Abstract
Background
An increasing incidence of parapneumonic effusion and pleural empyema (PPE/PE) has been reported in recent studies. As only few data on etiology of PPE/PE in Central Europe have been reported, we undertook a study on the etiology of PPE/PE in children, using both standard culture and molecular techniques.
Methods
This prospective study was conducted between June 2011 and December 2013. Consecutive children with PPE/PE complicating community acquired pneumonia, who required diagnostic/therapeutic thoracentesis were included. Blood and pleural fluid samples for microbiological cultures were collected. Molecular methods were applied to identify Streptococcus pneumonia, Haemophilus influenzae, Staphylococcus aureus, Streptococcus pyogenes, Mycoplasma pneumoniae, Chlamydophila pneumoniae, and respiratory viruses in pleural fluid.
Results
The study group included 64 children, median age 4 (1–15). Seven of 64 (10.9%) blood cultures and 11 of 64 (17.2%) pleural fluid cultures revealed bacterial growth. The most common bacteria detected was S. pneumoniae (13 blood and pleural fluid samples from 11/64 (17.2%) children). DNA sequences of typical bacteria were found in 29/64 (45.3%) pleural fluid samples. S. pneumoniae was identified in 90% of these samples. The most common serotypes were: serotype 6B in 9/26 (36.6%), 19A in 6/26 (23%), serotype 3 in 3/26 (11.5%), 6A and 23F (both in 2/26 i.e. 7.7%) patients. Molecular methods identified atypical bacteria in 8/58 (13.8%) and respiratory viruses in 12/58 (20.7%) pleural fluid samples.
Conclusions
S. pneumoniae, in particular serotype 6B and 19A, is the most common etiologic agent of PPE/PE in Polish children. The use of PCR significantly improves pathogen identification in pleural fluid.
Keywords: Pneumonia, Streptococcus pneumoniae, Pneumococcal serotypes, PCR, Pleural effusion
Highlights
-
•
Pleural empyema is challenging complication of childhood community acquired pneumonia.
-
•
S. pneumoniae is the most common pathogen responsible for pleural empyema in children.
-
•
Serotypes 6B and 19A were most commonly identified as causative pathogens.
-
•
The use of PCR improves the efficacy of pathogen identification in pleural fluid.
-
•
Molecular methods can help to detect atypical bacteria and viruses in pleural fluid.
1. Introduction
Community acquired pneumonia (CAP) remains one of the major causes of morbidity in children worldwide [1]. For years, Streptococcus pneumoniae has been invariably found the most common etiologic factor of CAP [2]. The introduction of the pneumococcal conjugate vaccine (PCV) had an important impact on both the incidence of CAP and its complication rate. The incidence of CAP has been reported to decline in some well developed countries, where PCV had been widely introduced, but at the same time, an increasing incidence of local complications was noted. Data from the United States collected between 1996 and 2007 showed that all-cause pneumonia hospitalization rate in children younger than 2 years decreased by 33%, and the number of children hospitalized due to community acquired pneumococcal pneumonia decreased even by 61%. However, this decline was associated with 2-fold increase of empyema. This phenomenon may have an important impact on the overall efficacy of CAP treatment [3].
Parapneumonic effusion and pleural empyema (PPE/PE) are the most common complications of CAP in children, with the annual incidence of hospitalization reaching 12.5 per 100 000 in some regions of the US [4]. S. pneumoniae remains the most common etiologic agent of PPE/PE in children, exceeding 60% of cases in some studies [5]. Streptococcus pyogenes and Staphylococcus aureus were reported as the second and the third most common species identified as the causes of PPE/PE in children. [3], [5], [6] Nevertheless, the spectrum of causative organisms is much wider and includes Haemophilus influenzae, Streptococcus milleri group, Stenotrophomonas maltophila, anaerobic and atypical bacteria [5], [6], [7]. Some viruses are also considered to play a potential role in the pathogenesis of PPE/PE in children [8].
The treatment of PPE/PE is challenging. Proper antibiotic therapy and an early local intervention are the cornerstones of the effective management. Since the results of individual microbiological studies are usually not available at the time of decision making on antibiotic therapy and the positive results of these studies can be expected in only minority of patients [5], [6], [9], the choice of antibiotics is based on local microbiological data and the results of epidemiological studies. In this context, studies on etiology of PPE and PE have an enormous impact on proper antibiotic treatment in children with local complications of CAP. It has been shown that molecular techniques are significantly more sensitive than standard microbiological cultures. Their use may be associated with even several fold increase in the detection rate of S. pneumoniae and other bacteria [5], [6], [10].
As some differences in etiology of PPE/PE were observed in earlier studies [5], [6], [9], [11], reliable local microbiological and epidemiological data seem to be a prerequisite for the proper choice of antibiotic therapy. To our knowledge no such data have been published for Poland. This issue seems to be particularly important in terms of increasing proportion of Polish children receiving PCV. Thus, we undertook a study aimed at the evaluation of the etiology of PPE/PE in children managed in the referral pulmonary center in Poland. The secondary goals of the study were: 1) comparison of the diagnostic yield of standard culture with molecular techniques in the determination of PPE/PE etiology in children, 2) comparison of microorganisms responsible for local complications of CAP in children vaccinated vs. non-vaccinated with PCV.
