Netherlands, and financially supported by TerumoBCT. The Canadian arm of the trial involved CBS producing Mirasol-treated pooled platelets at its Ottawa manufacturing site and several hospitals in Ontario. To account for the observed damage seen in PI techniques in other trials and to be consistent with the preparation of platelet pools in the Netherlands, Mirasol-treated platelets contain the donation of five donors, rather than four, which is the standard buffy coat platelet product prepared by CBS.
Although simply increasing the platelet dose in this setting is a straightforward solution to the issue of platelet damage seen with PI, Dr Schubert noted that the mechanisms of this PI-associated damage are unknown. Currently in the literature, there is debate regarding the clinical efficacy of PI platelets. A meta-analysis of bleeding complications in randomized controlled trials using the INTERCEPT system suggests an increased risk of clinically significant bleeding [101] while a meta-analysis from the Cochrane Collaboration suggests no difference in bleeding with PI platelets, although this conclusion is limited by significant heterogeneity between studies [102].
Thus, elucidating potential mechanisms is important to further fine-tune these systems and Dr Schubert provided an overview of the effect on quality parameters of platelet products by the three different PI systems. The common trend is a negative impact on routine quality measures as well as newer tools introduced to further characterize platelet quality and functionality; however, the magnitude of the effect can be different dependent on the PI technology. Amongst other effects, PI treatment increases metabolism, apoptosis development, and platelet activation. These features lead to decreased platelet responsiveness and in vitro clot formation [103]. Furthermore, the effect of PI treatment on novel aspects of platelet function has also been extensively investigated. These studies show PI-dependent effects on cytokine [104], mitochondrial DNA and microparticle release [105], [106], [107], generation of reactive oxygen species and initiation of signaling cascades [108], and expression profiles of mRNA and miRNA [109], [110]. These “novel” platelet features might assist with explaining PI-effects associated with increased inflammation, cell damage, and apoptosis, transfusion-related acute lung injury, and modulation of endothelial cell functions. In addition, studies of the effects of PI on the proteome of platelets could provide further insights into mechanisms, although the overall impact on the protein expression profile is relatively small [111], suggesting PI-triggered modulations of protein activities. Finally, some of Dr Schubert's own work on elucidating signaling cascades in Mirasol-treated platelets revealed a central role of p38MAPK in regulating platelet activation and apoptosis development [112], [113].
Besides platelet concentrates, PI-treatment of plasma is used routinely in some jurisdictions [94]; however, PI of RBC concentrates has been challenging due to their high optical density, requiring a large amount of UV light, accordingly leading to significant RBC damage. The Cerus S-303 system for PI of RBCs demonstrated similar 24-hour recovery to standard RBCs, but these products would not meet Canadian standards due to their decreased shelf life [114]. Dr Schubert then discussed how PI of whole blood may be a more efficient approach compared to PI of individual components. Potential benefits of whole blood PI include early removal of pathogens, protection against transfusion-associated graft versus host disease due to WBC inactivation, and increased safety of all blood product components. The application of the Mirasol technology on whole blood has demonstrated efficacy against HIV [115], Trypansoma cruzi [116], and B microti [117]. Most recently, a study using Mirasol for malaria inactivation in whole blood has been performed in Ghana [118]. However, reduction of TTIs must be balanced against the effect that whole blood PI has on blood component quality. A study of in vivo viability of stored RBCs derived from Mirasol-treated whole blood suggests decreased viability that correlates with quality variables such as hemolysis and ATP concentration [119]. This approach was complemented by a study by Schubert and colleagues further demonstrating that platelet product quality seems to be less affected when produced from whole blood illumination compared to platelet component treatment [120].
Dr Schubert concluded by acknowledging that there are two sides to the PI coin, and that a balance must be achieved between safety and quality. Further research is needed to understand molecular mechanisms that lead to changes in quality with PI and to balance potential reductions in component effectiveness against reduction in TTIs. Ultimately, clinical trials are essential when it comes to making decisions regarding the implementation of pathogen inactivation into blood banking.
Economic and Health Outcome Implications of Introducing New Pathogen Testing and Inactivation Technologies
Key Messages
-
•
In order to be cost-effective, implementation of broad spectrum interventions such as PI are likely to require discontinuation of some current interventions.
-
•
The Risk-Based Decision-Making framework is a useful set of guiding principles for health economic assessments of blood safety interventions.
To end the day, Dr Brian Custer, a Senior Investigator with Blood Systems Research Institute in San Francisco and an Adjunct Professor at the Department of Laboratory Medicine at UCSF, gave an informative lecture on the economic and health outcome implications of introducing new blood safety interventions with a focus on pathogen testing and inactivation technologies.
Dr Custer began his presentation by introducing the audience to the three main concepts that are the basis of economics in general, and of health economics in particular. The first concept, scarcity, stresses the current reality of limited resources and budgets, which may restrict the scope of services health care systems and practitioners are able to provide. The second concept, choice, has to do with choosing how to allocate the limited resources available. The third basic economic concept is opportunity cost, which relates to the next best alternative use of resources. In other words, opportunity cost is the benefit willingly forgone when we do not choose the next best alternative. These three notions summarize the challenge of the decision making process when it comes to economic information.
In most jurisdictions, system-level health care decisions are made using a different decision-making framework. District health authorities resolve problems based on the determinants of health priorities such as: national and regional targets; clinical and research data; health experts' opinions and views; and the public's participation. For example, the FDA annually tracks data on fatalities related to blood collection, transfusion reactions, and transmissible diseases (http://www.fda.gov). Microbial fatalities from transfusion are tracked using variables that experts deem important for focusing future pathogen testing and inactivation efforts such as by the type organism and blood product. These fatality reports help determine the relative burden of fatalities from different causes. As a result, it was shown that in the United States, platelet-related deaths are mainly caused by bacteria and red cell-related deaths are mainly attributable to Babesia. Data sources, such as the FDA fatality data, help health authorities to understand the most important infection risks blood recipients face.
