Abstract
Despite the recent advances in controlling some viral pathogens, most viral infections still lack specific treatment. Indeed, the need for effective therapeutic strategies to combat ‘old’, emergent, and re-emergent viruses is not paralleled by the approval of new antivirals. In the past years, drug repurposing combined with innovative approaches for drug validation, and with appropriate animal models, significantly contributed to the identification of new antiviral molecules and targets for therapeutic intervention. In this review, we describe the main strategies of drug repurposing in antiviral discovery, discuss the most promising candidates that could be repurposed to treat viral infections, and analyze the possible caveats of this trendy strategy of drug discovery.
Keywords: drug repurposing, antiviral drugs, phenotypic screening, mechanism-based screening, combination therapy, emerging viruses
Highlights
Repurposing existing drugs is an emerging strategy for expediting the approval of effective and safe therapeutics, such as for the treatment of orphan drug diseases.
New indications for antiviral activity can be identified for molecules of different origins showing repurposing potential by acting against a previously known target or a new antiviral target.
Innovative approaches for target validation (e.g., gene editing by CRISPR/Cas9) and new experimental models (e.g., organoids) allowed the identification of novel antiviral agents and the unraveling of molecular pathways underlying viral pathogenesis.
Drug repurposing has successfully identified promising candidate drugs that can open new therapeutic avenues to counteract current viral pathogens and possible emerging viruses.
Drug Repurposing in Antiviral Drug Discovery
The identification of new targets for chemotherapeutic intervention is often not paralleled by the development and licensing of new drugs. Antiviral agents for treating viral infectious diseases are not an exception. From 2012 to 2017, only 12 new antivirals have been approved by the Food and Drug Administration (FDA) in the USA, of which 8 are for the treatment of hepatitis C virus (HCV)-related pathologies and 2 are combinations of anti-human immunodeficiency virus (HIV) drugs (www.fda.gov). On the other hand, governments and the World Health Organization (WHO) have to deal with critical (re)emerging viruses, characterized by pandemic potential and responsible for alarming outbreaks in recent years, which still lack specific treatment, such as Zika virus (ZIKV), Ebola virus (EBOV), and Middle East respiratory syndrome coronavirus (MERS-CoV).
A drug discovery approach that has recently become very popular is drug repurposing (DR), which consists of giving old drugs a new indication by exploring new molecular pathways and targets for intervention 1, 2. DR applied to viral infectious diseases takes into account different strategies by integrating both screenings of bioactive small-molecule collections and computational methods (in silico screenings, mining of database with transcriptomic profiles, etc.) in order to find a molecule, a pathway, or a biological activity that could be recycled in fighting a viral pathogen. Beyond the unquestionable economic advantage derived from such an approach in the drug development process, repurposed drugs can quickly enter clinical trials or be employed for compassionate use, especially in the case of viral diseases lacking of specific treatment. Moreover, DR represents a constant source of new knowledge in virus biology as well as of molecules with previously undescribed antiviral properties that can be further used as molecular tools in uncovering molecular mechanisms of virus replication and pathogenesis. In many cases, DR points out previously unexplored cellular pathways, turning them into targets for new therapeutic strategies, even if the identified molecules cannot be introduced in clinical therapy. In this review, we describe the most meaningful results of this ‘from bed to bench’ approach, discuss the most promising drugs that could be repurposed to treat viral infections, and analyze the possible caveats of this trendy strategy of drug discovery, whose advantages and pitfalls are summarized in Table 1 .
Table 1.