2. Material and methods
2.1. Study design
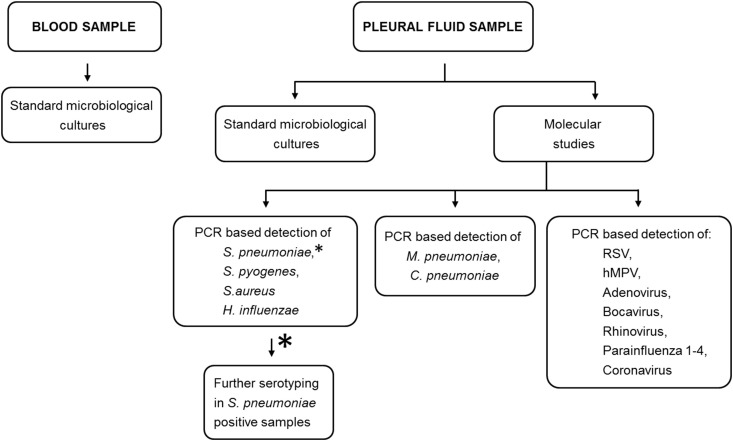
This prospective, observational study included all consecutive children with PPE/PE complicating CAP who were referred to our institution between June 2011 and December 2013, and who required a diagnostic/therapeutic thoracentesis or other more complex pleural intervention. In all patients, blood samples for standard microbiological cultures were collected on the day of admission. Pleural fluid samples were taken between the 1st and 5th day of hospital stay. The diagnostic algorithm used to identify microorganisms in blood and pleural fluid samples is presented in Fig. 1 . All children were treated with intravenous antibiotics. The type of local intervention i.e. therapeutic thoracentesis, chest tube drainage or video-assisted thoracoscopic surgery depended on the clinical course of the disease, the results of pleural fluid analysis and pleural space anatomy (the volume and loculation of pleural effusion).
Fig. 1.
PCR-based detection of S. pneumoniae, S. pyogenes, S. aureus, H. influenzae, M. pneumoniae, C. pneumoniae and respiratory viruses.
2.2. Patients
Children who met the following inclusion criteria were enrolled: age between 1 month and 18 years, CAP defined according to British Thoracic Society (BTS) guidelines [2], pleural effusion that required at least a diagnostic thoracentesis. The exclusion criteria were as follows: immunodeficiency, pleural effusion caused by diseases other than CAP.
2.3. Microbiological investigations
2.3.1. Culture
Culture of blood samples was processed using the BacT/Alert 3D60 platform (bioMérieux, France). Samples of pleural fluids were plated on Columbia agar with 5% sheep blood, Chocolate agar PolyVitex, MacConkey agar (bioMérieux, France), brain heart infusion broth (Grasso, Poland). The plates (Columbia agar, chocolate agar) were incubated at 37 °C in a 5% CO2 –atmosphere or under aerobic conditions (MacConkey, BHI broth) for 3 days. Bacterial isolates were identified using standard laboratory methods, including optochin susceptibility and bile solubility testing for S. pneumoniae, latex agglutination for group A antigen and bacitracin disk sensitivity for S. pyogenes, and latex clumping factor and protein A for S. aureus. All other isolates were identified with VITEK 2 system (bioMérieux, France), the isolates’ identification was confirmed only when probabilities of identification was ≥92%.
2.3.2. Molecular techniques
Pleural fluid samples were stored in temperature −70 °C for a maximum 6 months before testing.
-
a)
Nucleic acid extraction
DNA was extracted from pleural fluid samples with GeneProof PathogenFree DNA nucleic acid extraction kit (GeneProof, Brno, Czech Republic) or QIAamp DNA Mini Kit (Qiagen, Düsseldorf, Germany). RIBO-sorb (Amplisens, Moscow, Russia) nucleic acid extraction kit was used to isolate viral RNA from pleural fluid samples. Both tests were performed in accordance with the manufacturer’s protocols. In case of nucleic acids of Mycoplasma pneumoniae, Chlamydophila pneumoniae and all viruses, extraction from every assayed sample was carried out in the presence of internal control.
-
b)
cDNA synthesis
REVERTA-L reverse transcription kit (Amplisens, Moscow, Russia) was used for complementary DNA (cDNA) synthesis from isolated RNA. The reverse transcription procedure was carried out according to the manufacturer’s instruction.