With respect to health economics of the blood supply, Dr Custer suggested that the blood supply aims to serve the public good. Achieving this aim necessitates that scarce resources be allocated in ways that maximize social welfare. The objective enumeration of costs, benefits, and consequences of alternate health programs must be weighed in comparison to society's ethical beliefs and expectations. If resource allocation does not align well with the values that society allocates to certain health outcomes, this may lead to friction between the determinants of health priorities, and ultimately to difficulties in decision making [121].
The Risk-Based Decision-Making framework developed by the Alliance of Blood Operators is intended as a guide for the assessment of blood safety interventions. As an integral part of any blood system's risk-management program, the Risk-Based Decision-Making framework has two components as shown in Figure 1: policy foundations; and the decision making process. Adherence to the framework increases consistency in methods and results, and it integrates contextual issues (social concern, legal considerations) into the decision making process.
The concept of Quality Adjusted Life Years (QALY) has been developed to facilitate decision making by establishing a balance between public good (subjective qualitative outcome) and resource allocation (objective quantitative outcome). QALY is a tool that measures disease burden, including both the quality and the quantity of years lived. It is a measure of a medical intervention's “value for money”. Scientists may make various assumptions when measuring health resource outcomes, from choosing the wrong outcome to choosing an inappropriate test to measure it. These limitations, as well as an incomplete knowledge base, lead to uncertainty in data used for health economic analyses. When measuring health costs, one assumes that some expenditures are more important than others for the health care system; when measuring consequences from an intervention, one assumes that there is a sufficient understanding of the risks and benefits associated with that intervention. Cost per QALY provides a common denominator for comparing interventions, but cannot reduce uncertainty introduced by unavoidable assumptions.
Established thresholds for what is considered “cost-effective” in health care interventions are in the range of $50,000 to $100,000/QALY (all values in US dollars). Blood safety interventions, however, consistently fall well above these thresholds. Good examples are NAT for HIV, HCV, and HBV ($1,500,000-7,500,000/QALY; broad range reflects uncertainty and testing strategy used) serology and PCR for Babesia species ($251,000-6,582,000/QALY) and WNV NAT testing ($520,000-897,000/QALY), and T cruzi antibody testing for Chagas disease ($757,000-1,360,000/QALY).
When it comes to considering pathogen testing and PI, some outcomes of interest may not be available to aid in decision making. For example, QALY data on the utility of testing for CHIKV, dengue viruses, and hepatitis E virus in the blood safety context are not available to inform an economic analysis. Hence, understanding the budget impact of such tests becomes difficult. On the other hand, data regarding Babesia testing are available. Babesia screening in endemic American states was compared to universal screening using QALY and taking into account the number of deaths prevented and the testing methodology used [122].
There are also QALY data available to support the practice of bacterial testing of platelets, and pathogen inactivation of plasma [123]. The data estimate that platelet bacterial testing is more cost-effective ($91,000/QALY) than pathogen inactivation ($497,000/QALY) [123]. When implementation of a cost-effective blood donor test is needed, QALY has been applied to several patient populations and several interventions. Bell et al compared platelet PI using INTERCEPT in leukemia, lymphoma, orthopedic, and cardiac patients [124]. The authors found that the new method's cost effectiveness ($/QALY) is similar to other accepted blood safety interventions. This means the method can be considered cost-effective in the blood safety context while preserving patient quality of life. Dr Custer and colleagues have modeled the cost-effectiveness of PI as an addition to current pathogen testing based on Canadian data. Comparing Mirasol treatment of whole blood, and of platelet and plasma, costs per QALY of $1,276,000 and $1,423,000, respectively (CDN, in this instance), were shown [125]. A more recent cost-utility analysis of implementing PI in Poland suggest high costs, but better cost-effectiveness than found in previous analyses of PI and nucleic acid testing in North America [126].
PI may replace other interventions that currently incur large costs to the health care system such as blood irradiation, bacterial culture, and maintenance of CMV negative inventory [127]. If PI is implemented, removing costs associated with bacterial culture and irradiation could result in a reduction in the cost/QALY of PI by about 14 % [128], [129]. Other operational gains may further offset investment costs, including reduction in product wastage [130].
These analyses are based on assumptions, and while there is a high degree of uncertainty in the results, their potential usefulness, as blood operators move into the era of PI, is substantial. Health economic analysis allows quantification of alternatives and comparison of interventions in different areas of medicine. Broad implementation of PI for blood safety may thus prove unpalatable to funders without compensatory cost reductions in other aspect of component production. Either a more focused application of PI or lack of implementation until lower-cost alternatives are developed may ensue. Decision making based on health economics and on risk assessments is likely to become the new norm in blood transfusion. This would comprise a strikingly different approach to blood safety implementation and decision making than in the past, when novel interventions were added to older ones, sequentially increasing costs and overall blood safety budgets. This shift represents a move towards effectively and transparently managing process change, rather than the traditional approach of reactive change in response to an emerging situation or disaster.
Conclusion
Defending the blood supply from the threat of transfusion-transmissible infections remains a high priority for transfusion services in Canada and around the world. The residual risk from established pathogens is exceptionally small. The emerging agents of greatest concern in the North American setting are arboviruses and Babesia. The 2014 West African Ebola virus epidemic reinforced concerns about environmental change fueling the rapid emergence of pathogens not previously thought to have potential global significance. Pathogen inactivation of blood products may further reduce residual risk from established agents and provide protection from new agents in the medium term; however, the most likely driver for a paradigm shift to the use of pathogen inactivation is to reduce the ever-present risk of bacterial contamination of platelet products. Longer term, the possibility of donor-free cellular products and rapid next generation sequencing could drive down transfusion-transmission rates even further than the impressive safety margins now in place in Canada and other developed countries. Health economic analysis and risk-based decision making will be required to determine if anticipated benefits outweigh the considerable costs of these potential steps.