Advantages and Pitfalls of a Drug Repurposing Approach for Antiviral Drug Discovery
| Advantages | Pitfalls |
|---|---|
| Low cost and less time-consuming (essential for the development of drugs to treat neglected diseases) | Target identification can be circuitous, and identified drugs may show polypharmacology |
| Possibility to skip preclinical trials (no animal studies) and to directly enter phase 2 clinical trials | Due to the high doses employed in the screenings, toxic drugs can be initially misidentified as active |
| Potential for combination strategies with the possibility to delay or reduce resistance associated with monotherapy | Effective concentrations are often higher than the plasma levels achievable in humans |
| Often analogs (together with pharmacological information) are already available for testing | Medicinal chemistry to design more potent analogs is not applicable without losing repurposing potential |
| Academic/small laboratories can be determinant in the drug-discovery process | Identified drugs are often under intellectual property and/or programs that make them unavailable or unattractive for other pharmaceutical companies that could take over the further development and costs of clinical trials |
| Formulations and manufacturing chains are already established for the large-scale production (launching costs are avoided) |
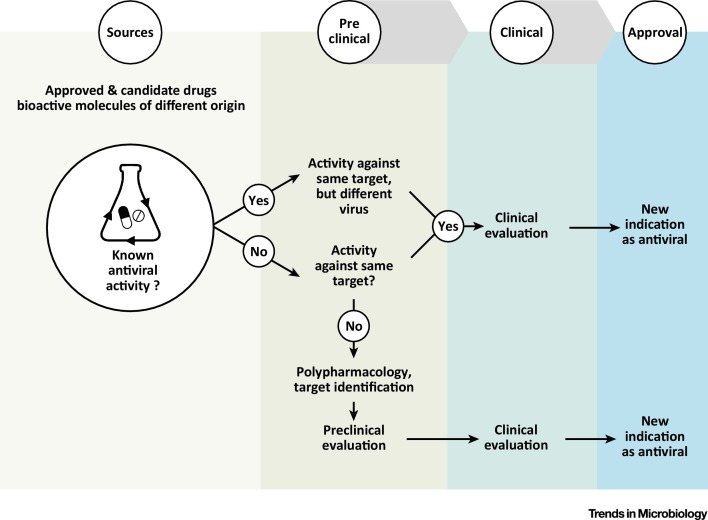
Three different scenarios of antiviral DR can be identified depending on whether the repurposed molecule has previously known antiviral activity (Figure 1 , Key Figure):
Figure 1.
Key Figure: Possible Strategies of Drug Repurposing in Antiviral Drug Discovery
Repurposing of existing drugs, candidate drugs, and bioactive molecules of different origin can be pursued by three main strategies. If the molecule has a previous known antiviral activity, efficacy against other viruses can be hypothesized and tested based on similarity of the target or dependence on common pathways. In the absence of previously identified antiviral activity, the repurposed molecule can either act against the same target or show polypharmacology and interfere with a different function (cellular or viral). In this latter case, target validation cannot be avoided. Skipping preclinical and phase 1 clinical studies when the first two approaches are followed is conceivable; however, it may not be possible if the repurposed molecule shows polypharmacology, and testing the efficacy in suitable animal models remains an essential step.
(i) Same target – new virus. The first option is when an antiviral drug that is known to target a specific viral or cellular function/pathway is found to possess activity against other viruses. The antiviral activity relies on structural homology and common enzymatic features of the viral target or on shared pathways exploited for virus replication. Viral RNA polymerase inhibitors such as favipiravir and sofosbuvir (approved for the treatment of influenza virus and HCV infections, respectively) showed repurposing potential against EBOV and ZIKV (see below). Another example is represented by drugs (e.g., chloroquine) that interfere with the late-stage entry process of viruses, such as filoviruses and coronaviruses, which exploit cellular endocytic pathways to enter the host cell.
(ii) Same target – new indication. This occurs when a pharmacological target (i.e., a protein or a pathway that can be modulated by an approved drug) is found to be essential in a pathogenic process associated with a viral infection. In this case, the approved drug can be exploited also as an antiviral therapeutic agent (new indication). The case is exemplified by the anticancer drug imatinib that inhibits cellular Abelson (ABL) kinase and was found to be also active against pathogenic coronaviruses [3].
(iii) New target – new indication. This occurs when an approved drug with established bioactivity in a specific pathway or mechanism is found to have a new molecular target (i.e., it shows polypharmacology, see Glossary) which is essential for virus replication. Examples are antimicrobial agents (e.g., teicoplanin, ivermectin, itraconazole, and nitazoxanide) that were found to have a target also in virus-infected cells, whose inhibition has detrimental effects on viral replication 4, 5.