-
c)
Polymerase chain reaction (PCR)-based detection of C. pneumoniae, M pneumoniae
For detection of the DNA of C. pneumoniae, M. pneumoniae AmpliSens M. pneumoniae/C. pneumoniae - FRT qualitative PCR kit (AmpliSens, Moscow, Russia) was applied. The test was performed according to the manufacturer’s instruction on Rotor-Gene Q instrument (Qiagen, Germany). The real-time hybridization-fluorescence and following amplification conditions were applied: pre-denaturation 95 °C 5 min, I stage denaturation 95 °C 10 s, annealing 63 °C 30 s, elongation 72 °C 10 s, II stage denaturation 95 °C 10 s, annealing 60 °C 30 s, elongation 72 °C 10 s. The human prothrombin gene fragment was used as an endogenous internal control fragment for extraction and amplification stages. Analysis of results measured in three channels (FAM, ROX, JOE) was performed by the software of Rotor-Gene Q instrument.
-
d)
PCR-based detection of respiratory viruses
Multiplex real-time PCR with hybridization-fluorescence detection was applied for detection of seven viral pathogens that cause acute respiratory viral infections. AmpliSens® ARVI-screen-FRT PCR kit was applied for identification of specific nucleic acid fragments of Respiratory Syncytial virus (RSV), human Metapneumovirus (hMPV), Parainfluenza virus-1-4, Coronavirus, Rhinovirus, Adenovirus and Bocavirus. Hot-start technology was used. The total reaction volume was 25 μl, the volume of DNA sample 10 μl. Amplification was carried out on Rotor-Gene Q instrument (Germany) according to the manufacturer’s settings. Data analysis for each PCR-mix was performed individually for each pathogen. The results were interpreted by the software of Rotor-Gene Q. The results of analysis were considered reliable only if the results obtained for positive and negative controls of amplification and negative control of extraction were correct.
-
e)
Detection of S. pneumoniae, H. influenzae, S. aureus, S. pyogenes.
Detection of DNA of S. pneumoniae, H. influenzae, S. aureus, S. pyogenes was performed by PCR with primers specific for the S. pneumoniae ply gene (5′-CAGTCGCCTCTATCCTGGAG and 5′-CTTAGCCAACAAATCGTTTACCGC), H. influenzae hpd gene (5′-CTTTACCATTTTGATGGTGGAACC and 5′-GAAACCAAAGATGGCAAACAAGC), S. aureus species-specific NAPDH reductase gene (5′-GATTCGTTTCGTCGATCAGTTG and 5′-CGGTAAGCCAACACCATCTTG), S. pyogenes spy gene (5′-CTTACCTCAAATTTCCGCAACTC and 5′-AATATCGGCAACCTCTTCAGTG) [12], [13], [14], [15]. DNAs of clinical strains of these species, available from the National Medicines Institute collection were used as positive controls. S. pneumoniae serotypes/serogroups were established by multiplex PCR using the protocol of Siira et al. [16]. Further analysis of serotypes within serogroup 6 was performed by sequencing of the wciP gene fragment with the use of primer pair described by Pai et al. [17] for the first PCR and primer pair 5′-TGGGGGTGTACTGCAGGTT and 5′-CAGGGCAGAACAACACCTTC for the subsequent nested PCR and sequencing.
2.4. Data presentation and statistical analysis
Continuous variables are presented as median and ranges. Data on the results of different microbiological studies of blood and pleural fluid samples are showed as numbers and percentages. Statistical analysis was performed using Statistica 10.0 (StatSoft Inc., Tulsa, USA) and MedCalc Statistical Software version 13.2.2 (MedCalc Software bvba, Ostend, Belgium). The differences between categorical variables (the proportions of positive results of different microbiological studies) were tested using Fischer’s exact test. Cohen kappa coefficient was used to assess the agreement between the results of different microbiological methods. Statistical significance was accepted at p-value less than 0.05.
2.5. Ethical considerations
The protocol of the study was accepted by the Institutional Review Board of Medical University of Warsaw. Informed written consent was obtained from parents or guardians.
3. Results
Sixty four children with PPE/PE (36 boys and 28 girls) median age 4 years (range 1–15 years) were enrolled. The majority of children were treated with antibiotics prior to the collection of blood and pleural fluid samples culture (55 and 64 children, respectively).
Blood and pleural fluid cultures, as well as molecular studies identifying DNA of S. pneumoniae, S. pyogenes, S. aureus and H. influenzae were performed in all 64 patients. Molecular tests detecting specific nucleic acid sequences of respiratory viruses and atypical pathogens were done in 58 (90.6%) patients. The results of microbiological studies (cultures and molecular diagnostics) are summarized in Table 1 .
Table 1.