Disclosures
The 13th Annual International Symposium received funding from the Canadian Blood Services Centre for Innovation. Among the authors of this report, Dr Geraldine Walsh, Dr Mia Golder, Dr Margaret Fearon (author and speaker), and Dr William Sheffield have no actual or potential conflict of interest in the context of the subject of this program over the past five years. Dr Peter Schubert, author and speaker, discloses grant/research support from TerumoBCT and Macopharma. Among the speakers, Dr Allison McGeer, Nancy Angus, and Dr Samia Naccache have no actual or potential conflict of interest in the context of the subject of this program over the past five years. The following relationships that could be perceived as a related or apparent conflict of interest in the context of the subject of this program over the past five years are disclosed by Dr Roger Dodd: grant/research support Grifols and Cerus; consultant with MosaiQ and Roche; Dr Marc Turner: grant/research support from the Wellcome Trust; Dr Brian Custer: grant/research support from Grifols (formerly Novartis), Hologic (formerly Gen-Probe) and Macopharma, and member of the Speakers' Bureau with TerumoBCT. Among members of the planning committee who were not speakers at the event, Dr Kathryn Webert, Dr Robert Skeate, Dr Sophie Chargé, Ahmed Coovadia, and Sue Gregoire have no actual or potential conflict of interest in relation to this program over the past five years. Dr Ed Pryzdial discloses grant/research support from Biogen.
Acknowledgements
We would like to acknowledge the 2015 Planning Committee Members who are not authors on this publication: Dr Kathryn Webert (Co-chair), Medical Director, Utilization, Canadian Blood Services, and Associate Professor, McMaster University; Dr Robert Skeate (Course Director), Associate Medical Director, Medical Services, Canadian Blood Services, and Assistant Professor, University of Toronto; Dr Ed Pryzdial, Scientist, Centre for Innovation, Canadian Blood Services, and Clinical Professor, University of British Columbia; Dr Sophie Chargé, Associate Director, Centre for Innovation, Canadian Blood Services; Mr Ahmed Coovadia, Hospital Liaison Specialist, Medical Services and Utilization, Canadian Blood Services; Miss Sue Gregoire, Senior Administrative Assistant, Centre for Innovation, Canadian Blood Services. Please note, the following authors were also members of the 2015 Planning Committee: Dr William Sheffield (Co-chair), Associate Director and Senior Scientist, Centre for Innovation, Canadian Blood Services, and Professor, McMaster University; Dr Margaret Fearon, Medical Director, Medical Microbiology, Canadian Blood Services, and Assistant Professor, University of Toronto; Dr Mia Golder, Coordinator, Centre for Innovation, Canadian Blood Services; Dr Geraldine Walsh, Scientific Writer, Centre for Innovation, Canadian Blood Services. The program was developed in collaboration with Continuing Education and Professional Development, Faculty of Medicine, University of Toronto. The event was an accredited group learning activity (Section 1) as defined by the Maintenance of Certification program of the Royal College of Physicians and Surgeons of Canada.
References
- 1.Lin Y., Pavenski K., Saidenberg E., Branch D.R. Blood group antigens and normal red blood cell physiology: A Canadian Blood Services Research and Development symposium. Transfus Med Rev. 2009;23:292–309. doi: 10.1016/j.tmrv.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Pavenski K., Saidenberg E., Lavoie M., Tokessy M., Branch D.R. Red blood cell storage lesions and related transfusion issues: A Canadian Blood Services Research and Development symposium. Transfus Med Rev. 2012;26:68–84. doi: 10.1016/j.tmrv.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Saidenberg E., Petraszko T., Semple E., Branch D.R. Transfusion-related acute lung injury (TRALI): A Canadian Blood Services Research and Development Symposium. Transfus Med Rev. 2010;24:305–324. doi: 10.1016/j.tmrv.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Sheffield W.P., Tinmouth A., Branch D.R. Blood group biochemistry: A Canadian Blood Services Research and Development Symposium. Transfus Med Rev. 2005;19:295–307. doi: 10.1016/j.tmrv.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Shehata N., Lin Y., Pendergrast J., Branch D.R. Cellular therapies: A Canadian Blood Services Research and Development Symposium. Transfus Med Rev. 2007;21:317–336. doi: 10.1016/j.tmrv.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Tinmouth A.T., Semple E., Shehata N., Branch D.R. Platelet immunopathology and therapy: A Canadian Blood Services Research and Development Symposium. Transfus Med Rev. 2006;20:294–314. doi: 10.1016/j.tmrv.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Webert K.E., Alam A.Q., Charge S.B., Sheffield W.P. Platelet utilization: A Canadian Blood Services Research and Development Symposium. Transfus Med Rev. 2014;28:84–97. doi: 10.1016/j.tmrv.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Zeller M.P., Al-Habsi K.S., Golder M., Walsh G.M., Sheffield W.P. Plasma and Plasma Protein Product Transfusion: A Canadian Blood Services Centre for Innovation Symposium. Transfus Med Rev. 2015;29:181–194. doi: 10.1016/j.tmrv.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 9.O'Brien S.F., Yi Q.L., Fan W., Scalia V., Fearon M.A., Allain J.P. Current incidence and residual risk of HIV, HBV and HCV at Canadian Blood Services. Vox Sang. 2012;103:83–86. doi: 10.1111/j.1423-0410.2012.01584.x. [DOI] [PubMed] [Google Scholar]
- 10.Montgomery S.P., Brown J.A., Kuehnert M., Smith T.L., Crall N., Lanciotti R.S. Transfusion-associated transmission of West Nile virus, United States 2003 through 2005. Transfusion. 2006;46:2038–2046. doi: 10.1111/j.1537-2995.2006.01030.x. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien S.F., Scalia V., Zuber E., Hawes G., Alport E.C., Goldman M. West Nile virus in 2006 and 2007: the Canadian Blood Services' experience. Transfusion. 2010;50:1118–1125. doi: 10.1111/j.1537-2995.2009.02550.x. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization Chagas disease (American trypanosomiasis). Fact sheet N°340. 2015. http://www.who.int/mediacentre/factsheets/fs340/en/ URL: [Last accessed 16th Dec 2015]
- 13.Bern C., Montgomery S.P. An estimate of the burden of Chagas disease in the United States. Clin Infect Dis. 2009;49:e52–e54. doi: 10.1086/605091. [DOI] [PubMed] [Google Scholar]
- 14.AABB Chagas Biovigilance Network. 2015. http://www.aabb.org/research/hemovigilance/Pages/chagas.aspx URL: [Last accessed 16th Dec 2015]
- 15.O'Brien S.F., Scalia V., Goldman M., Fan W., Yi Q.L., Huang M. Evaluation of selective screening of donors for antibody to Trypanosoma cruzi: Seroprevalence of donors who answer “no” to risk questions. Transfusion. 2014;54:863–869. doi: 10.1111/trf.12219. [DOI] [PubMed] [Google Scholar]
- 16.Vannier E., Krause P.J. Human babesiosis. N Engl J Med. 2012;366:2397–2407. doi: 10.1056/NEJMra1202018. [DOI] [PubMed] [Google Scholar]
- 17.Food and Drug Administration Biological Product and HCT/P Deviation Reports – Annual Summary for Fiscal Year 2014. 2015. http://www.fda.gov/downloads/BiologicsBloodVaccines/SafetyAvailability/ReportaProblem/BiologicalProductDeviations/UCM440635.pdf URL: [Last accessed 16th Dec 2015]
- 18.Herwaldt B.L., Linden J.V., Bosserman E., Young C., Olkowska D., Wilson M. Transfusion-associated babesiosis in the United States: A description of cases. Ann Intern Med. 2011;155:509–519. doi: 10.7326/0003-4819-155-8-201110180-00362. [DOI] [PubMed] [Google Scholar]
- 19.Hewitt P.E., Ijaz S., Brailsford S.R., Brett R., Dicks S., Haywood B. Hepatitis E virus in blood components: A prevalence and transmission study in southeast England. Lancet. 2014;384:1766–1773. doi: 10.1016/S0140-6736(14)61034-5. [DOI] [PubMed] [Google Scholar]
- 20.Leparc-Goffart I., Nougairede A., Cassadou S., Prat C., de Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383:514. doi: 10.1016/S0140-6736(14)60185-9. [DOI] [PubMed] [Google Scholar]
- 21.Gabriele-Rivet V., Arsenault J., Badcock J., Cheng A., Edsall J., Goltz J. Different Ecological Niches for Ticks of Public Health Significance in Canada. PLoS One. 2015;10:e0131282. doi: 10.1371/journal.pone.0131282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Brien S.F., Delage G., Scalia V., Lindsay R., Bernier F., Dubuc S. Seroprevalence of Babesia microti infection in Canadian blood donors. Transfusion. 2016;56:237–243. doi: 10.1111/trf.13339. [DOI] [PubMed] [Google Scholar]
- 23.Leach Bennett J., Blajchman M.A., Delage G., Fearon M., Devine D. Proceedings of a consensus conference: Risk-Based Decision Making for Blood Safety. Transfus Med Rev. 2011;25:267–292. doi: 10.1016/j.tmrv.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Alliance of Blood Operators The risk-based decision-making framework. 2014. https://allianceofbloodoperators.org/abo-resources/risk-based-decision-making/rbdm-framework.aspx URL: [Last accessed 16th Dec 2015]
- 25.Stein J., Besley J., Brook C., Hamill M., Klein E., Krewski D. Risk-based decision-making for blood safety: preliminary report of a consensus conference. Vox Sang. 2011;101:277–281. doi: 10.1111/j.1423-0410.2011.01526.x. [DOI] [PubMed] [Google Scholar]
- 26.Menitove J.E., Leach Bennett J., Tomasulo P., Katz L.M. How safe is safe enough, who decides and how? From a zero-risk paradigm to risk-based decision making. Transfusion. 2014;54:753–757. doi: 10.1111/trf.12569. [DOI] [PubMed] [Google Scholar]
- 27.Dodd R.Y. Emerging pathogens and their implications for the blood supply and transfusion transmitted infections. Br J Haematol. 2012;159:135–142. doi: 10.1111/bjh.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diack A.B., Head M.W., McCutcheon S., Boyle A., Knight R., Ironside J.W. Variant CJD. 18 years of research and surveillance. Prion. 2014;8:286–295. doi: 10.4161/pri.29237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berry M., Gamieldien J., Fielding B.C. Identification of new respiratory viruses in the new millennium. Viruses. 2015;7:996–1019. doi: 10.3390/v7030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sridhar S., Lau S.K., Woo P.C. Hepatitis E: A disease of reemerging importance. J Formos Med Assoc. 2015;114:681–690. doi: 10.1016/j.jfma.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen J.R. Transmission of human immunodeficiency virus (HIV) by blood and blood components. In: Moore S.B., editor. Transfusion-transmitted viral diseases. American Association of Blood Banks; Arlington, VA: 1987. pp. 37–51. [Google Scholar]
- 33.Dodd R.Y., Foster G.A., Stramer S.L. Keeping Blood Transfusion Safe From West Nile Virus: American Red Cross Experience, 2003 to 2012. Transfus Med Rev. 2015;29:153–161. doi: 10.1016/j.tmrv.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Stramer S.L., Hollinger F.B., Katz L.M., Kleinman S., Metzel P.S., Gregory K.R. Emerging infectious disease agents and their potential threat to transfusion safety. Transfusion. 2009;49(Suppl. 2):1S–29S. doi: 10.1111/j.1537-2995.2009.02279.x. [DOI] [PubMed] [Google Scholar]
- 35.Stramer S.L., Dodd R.Y. AABB Transfusion-Transmitted Diseases Emerging Infectious Diseases Subgroup: Transfusion-transmitted emerging infectious diseases: 30 years of challenges and progress. Transfusion. 2013;53:2375–2383. doi: 10.1111/trf.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaunmuktane Z., Mead S., Ellis M., Wadsworth J.D., Nicoll A.J., Kenny J. Evidence for human transmission of amyloid-beta pathology and cerebral amyloid angiopathy. Nature. 2015;525:247–250. doi: 10.1038/nature15369. [DOI] [PubMed] [Google Scholar]