Microorganisms detected by blood and pleural fluid culture and by pleural fluid PCR in children with parapneumonic effusion/pleural empyema; (−) not applicable.
| Microorganisms | Blood culture n (%) | Pleural fluid |
|
|---|---|---|---|
| Culture n (%) | PCR n (%) | ||
| S. pneumoniae | 3/64 (4.7) | 10/64 (15.6) | 26/64 (40.6) |
| S. pyogenes | 1/64 (1.6) | 1/64 (1.6) | 3/64 (4.7) |
| S. aureus | 0 | 0 | 0 |
| H. influenzae | 0 | 0 | 0 |
| S. hominis | 2/64 (3.1) | 0 | – |
| S. maltophila | 1/64 (1.6) | 0 | – |
| M. pneumoniae | – | – | 3/58 (5.2) |
| C. pneumoniae | – | – | 5/58 (8.6) |
| RSV | – | – | 1/58 (1.7) |
| hMPV | – | – | 1/58 (1.7) |
| Adenovirus | – | – | 8/58 (13.8) |
| Bocavirus | – | – | 1/58 (1.7) |
| Rhinovirus | – | – | 1/58 (1.7) |
| Parainfluenza 1 | – | – | 0 |
| Parainfluenza 2 | – | – | 0 |
| Parainfluenza 3 | – | – | 0 |
| Parainfluenza 4 | – | – | 0 |
| Coronavirus | – | – | 0 |
Seven of 64 (10.9%) blood cultures and 11 of 64 (17.2%) pleural fluid cultures revealed bacterial pathogen growth. The number of positive blood cultures was not significantly different than the number of positive pleural fluid cultures (NS, p = 0.45) and there was only slight agreement between the results of blood and pleural fluid cultures (Cohen’s kappa coefficient = 0.104; 95% CI −0.174–0.382). In all positive pleural fluid samples gram positive cocci that are known to cause pulmonary infections were identified. Specific DNA sequences of typical bacteria were found in 29/64 (45.3%) pleural fluid samples. Thus, the number of positive PCR tests showing bacterial DNA sequences of S. pneumoniae, S. pyogenes, S. aureus and H. influenzae in pleural effusion was significantly higher than the number of positive pleural fluid cultures (p = 0.001). Moderate agreement between the results of pleural fluid culture and PCR test detecting bacterial DNA was demonstrated (Cohen’s kappa coefficient = 0.41; 95% CI 0.216–0.592). Additionally, molecular methods allowed to detect respiratory viruses and atypical bacteria in 12/58 (20.7%) pleural fluid samples. In 22/64 (34%) children all microbiological tests in blood and pleural effusion gave negative results.
S. pneumoniae was the most common bacteria detected by both standard culture as well as PCR. This pathogen was identified in 13 blood and pleural fluid samples collected from 11/64 (17.2%) children (in two children S. pneumoniae was cultured from both specimens). Thus, S. pneumoniae was responsible for 42.8% and 91% of all positive blood and pleural fluid cultures, respectively. No significant difference was found between the number of S. pneumoniae positive blood and pleural fluid cultures (p = 0.076) and there was only fair agreement between S. pneumoniae positive blood and pleural fluid samples (Cohen’s kappa coefficient = 0.255; 95% CI −0.066–0.576). On the other hand, molecular tests revealed 2.6 fold more patients with pneumococcal pleural infection as compared to those identified by standard pleural fluid cultures (26 vs. 10). This difference was statistically significant (p = 0.003). A similar proportion was found for S. pyogenes (1 vs. 3). There was a moderate agreement between pleural fluid cultures and pleural fluid PCR tests in terms of S. pneumoniae detection (Cohen’s kappa coefficient = 0.426; 95% CI 0.227–0.625). Based on the results of culture and PCR tests, S. pneumoniae was found responsible for 40.6% of all pleural infections. In all S. pneumoniae and S. pyogenes positive blood and pleural fluid samples specific DNA sequences of these pathogens has also been found by PCR method.
Pneumococcal serotypes were studied in all 26 children with pneumococcal infection. Serogroup 6 was the most common and was found in 11/26 (40.3%) patients. Partial sequencing of the wciP gene from these samples revealed the presence of nucleotide variant characteristic for 6B serotypes in nine cases and the variant associated with 6A serotypes for two remaining patients. Other identified serotypes were as follows: 19A in 6/26 (23%) pts, serotype 3 in 3/26 (11.5%) pts, serotype 23F in 2/26 (7.7%) pts, serotype 1 in 1/26 (3.8%) pts and serotype 14 in 1/26 (3.8%) pts. In 2/26 (7.7%) samples serotype could not have been determined (ND).
Forty eight children (75%) in the whole study group were not vaccinated against S. pneumoniae, 12 (18.7%) were fully vaccinated and 2 (3.1%) were partially vaccinated. In two children, the vaccination status was unknown. The vaccination status in the subgroup with pneumococcal pleural infection was as follows: 17 pts not vaccinated, 9 pts vaccinated. Details on vaccination status and pneumococcal serotypes found in children with pneumococcal pleural infection are presented in Table 2 .
Table 2.