- 37.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stramer S.L., Linnen J.M., Carrick J.M., Foster G.A., Krysztof D.E., Zou S. Dengue viremia in blood donors identified by RNA and detection of dengue transfusion transmission during the 2007 dengue outbreak in Puerto Rico. Transfusion. 2012;52:1657–1666. doi: 10.1111/j.1537-2995.2012.03566.x. [DOI] [PubMed] [Google Scholar]
- 39.Petersen L.R., Epstein J.S. Chikungunya virus: new risk to transfusion safety in the Americas. Transfusion. 2014;54:1911–1915. doi: 10.1111/trf.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerus Corporation First patient enrolled in Cerus' TRUE study with the American Red Cross to address chikungunya and dengue blood safety risks with pathogen reduced platelets. 2015. http://www.cerus.com/Investors/Press-Releases/Press-Release-Details/2015/First-Patient-Enrolled-in-Cerus-TRUE-Study-With-the-American-Red-Cross-to-Address-Chikungunya-and-Dengue-Blood-Safety-Risks-With-Pathogen-Reduced-Platelets/default.aspx URL: [Last accessed 16th Dec 2015]
- 41.Gonzalez J.P., Herbreteau V., Morvan J., Leroy E.M. Ebola virus circulation in Africa: A balance between clinical expression and epidemiological silence. Bull Soc Pathol Exot. 2005;98:210–217. [PubMed] [Google Scholar]
- 42.Martines R.B., Ng D.L., Greer P.W., Rollin P.E., Zaki S.R. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- 43.Ansari A.A. Clinical features and pathobiology of Ebolavirus infection. J Autoimmun. 2014;55:1–9. doi: 10.1016/j.jaut.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Shears P., O'Dempsey T.J. Ebola virus disease in Africa: epidemiology and nosocomial transmission. J Hosp Infect. 2015;90:1–9. doi: 10.1016/j.jhin.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Lanini S., Zumla A., Ioannidis J.P., Di Caro A., Krishna S., Gostin L. Are adaptive randomised trials or non-randomised studies the best way to address the Ebola outbreak in west Africa? Lancet Infect Dis. 2015;15:738–745. doi: 10.1016/S1473-3099(15)70106-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.World Health Organization WHO global health observatory data repository. http://www.who.int/countries/en/ URL: [Last accessed 16th Dec. 2015]
- 47.Belluz J. Vox Healthcare; 2014. Why is Ebola less deadly in America than in Africa? [URL: http://www.vox.com/2014/10/24/7059743/why-is-ebola-virus-outbreak-american-africa-nina-pham. Last accessed 16th Dec 2015] [Google Scholar]
- 48.Ginsburg J.A. The Guardian; 2014. How saving West African forests might have prevented the Ebola epidemic. [URL: http://www.theguardian.com/vital-signs/2014/oct/03/ebola-epidemic-bats-deforestation-west-africa-guinea-sierra-leone-liberia. Last accessed 16th Dec 2015] [Google Scholar]
- 49.Gostin L.O., Friedman E.A. A retrospective and prospective analysis of the west African Ebola virus disease epidemic: Robust national health systems at the foundation and an empowered WHO at the apex. Lancet. 2015;385:1902–1909. doi: 10.1016/S0140-6736(15)60644-4. [DOI] [PubMed] [Google Scholar]
- 50.Nyenswah T., Blackley D.J., Freeman T., Lindblade K.A., Arzoaquoi S.K., Mott J.A. Community quarantine to interrupt Ebola virus transmission - Mawah Village, Bong County, Liberia, August-October, 2014. MMWR Morb Mortal Wkly Rep. 2015;64:179–182. [PMC free article] [PubMed] [Google Scholar]
- 51.Henao-Restrepo A.M., Longini I.M., Egger M., Dean N.E., Edmunds W.J., Camacho A. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: Interim results from the Guinea ring vaccination cluster-randomised trial. Lancet. 2015;386:857–866. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- 52.World Health Organization WHO commends Sierra Leone for stopping Ebola virus transmission. 2015. http://www.afro.who.int/en/sierra-leone/press-materials/item/8139-who-commends-sierra-leone-for-stopping-ebola-virus-transmission.html URL: [Last accessed 16th Dec 2015]
- 53.Alexander K.A., Sanderson C.E., Marathe M., Lewis B.L., Rivers C.M., Shaman J. What factors might have led to the emergence of Ebola in West Africa? PLoS Negl Trop Dis. 2015;9:e0003652. doi: 10.1371/journal.pntd.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woolhouse M.E., Rambaut A., Kellam P. Lessons from Ebola: Improving infectious disease surveillance to inform outbreak management. Sci Transl Med. 2015;7:307rv305. doi: 10.1126/scitranslmed.aab0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fineberg H.V. The paradox of disease prevention: celebrated in principle, resisted in practice. JAMA. 2013;310:85–90. doi: 10.1001/jama.2013.7518. [DOI] [PubMed] [Google Scholar]
- 56.Fallah M., Skrip L.A., d'Harcourt E., Galvani A.P. Strategies to prevent future Ebola epidemics. Lancet. 2015;386:131. doi: 10.1016/S0140-6736(15)61233-8. [DOI] [PubMed] [Google Scholar]
- 57.Bennett J.E., Dolin R., Blaser M.J. 8th ed. Elsevier Health Sciences; 2015. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. [Google Scholar]
- 58.McCulloch E.A., Till J.E. The radiation sensitivity of normal mouse bone marrow cells, determined by quantitative marrow transplantation into irradiated mice. Radiat Res. 1960;13:115–125. [PubMed] [Google Scholar]
- 59.Gregory C.J., Eaves A.C. Three stages of erythropoietic progenitor cell differentiation distinguished by a number of physical and biologic properties. Blood. 1978;51:527–537. [PubMed] [Google Scholar]
- 60.Mountford J.C., Turner M. In vitro production of red blood cells. Transfus Apher Sci. 2011;45:85–89. doi: 10.1016/j.transci.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 61.Fibach E., Manor D., Oppenheim A., Rachmilewitz E.A. Proliferation and maturation of human erythroid progenitors in liquid culture. Blood. 1989;73:100–103. [PubMed] [Google Scholar]
- 62.Fibach E., Manor D., Treves A., Rachmilewitz E.A. Growth of human normal erythroid progenitors in liquid culture: A comparison with colony growth in semisolid culture. Int J Cell Cloning. 1991;9:57–64. doi: 10.1002/stem.5530090108. [DOI] [PubMed] [Google Scholar]
- 63.Douay L., Giarratana M.C. Ex vivo generation of human red blood cells: A new advance in stem cell engineering. Methods Mol Biol. 2009;482:127–140. doi: 10.1007/978-1-59745-060-7_8. [DOI] [PubMed] [Google Scholar]
- 64.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 65.Kaufman D.S., Hanson E.T., Lewis R.L., Auerbach R., Thomson J.A. Hematopoietic colony-forming cells derived from human embryonic stem cells. PNAS. 2001;98:10716–10721. doi: 10.1073/pnas.191362598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi K., Okita K., Nakagawa M., Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 67.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 68.Lengerke C., Grauer M., Niebuhr N.I., Riedt T., Kanz L., Park I.H. Hematopoietic development from human induced pluripotent stem cells. Ann N Y Acad Sci. 2009;1176:219–227. doi: 10.1111/j.1749-6632.2009.04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi K.D., Yu J., Smuga-Otto K., Salvagiotto G., Rehrauer W., Vodyanik M. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobari L., Yates F., Oudrhiri N., Francina A., Kiger L., Mazurier C. Human induced pluripotent stem cells can reach complete terminal maturation: In vivo and in vitro evidence in the erythropoietic differentiation model. Haematologica. 2012;97:1795–1803. doi: 10.3324/haematol.2011.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang X., Shah S., Wang J., Ye Z., Dowey S.N., Tsang K.M. Extensive ex vivo expansion of functional human erythroid precursors established from umbilical cord blood cells by defined factors. Mol Ther. 2014;22:451–463. doi: 10.1038/mt.2013.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang K.H., Bonig H., Papayannopoulou T. Generation and characterization of erythroid cells from human embryonic stem cells and induced pluripotent stem cells: An overview. Stem Cells Int. 2011;2011:791604. doi: 10.4061/2011/791604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mazurier C., Douay L., Lapillonne H. Red blood cells from induced pluripotent stem cells: Hurdles and developments. Curr Opin Hematol. 2011;18:249–253. doi: 10.1097/MOH.0b013e3283476129. [DOI] [PubMed] [Google Scholar]
- 74.Trakarnsanga K., Wilson M.C., Lau W., Singleton B.K., Parsons S.F., Sakuntanaga P. Induction of adult levels of beta-globin in human erythroid cells that intrinsically express embryonic or fetal globin by transduction with KLF1 and BCL11A-XL. Haematologica. 2014;99:1677–1685. doi: 10.3324/haematol.2014.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurita R., Suda N., Sudo K., Miharada K., Hiroyama T., Miyoshi H. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8:e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cambot M., Mazurier C., Canoui-Poitrine F., Hebert N., Picot J., Clay D. In vitro generated Rh(null) red cells recapitulate the in vivo deficiency: A model for rare blood group phenotypes and erythroid membrane disorders. Am J Hematol. 2013;88:343–349. doi: 10.1002/ajh.23414. [DOI] [PubMed] [Google Scholar]
- 77.Giarratana M.C., Rouard H., Dumont A., Kiger L., Safeukui I., Le Pennec P.Y. Proof of principle for transfusion of in vitro-generated red blood cells. Blood. 2011;118:5071–5079. doi: 10.1182/blood-2011-06-362038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mittra J., Tait J., Mastroeni M., Turner M.L., Mountford J.C., Bruce K. Identifying viable regulatory and innovation pathways for regenerative medicine: A case study of cultured red blood cells. N Biotechnol. 2015;32:180–190. doi: 10.1016/j.nbt.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 79.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. PNAS. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen C.Y. DNA polymerases drive DNA sequencing-by-synthesis technologies: both past and present. Front Microbiol. 2014;5:305. doi: 10.3389/fmicb.2014.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 82.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 83.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 84.Pickrell W.O., Rees M.I., Chung S.K. Next generation sequencing methodologies--an overview. Adv Protein Chem Struct Biol. 2012;89:1–26. doi: 10.1016/B978-0-12-394287-6.00001-X. [DOI] [PubMed] [Google Scholar]
- 85.Pareek C.S., Smoczynski R., Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet. 2011;52:413–435. doi: 10.1007/s13353-011-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rusk N. MinION takes center stage. Nat Methods. 2015;12:12–13. doi: 10.1038/nmeth.3244. [DOI] [PubMed] [Google Scholar]
- 87.Naccache S.N., Federman S., Veeraraghavan N., Zaharia M., Lee D., Samayoa E. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilson M.R., Naccache S.N., Samayoa E., Biagtan M., Bashir H., Yu G. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naccache S.N., Peggs K.S., Mattes F.M., Phadke R., Garson J.A., Grant P. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis. 2015;60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Greninger A.L., Naccache S.N., Federman S., Yu G., Mbala P., Bres V. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu B., Zhi N., Hu G., Wan Z., Zheng X., Liu X. Hybrid DNA virus in Chinese patients with seronegative hepatitis discovered by deep sequencing. PNAS. 2013;110:10264–10269. doi: 10.1073/pnas.1303744110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 92.Naccache S.N., Greninger A.L., Lee D., Coffey L.L., Phan T., Rein-Weston A. The perils of pathogen discovery: Origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol. 2013;87:11966–11977. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pietersz R.N., Reesink H.W., Panzer S., Oknaian S., Kuperman S., Gabriel C. Bacterial contamination in platelet concentrates. Vox Sang. 2014;106:256–283. doi: 10.1111/vox.12098. [DOI] [PubMed] [Google Scholar]