Pneumococcal serotypes and vaccination status of children with parapneumonic effusion/pleural empyema. ND – not determined, PCV - pneumococcal conjugate vaccine.
| Serotype | Vaccination status | |
|---|---|---|
| 1. | 1 | Not vaccinated |
| 2. | 3 | Not vaccinated |
| 3. | 3 | Not vaccinated |
| 4. | 3 | Fully vaccinated PCV 10 (3 + 1 <1 yr) |
| 5. | 6A | Not vaccinated |
| 6. | 6A | Not vaccinated |
| 7. | 6B | Not vaccinated |
| 8. | 6B | Not vaccinated |
| 9. | 6B | Not vaccinated |
| 10. | 6B | Not vaccinated |
| 11. | 6B | Not vaccinated |
| 12. | 6B | Not vaccinated |
| 13. | 6B | Not vaccinated |
| 14. | 6B | Fully vaccinated PCV 10 (3 + 1 < 1 yr) |
| 15. | 6B | Fully vaccinated PCV 10 (2 doses 1–2 yrs) |
| 16. | 14 | Not vaccinated |
| 17. | 19A | Not vaccinated |
| 18. | 19A | Not vaccinated |
| 19. | 19A | Not vaccinated |
| 20. | 19A | Fully vaccinated PCV 7 (1 dose > 2 yrs) |
| 21. | 19A | Fully vaccinated PCV 13 (1 dose > 2 yrs) |
| 22. | 19A | Fully vaccinated PCV 13 (1 dose > 2 yrs) |
| 23. | 23F | Not vaccinated |
| 24. | 23F | Not fully vaccinated PCV 7 (1 dose 1–2 yrs) |
| 25. | ND | Fully vaccinated PCV 13 (1 dose >2 yrs) |
| 26. | ND | Fully vaccinated PCV 7 (3 + 1 < 1 yr) |
C. pneumoniae was the most common atypical bacteria detected in 5/58 (8.6%) pts. Among viruses, adenovirus was the leading pathogen 8/64 (13.8%) whereas RSV, hMPV, rhino and bocaviruses were found only in single cases. In 8 children, more than one pathogen was detected. Coexistence of S. pneumoniae and adenovirus was found in 3 cases, S. pneumoniae and rhinovirus in 1 case, S. pneumoniae and hMPV in 1 case, both M. pneumoniae and C. pneumoniae were detected in 2 children. S. pneumoniae, adenovirus and RSV was found in one child.
4. Discussion
Our study showed that the most common etiologic factor of PPE/PE in Polish children is S. pneumoniae with significant predominance of serogroup 6 (in particular serotype 6B) and serotype 19A. It should be noted that PCR method applied to pleural fluid samples identified significantly more different microorganisms responsible for PPE/PE than did standard pleural fluid and blood cultures. To our knowledge, this is one of only very few studies on causes of children PPE/PE conducted in recent years in this part of Europe. We believe the strength of our study is the relatively large study group and the use of both conventional cultures as well as modern molecular techniques to identify a wide spectrum of microorganisms (typical and atypical bacteria and respiratory viruses) responsible for pleural infections.
Although data on the etiology of PPE/PE in children vary in different countries, S. pneumoniae has been invariably reported as a leading pathogen [3], [5], [6]. It could have been expected that the introduction of PCV would decrease the proportion of children with pneumococcal pleural infections in favor of other pathogens, e.g. S. pyogenes and S. aureus. Data on such phenomenon are rather equivocal. Indeed, in some countries an increase in the proportion of staphylococcal pleural infections has been observed [3], [18], [19]. In the United States, empyema caused by S. aureus in children less than 2 years old increased 4 - fold between 1996–1998 and 2005–2007 [3]. On the other hand, in a Canadian study by Pernica et al. performed between January 2009 and March 2011 not even a single case of staphylococcal infection was found [5]. At this time all children in Canada were routinely vaccinated against pneumococcal infections. Thus, although we were surprised not to identify any patient with PPE/PE caused by S. aureus, our results are in agreement with the above mentioned results of the Canadian study. In both these studies, the two most common pathogens responsible for pleural infections were S. pneumoniae and S. pyogenes. However, significantly fewer patients with unknown cause of PPE/PE were reported in the study by Pernica et al. than in our study. S. pneumoniae and S. pyogenes were responsible for 62% and 16% of all pleural infections in Canada and for 41% and 5% in Poland, respectively.
Another interesting issue is the changing role of different pneumococcal serotypes in children pleural infections related to PCV introduction. A shift between the serotypes causing PPE/PE has been observed after vaccination with PCV7 [20]. The predominant serotypes responsible for pleural infections in countries where PCV vaccination is common are serotypes 1 and 19A. Serotype 1 infections have been frequently reported in Spain and Turkey [10], [21]. In contrast, in our study S. pneumoniae serotype 1 was detected in only one pleural fluid sample. The second most common serotype responsible for the replacement phenomenon observed after introduction of PCV7 is serotype 19A [6], [22]. Our study showed that, contrary to serotype 1, serotype 19A is also a significant problem in country, where the percentage of vaccinated children is low. This serotype was the second most common serotype in our study - 6/26 (23%). Four children infected with serotype 19A were either not vaccinated or received PCV7, while 2 were vaccinated with PCV13. Both of these children were vaccinated in the third year of life (1 dose). As the immunogenity after a single dose vaccination with serotype 19A has not been reported to be impaired (as was with serotype 3), the serious infections with 19A serotype seem to be somewhat surprising [23]. The most common serogroup in our study, responsible for 40% of all infections was serogroup 6. This could have been expected, as only approximately 20% of our patients received PCV. Of 11 children infected with serogroup 6 (serotype 6A in 2 cases and serotype 6B 9 cases), two had earlier received PCV10. Despite that, these two children had PPE/PE caused by serotype 6B S. pneumoniae, which is covered by PCV10. In contrast to our observation, the studies performed in other countries where the percentage of vaccinated children in the study groups was relatively low (less than 50%) the infections with serogroup 6 were rather rare [10], [21]. Serotype 3 was the third most common serotype found in our study. All cases occurred in non-immunized children: 2 not vaccinated and 1vaccinated with PCV10, while serotype 3 is covered only by PCV13. However, alarming reports on serotype 3 PPE/PE occurring in children after PCV 13 have been published recently [24], [25].
Further changes in pneumococcal serotypes responsible for pleural empyema in children might be expected in the near future. This will be due to wider introduction of PCV 13. For example, in Germany, the rate of increase in the number of invasive pneumococcal disease caused by one of the most emerging serotypes - 19A declined after 2010 [26]. In a recently published study from Scotland, the overall incidence of PPE/PE has fallen since 2010 [27].
Our study showed that other pathogens play only a marginal role as the cause of pleural infections in children. Similarly to other studies, H. influenzae was not found to be present in pleural infections [5], [6], [10]. This may be a consequence of the widespread use of H. influenzae type b vaccine. S. maltophila was detected in blood sample of one patient. However, according to previous studies the role of this organism in pulmonary infection is unclear. There are opinions that S maltophila is contaminant rather than a true pathogen, particularly in immunocompetent patients [28]. On the other hand, there are studies showing that S. maltophila is an important etiologic factor of PPE/PE in immunocompromised adults. Surgery or chest tube drainage as well as broncho-pleural and esophageal-pleural fistulas seem to be risk factors for empyema caused by this pathogen [28]. The similar concerns - true pathogen or contamination – refer to coagulase-negative staphylococci (Staphylococcus hominis identified in blood cultures of two children in our study).
The role of atypical pathogens in children pleural infection is obscure. Although M. pneumoniae was detected in pleural fluid in some studies, it usually represented a small percentage of PPE/PE etiologic factors [29]. In our study, M. pneumoniae and C. pneumoniae DNA was found in 3/58 (5.2%) and 5/58 (8.6%) pleural fluid specimens, respectively. Noteworthy is that in none of our patients M. pneumoniae was isolated as single pathogen (2 pleural fluid samples were also positive for C. pneumoniae and one for S. pneumoniae). There are few studies aimed to detect C. pneumoniae in children with PPE/PE. In a study by Strachan et al., C. pneumoniae was found in one specimen of pleural fluid (1/145–0.7%) and was accompanied by S. pneumoniae [6]. In our study group, DNA of C. pneumoniae was found in 5 pleural fluid samples. Two of these samples were also positive for M. pneumoniae but in 3 patients C. pneumoniae was the only pathogen. In fact, C. pneumoniae was the second most common bacteria found in our patients with pleural infections.
Viruses are rarely considered as a potential etiologic factor of PPE/PE in immunocompetent children and data on the prevalence of viral pleural infection or co-infection are scarce. In our study group, adenovirus was the most common viral agent detected in pleural fluid. This seems to be consistent with data cited by the BTS guidelines [30]. Interestingly, in four of our patients, adenoviruses accompanied pleural infection caused by S. pneumoniae. An increased incidence of PPE/PE during influenza virus pandemic was observed in different parts of the world [10], [31], [32]. In southern Spain, 36% of children with pleural empyema admitted to the hospital during influenza A virus (H1N1) pandemic had influenza co-infection. Single cases of empyema caused by other viruses were also reported, e.g. cytomegalovirus in an immunocompromised child [33]. Recently Games et al. published a study aimed to establish the potential role of torque teno mini viruses (TTMV) in the pathogenesis of parapneumonic empyema in children. The authors showed that TTMV can deeply colonize lungs and are able to modulate the innate immune response of pulmonary cells by inducing the production of inflammatory mediators [34].
We are aware of several, true or potential, limitations of our study. First, in the majority of children antibiotic therapy had been initiated before blood and pleural fluid samples were collected for microbiological studies. Undoubtedly, this could have influenced the results declining the percentage of children with positive blood or pleural fluid cultures. Second, as the microbiological data on the etiology of PPE/PE come from one referral children hospital where mainly patients from Warsaw or surrounding area are treated, they may not ideally reflect epidemiological situation in other regions. Third, although relatively wide range of molecular primers was used to detect the major pathogens that are known to be involved in children pleural infections, it cannot be excluded that some less common microorganisms have not been detected.