- 94.AABB Listing of countries in which pathogen reduction technology systems and products are in use. 2015. https://www.aabb.org/tm/eid/Documents/prt-systems-in-use-country-listing.pdf URL: [Last accessed 16th Dec 2015]
- 95.Prowse C.V. Component pathogen inactivation: a critical review. Vox Sang. 2013;104:183–199. doi: 10.1111/j.1423-0410.2012.01662.x. [DOI] [PubMed] [Google Scholar]
- 96.van Rhenen D., Gulliksson H., Cazenave J.P., Pamphilon D., Ljungman P., Kluter H. Transfusion of pooled buffy coat platelet components prepared with photochemical pathogen inactivation treatment: The euroSPRITE trial. Blood. 2003;101:2426–2433. doi: 10.1182/blood-2002-03-0932. [DOI] [PubMed] [Google Scholar]
- 97.Snyder E., McCullough J., Slichter S.J., Strauss R.G., Lopez-Plaza I., Lin J.S. Clinical safety of platelets photochemically treated with amotosalen HCl and ultraviolet A light for pathogen inactivation: The SPRINT trial. Transfusion. 2005;45:1864–1875. doi: 10.1111/j.1537-2995.2005.00639.x. [DOI] [PubMed] [Google Scholar]
- 98.Mirasol Clinical Evaluation Study Group A randomized controlled clinical trial evaluating the performance and safety of platelets treated with Mirasol pathogen reduction technology. Transfusion. 2010;50:2362–2375. doi: 10.1111/j.1537-2995.2010.02694.x. [DOI] [PubMed] [Google Scholar]
- 99.McCullough J., Vesole D.H., Benjamin R.J., Slichter S.J., Pineda A., Snyder E. Therapeutic efficacy and safety of platelets treated with a photochemical process for pathogen inactivation: The SPRINT Trial. Blood. 2004;104:1534–1541. doi: 10.1182/blood-2003-12-4443. [DOI] [PubMed] [Google Scholar]
- 100.Sanquin The PREPAReS study. 2013. http://www.sanquin.nl/en/research/departments/clinical-transfusion-research/prepares-study/ URL: [Last accessed 16th Dec 2015]
- 101.Vamvakas E.C. Meta-analysis of the studies of bleeding complications of platelets pathogen-reduced with the Intercept system. Vox Sang. 2012;102:302–316. doi: 10.1111/j.1423-0410.2011.01555.x. [DOI] [PubMed] [Google Scholar]
- 102.Butler C., Doree C., Estcourt L.J., Trivella M., Hopewell S., Brunskill S.J. Pathogen-reduced platelets for the prevention of bleeding. Cochrane Database Syst Rev. 2013;3:Cd009072. doi: 10.1002/14651858.CD009072.pub2. [DOI] [PubMed] [Google Scholar]
- 103.Van Aelst B., Devloo R., Vandekerckhove P., Compernolle V., Feys H.B. Ultraviolet C light pathogen inactivation treatment of platelet concentrates preserves integrin activation but affects thrombus formation kinetics on collagen in vitro. Transfusion. 2015;55:2404–2414. doi: 10.1111/trf.13137. [DOI] [PubMed] [Google Scholar]
- 104.Tauszig M.E., Picker S.M., Gathof B.S. Platelet derived cytokine accumulation in platelet concentrates treated for pathogen reduction. Transfus Apher Sci. 2012;46:33–37. doi: 10.1016/j.transci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 105.Bakkour S., Chafets D.M., Wen L., van der Meer P.F., Mundt J.M., Marschner S. Development of a mitochondrial DNA real-time polymerase chain reaction assay for quality control of pathogen reduction with riboflavin and ultraviolet light. Vox Sang. 2014;107:351–359. doi: 10.1111/vox.12173. [DOI] [PubMed] [Google Scholar]
- 106.Bashir S., Cookson P., Wiltshire M., Hawkins L., Sonoda L., Thomas S. Pathogen inactivation of platelets using ultraviolet C light: Effect on in vitro function and recovery and survival of platelets. Transfusion. 2013;53:990–1000. doi: 10.1111/j.1537-2995.2012.03854.x. [DOI] [PubMed] [Google Scholar]
- 107.Picker S.M., Steisel A., Gathof B.S. Cell integrity and mitochondrial function after Mirasol-PRT treatment for pathogen reduction of apheresis-derived platelets: Results of a three-arm in vitro study. Transfus Apher Sci. 2009;40:79–85. doi: 10.1016/j.transci.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 108.Johnson L., Marks D. Treatment of Platelet Concentrates with the Mirasol Pathogen Inactivation System Modulates Platelet Oxidative Stress and NF-kappaB Activation. Transfus Med Hemother. 2015;42:167–173. doi: 10.1159/000403245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Osman A., Hitzler W.E., Ameur A., Provost P. Differential Expression Analysis by RNA-Seq Reveals Perturbations in the Platelet mRNA Transcriptome Triggered by Pathogen Reduction Systems. PLoS One. 2015;10:e0133070. doi: 10.1371/journal.pone.0133070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Osman A., Hitzler W.E., Meyer C.U., Landry P., Corduan A., Laffont B. Effects of pathogen reduction systems on platelet microRNAs, mRNAs, activation, and function. Platelets. 2015;26:154–163. doi: 10.3109/09537104.2014.898178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prudent M., D'Alessandro A., Cazenave J.P., Devine D.V., Gachet C., Greinacher A. Proteome changes in platelets after pathogen inactivation--an interlaboratory consensus. Transfus Med Rev. 2014;28:72–83. doi: 10.1016/j.tmrv.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 112.Chen Z., Schubert P., Culibrk B., Devine D.V. p38MAPK is involved in apoptosis development in apheresis platelet concentrates after riboflavin and ultraviolet light treatment. Transfusion. 2014;55:848–857. doi: 10.1111/trf.12905. [DOI] [PubMed] [Google Scholar]
- 113.Schubert P., Coupland D., Culibrk B., Goodrich R.P., Devine D.V. Riboflavin and ultraviolet light treatment of platelets triggers p38MAPK signaling: inhibition significantly improves in vitro platelet quality after pathogen reduction treatment. Transfusion. 2013;53:3164–3173. doi: 10.1111/trf.12173. [DOI] [PubMed] [Google Scholar]