Our study confirms the role of new molecular methods in microbiological diagnostics of pleural infections in children. Although some studies showed that PCR only slightly increases the possibility of establishing etiology [35], the majority of publications reported a significant improvement. In one Australian study aimed to determine whether the identification of pneumococcal serotypes by nucleic acid testing in culture negative pleural fluid samples provides additional information, culture of pleural fluid was positive in 5.8% while PCR in 54.4%. Data from Spain and Canada show that introduction of molecular methods increases the detection rate of pathogenic microorganisms from 21% to 79% and 25%–82%, respectively [5], [10]. Also in our study, PCR method allowed an almost 3-fold increase in detection rate of microorganisms in PPE/PE.
5. Conclusions
The most common etiologic agent of PPE/PE in Polish children is S. pneumoniae with significant predominance of serogroup 6 (serotype 6B) and serotype 19A.The use of PCR improves the efficacy of pathogen identification in pleural fluid from children with empyema.
References
- 1.Rudan I., Boschi-Pinto C., Biloglav Z., Mulholland K., Campbell H. Epidemiology and etiology of childhood pneumonia. Bull. World Health Organ. 2008;86:408–416. doi: 10.2471/BLT.07.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris M., Clark J., Coote N. British thoracic society guidelines for the management of community acquired pneumonia in children: update 2011. Thorax. 2011;66(suppl 2):ii1–ii23. doi: 10.1136/thoraxjnl-2011-200598. [DOI] [PubMed] [Google Scholar]
- 3.Grijalva C.G., Nuorti J.P., Zhu Y., Griffin M.R. Increasing incidence of empyema complicating childhood community-acquired pneumonia in the United States. Clin. Infect. Dis. 2010;50:805–813. doi: 10.1086/650573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byington C.L., Hulten K.G., Ampofo K. Molecular epidemiology of pediatric pneumococcal empyema from 2001 to 2007 in Utah. J. Clin. Microbiol. 2010;48:520–525. doi: 10.1128/JCM.01200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pernica J.M., Moldovan I., Chan F., Slinger R. Real-time polymerase chain reaction for microbiological diagnosis of parapneumonic effusions in Canadian children. Can. J. Infect. Dis. Med. Microbiol. 2014;25:151–154. doi: 10.1155/2014/757963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strachan R.E., Cornelius A., Gilbert G.L. Bacterial causes of empyema in children, Australia, 2007–2009. Emerg. Infect. Dis. 2011;17:1839–1845. doi: 10.3201/eid1710.101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nyambat B., Kilgore P.E., Yong D.E. Survey of childhood empyema in Asia: implications for detecting the unmeasured burden of culture-negative bacterial disease. BMC Infect. Dis. 2008;8:90. doi: 10.1186/1471-2334-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galmes J., Li Y., Rajoharison A. Potential implication of new torque teno mini viruses in parapneumonic empyema in children. Eur. Respir. J. 2013;42:470–479. doi: 10.1183/09031936.00107212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eastham K.M., Freeman R., Kearns A.M. Clinical features, aetiology and outcome of empyema in children in the north east of England. Thorax. 2004;59:522–525. doi: 10.1136/thx.2003.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obando I., Camacho-Lovillo M.S., Porras A. Sustained high prevalence of pneumococcal serotype 1 in paediatric parapneumonic empyema in southern Spain from 2005 to 2009. Clin. Microbiol. Infect. 2012;18:763–768. doi: 10.1111/j.1469-0691.2011.03632.x. [DOI] [PubMed] [Google Scholar]
- 11.Baranwal A.K., Singh M., Marwaha R.K., Kumar L. Empyema thoracis: a 10-year comparative review of hospitalised children from south Asia. Arch. Dis. Child. 2003;88:1009–1014. doi: 10.1136/adc.88.11.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salo P., Örtqvist A., Leinonen M. Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J. Infect. Dis. 1995;171:479–482. doi: 10.1093/infdis/171.2.479. [DOI] [PubMed] [Google Scholar]
- 13.Wang X., Theodore M.J., Mair R. Clinical validation of multiplex real-time PCR assays for detection of bacterial meningitis pathogens. J. Clin. Microbiol. 2012;50:702–708. doi: 10.1128/JCM.06087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas L.C., Gidding H.F., Ginn A.N. Development of a realtime Staphylococcus aureus and MRSA (sam-) PCR for routine bloodculture. J. Microbiol. Methods. 2007;68:296–302. doi: 10.1016/j.mimet.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza a (h1n1) – United States, May – August 2009. MMWR Morb. Mortal. Wkly. Rep. 2009;58:1071–1074. [PubMed] [Google Scholar]