- 114.Henschler R., Seifried E., Mufti N. Development of the S-303 Pathogen Inactivation Technology for Red Blood Cell Concentrates. Transfus Med Hemother. 2011;38:33–42. doi: 10.1159/000324458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ruane P.H., Edrich R., Gampp D., Keil S.D., Leonard R.L., Goodrich R.P. Photochemical inactivation of selected viruses and bacteria in platelet concentrates using riboflavin and light. Transfusion. 2004;44:877–885. doi: 10.1111/j.1537-2995.2004.03355.x. [DOI] [PubMed] [Google Scholar]
- 116.Tonnetti L., Thorp A.M., Reddy H.L., Keil S.D., Goodrich R.P., Leiby D.A. Evaluating pathogen reduction of Trypanosoma cruzi with riboflavin and ultraviolet light for whole blood. Transfusion. 2012;52:409–416. doi: 10.1111/j.1537-2995.2011.03285.x. [DOI] [PubMed] [Google Scholar]
- 117.Tonnetti L., Thorp A.M., Reddy H.L., Keil S.D., Goodrich R.P., Leiby D.A. Riboflavin and ultraviolet light reduce the infectivity of Babesia microti in whole blood. Transfusion. 2013;53:860–867. doi: 10.1111/j.1537-2995.2012.03791.x. [DOI] [PubMed] [Google Scholar]
- 118.Owusu-Ofori S., Kusi J., Owusu-Ofori A., Freimanis G., Olver C., Martinez C.R. Treatment of Whole Blood With Riboflavin and UV Light: Impact on Malaria Parasite Viability and Whole Blood Storage. Shock. 2015;44(Suppl. 1):33–38. doi: 10.1097/SHK.0000000000000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cancelas J.A., Rugg N., Fletcher D., Pratt P.G., Worsham D.N., Dunn S.K. In vivo viability of stored red blood cells derived from riboflavin plus ultraviolet light-treated whole blood. Transfusion. 2011;51:1460–1468. doi: 10.1111/j.1537-2995.2010.03027.x. [DOI] [PubMed] [Google Scholar]
- 120.Schubert P., Culibrk B., Karwal S., Serrano K., Levin E., Bu D. Whole blood treated with riboflavin and ultraviolet light: Quality assessment of all blood components produced by the buffy coat method. Transfusion. 2014;55:815–823. doi: 10.1111/trf.12895. [DOI] [PubMed] [Google Scholar]
- 121.Custer B., Janssen M.P. Alliance of Blood Operators Risk-Based Decision-Making I. Health economics and outcomes methods in risk-based decision-making for blood safety. Transfusion. 2015;55:2039–2047. doi: 10.1111/trf.13080. [DOI] [PubMed] [Google Scholar]
- 122.Goodell A.J., Bloch E.M., Krause P.J., Custer B. Costs, consequences, and cost-effectiveness of strategies for Babesia microti donor screening of the US blood supply. Transfusion. 2014;54:2245–2257. doi: 10.1111/trf.12805. [DOI] [PubMed] [Google Scholar]
- 123.Janssen M.P., van der Poel C.L., Buskens E., Bonneux L., Bonsel G.J., van Hout B.A. Costs and benefits of bacterial culturing and pathogen reduction in the Netherlands. Transfusion. 2006;46:956–965. doi: 10.1111/j.1537-2995.2006.00828.x. [DOI] [PubMed] [Google Scholar]
- 124.Bell C.E., Botteman M.F., Gao X., Weissfeld J.L., Postma M.J., Pashos C.L. Cost-effectiveness of transfusion of platelet components prepared with pathogen inactivation treatment in the United States. Clin Ther. 2003;25:2464–2486. doi: 10.1016/S0149-2918(03)80288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Custer B., Agapova M., Martinez R.H. The cost-effectiveness of pathogen reduction technology as assessed using a multiple risk reduction model. Transfusion. 2010;50:2461–2473. doi: 10.1111/j.1537-2995.2010.02704.x. [DOI] [PubMed] [Google Scholar]
- 126.Agapova M., Lachert E., Brojer E., Letowska M., Grabarczyk P., Custer B. Introducing Pathogen Reduction Technology in Poland: A Cost-Utility Analysis. Transfus Med Hemother. 2015;42:158–165. doi: 10.1159/000371664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murphy W.G. Pathogen reduction: State of reflection in Ireland. Transfus Clin Biol. 2011;18:488–490. doi: 10.1016/j.tracli.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 128.Berger K., Bauer M., Schopohl D., Henschler R., Ostermann H. Model calculations to quantify clinical and economic effects of pathogen inactivation in platelet concentrates. Onkologie. 2013;36:53–59. doi: 10.1159/000346309. [DOI] [PubMed] [Google Scholar]
- 129.Girona-Llobera E., Jimenez-Marco T., Galmes-Trueba A., Muncunill J., Serret C., Serra N. Reducing the financial impact of pathogen inactivation technology for platelet components: Our experience. Transfusion. 2014;54:158–168. doi: 10.1111/trf.12232. [DOI] [PubMed] [Google Scholar]
- 130.Veihola M., Aroviita P., Linna M., Sintonen H., Kekomaki R. Variation of platelet production and discard rates in 17 blood centers representing 10 European countries from 2000 to 2002. Transfusion. 2006;46:991–995. doi: 10.1111/j.1537-2995.2006.00832.x. [DOI] [PubMed] [Google Scholar]
- 131.Cerus Corporation The INTERCEPT blood system: One system - two components, Cerus Corporation. http://www.interceptbloodsystem.com/product-overview/index URL: [Last accessed 16th Dec 2015]
- 132.TerumoBCT . TerumoBCT; 2012. Mirasol pathogen reduction technology system. [URL: https://www.terumobct.com/location/asia-pacific/Documents/306690231B_plateletEfficacy_WEB.pdf. Last accessed 16th Dec 2015] [Google Scholar]
- 133.Seghatchian J., Struff W.G., Reichenberg S. Main Properties of the Theraflex MB-Plasma System for Pathogen Reduction. Transfus Med Hemother. 2011;38:55–64. doi: 10.1159/000323786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Macopharma The Theraflex MB-plasma system is able to efficiently remove bacteria and bacterial spores from plasma. 2013. http://blood-safety.macopharma.com/en/the-theraflex-mb-plasma-system-is-able-to-efficiently-remove-bacteria-and-bacterial-spores-from-plasma/ URL: [Last accessed 16th Dec 2015]
- 135.Canadian Agency for Drugs and Technologies in Health . Use of Solvent/Detergent-Treated Human Plasma (Octaplas): Pilot Project. Canadian Agency for Drugs and Technologies in Health; Ottawa (ON): 2011. CADTH Optimal Use Reports. [PubMed] [Google Scholar]
- 136.Seghatchian J., Tolksdorf F. Characteristics of the Theraflex UV-Platelets pathogen inactivation system - An update. Transfus Apher Sci. 2012;46:221–229. doi: 10.1016/j.transci.2012.01.008. [DOI] [PubMed] [Google Scholar]