- 16.Siira L., Kaijalainen T., Lambertsen L., Nahm M.H., Toropainen M., Virolainen A. From Quellung to multiplex PCR, and back when needed, in pneumococcal serotyping. J. Clin. Microbiol. 2012;50:2727–2731. doi: 10.1128/JCM.00689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pai R., Limor J., Beall B. Use of pyrosequencing to differentiate Streptococcus pneumoniae serotypes 6A and 6B. J. Clin. Microbiol. 2005;43:4820–4822. doi: 10.1128/JCM.43.9.4820-4822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrillo-Marquez M.A., Hulten K.G., Hammerman W. Staphylococcus aureus pneumonia in children in the era of community-acquired methicillin-resistance at Texas children’s hospital. Pediatr. Infect. Dis. J. 2011;30:545–550. doi: 10.1097/INF.0b013e31821618be. [DOI] [PubMed] [Google Scholar]
- 19.Schultz K.D., Fan L.L., Pinsky J. The changing face of pleural empyemas in children: epidemiology and management. Pediatrics. 2004;113:1735–1740. doi: 10.1542/peds.113.6.1735. [DOI] [PubMed] [Google Scholar]
- 20.Raymond F., Boucher N., Allary R. Serotyping of Streptococcus pneumoniae based on capsular genes polymorphisms. PLOS ONE. 2013;8:e76197. doi: 10.1371/journal.pone.0076197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceyhan M., Ozsurekci Y., Gurler N. Distribution of Streptococcus pneumoniae serotypes that cause parapneumonic empyema in Turkey. Clin. Vaccine Immunol. 2013;20:972–976. doi: 10.1128/CVI.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas M.F., Sheppard C.L., Guiver M. Emergence of pneumococcal 19A empyema in UK children. Arch. Dis. Child. 2012;97:1070–1072. doi: 10.1136/archdischild-2012-301790. [DOI] [PubMed] [Google Scholar]
- 23.Frenck R., Jr., Thompson A., Yeh S.H. Immunogenicity and safety of 13-valent pneumococcal conjugate vaccine in children previously immunized with 7-valent pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 2011;30:1086–1091. doi: 10.1097/INF.0b013e3182372c6a. [DOI] [PubMed] [Google Scholar]
- 24.Antachopoulos C., Tsolia M.N., Tzanakaki G. Parapneumonic pleural effusions caused by Streptococcus pneumoniae serotype 3 in children immunized with 13-valent conjugated pneumococcal vaccine. Pediatr. Infect. Dis. J. 2014;33:81–83. doi: 10.1097/INF.0000000000000041. [DOI] [PubMed] [Google Scholar]
- 25.Madhi F., Godot C., Bidet P., Bahuaud M., Epaud R., Cohen R. Serotype 3 pneumococcal pleural empyema in an immunocompetent child after 13-valent pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 2014;33:545–546. doi: 10.1097/INF.0000000000000252. [DOI] [PubMed] [Google Scholar]
- 26.Weil-Olivier C., van der Linden M., de Schutter I., Dagan R., Mantovani L. Prevention of pneumococcal diseases in the post-seven valent vaccine era: a European perspective. BMC Infect. Dis. 2012;12:207. doi: 10.1186/1471-2334-12-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nath S., Thomas M., Spencer D., Turner S. Has the incidence of empyema in Scottish children continued to increase beyond 2005? Arch. Dis. Child. 2015;100:255–258. doi: 10.1136/archdischild-2014-306525. [DOI] [PubMed] [Google Scholar]
- 28.Lee M.R., Wang H.C., Yang C.Y. Clinical characteristics and outcomes of patients with pleural infections due to Stenotrophomonas maltophilia at a medical center in Taiwan, 2004-2012. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:1143–1148. doi: 10.1007/s10096-014-2060-8. [DOI] [PubMed] [Google Scholar]
- 29.Nyambat B., Kilgore P.E., Yong D.E. Survey of childhood empyema in Asia: implications for detecting the unmeasured burden of culture-negative bacterial disease. BMC Infect. Dis. 2008;8:90. doi: 10.1186/1471-2334-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balfour-Lynn I.M., Abrahamson E., Cohen G. BTS guidelines for the management of pleural infection in children. Thorax. 2005;60(Suppl I):i1–i21. doi: 10.1136/thx.2004.030676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.See H., Blonde´ R., Mariani P. Increased incidence of parapneumonic empyema in children at a French pediatric tertiary care center during the 2009 influenza A (H1N1) virus pandemic. Pediatr. Infect. Dis. J. 2010;29:786–787. doi: 10.1097/INF.0b013e3181e6c317. [DOI] [PubMed] [Google Scholar]
- 32.Ampofo K., Herbener A., Blaschke A.J. Association of 2009 pandemic influenza A (H1N1) infection and increased hospitalization with parapneumonic empyema in children in Utah. Pediatr. Infect. Dis. J. 2010;29:905–909. doi: 10.1097/INF.0b013e3181df2c70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eastham K.M., Freeman R., Kearns A.M. Clinical features, aetiology and outcome of empyema in children in the north east of England. Thorax. 2004;59:522–525. doi: 10.1136/thx.2003.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galmes J., Li Y., Rajoharison A. Potential implication of new torque teno mini viruses in parapneumonic empyema in children. Eur. Respir. J. 2013;42:470–479. doi: 10.1183/09031936.00107212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gollomp K., Rankin S.C., White C. Broad-range bacterial polymerase chain reaction in the microbiologic diagnosis of complicated pneumonia. J. Hosp. Med. 2012;7:8–13. doi: 10.1002/jhm.911. [DOI] [PubMed] [Google Scholar]