Drugs with repurposing potential against viral diseases have been identified mostly by the screening of small-molecule libraries consisting of drugs, both approved and developmental, and other compounds with known bioactivity, including compounds of natural origin. Most of these libraries are available at public institutions or commercially; however, a number of proprietary or in-house-made collections also exist, often consisting of only one class of drugs (e.g., kinase or apoptosis inhibitors, etc.) and can be useful for target deconvolution. Table 2 lists the drug collections most widely used in antiviral DR. In the past 5–10 years, partnerships between public and private institutions have been strongly encouraged by governments to expedite the identification of new candidate drugs, in particular for orphan diseases, and several pharmaceutical companies have opened their proprietary collections to collaboration programs with academic research groups [6].
Table 2.
Small-Molecule Libraries Used in Antiviral Drug Repurposing
| Library (Vendor) | Descriptiona | Refs |
|---|---|---|
| SCREEN-WELL FDA-Approved drugs Library (Enzo Life Sciences) | 774 approved drugs | 8, 47, 69 |
| Library Of Pharmacologically Active Compounds (LOPAC, Sigma-Aldrich) | 1280 bioactive compounds including FDA-approved drugs | 9, 66 |
| Bioactive Compound Library (Selleck Chemicals) |
>2000 bioactive compounds including FDA-approved drugs | 13, 25, 47 |
| Prestwick Library | 1280 bioactive compounds including FDA-approved drugs and candidate drugs | [14] |
| Spectrum Collection (Microsource) |
2320–2560 bioactive compounds including FDA-approved drugs | 48, 62, 63 |
| UCSF Small Molecule Discovery Center Library | 2177 bioactive compounds including FDA-approved drugs | [10] |
| National Institute of Health (NIH) Clinical Collection Library and Chemical Genomics Center (NCCGC) | >7600 bioactive compounds including FDA-approved drugs and candidate drugs | 5, 16, 26, 37, 75 |
A more detailed description of some of these libraries can be found in [1].
Despite the restricted number of available collections, and the fact that often the same drugs are present in various libraries, in several cases different outcomes for the same viral pathogen have been obtained. A possible explanation is the different experimental setup used, including differences in cell type, virus strain, dose, readout method, and so on. Conversely, by screening different libraries, the same drugs were independently identified in some cases (e.g., for MERS-CoV, see below). Finally, virtual screening of chemical structures can also be performed to find new potential antiviral drugs; however, this approach is dependent upon the availability of structural information and a priori knowledge of the target.
Target Validation and Screening Strategies for Antiviral Drug Repurposing
It is not usually required in DR screenings aimed at the discovery of new antiviral drugs to validate a target before the screening; in fact, an ex-post target validation, and even identification, is possible once an active molecule has been identified. Thus, DR screenings can mainly undertake two different experimental approaches: (i) unknown target – phenotypic assay, or (ii) known target − mechanism-based assay. In the first case, the target is generally unknown, and a phenotypic assay is employed for the identification of hit compounds. Phenotypic assays are based on a readout that is synonymous with antiviral activity and can be easily monitored in a high-throughput format. For example, the protection of infected cell monolayers by virus-induced cytopathic effect or the detection of a late-expressed, structural viral protein (envelope protein in most cases) are the most frequently exploited. After hit identification by phenotypic assay, the next step is the identification and validation of the target (a protein, a pathway, etc., which sometimes can be deduced from the mechanism of action of the identified hit compound). The validation is required to demonstrate that the target is indeed essential for virus replication and/or pathogenesis and is subordinated to the demonstration that its modulation leads to protection from virus infection and inhibition of virus-associated pathogenic effects. Experimental approaches for antiviral target validation can be exemplified by genetic knockdown through RNA interference, gene editing by CRISPR/Cas9 system, mutational analysis, and the use of pharmacological inhibitors (if available). All of these can be evaluated in cultured cells, in organoids, in ex vivo experimental models, as well as in animal models. Finally, when possible, a clinical validation on patient-derived samples or data can also be performed. Indeed, when a routinely employed drug is found to be potentially effective in the treatment of a viral infection, a possible positive correlation between drug administration in target patients and inhibition of the viral infection can be retrospectively analyzed in clinical studies.
Alternatively, in mechanism-based DR screenings, target validation is mandatory prior to the screening design. Most frequent targets are druggable viral or cellular enzymes or receptors. The experimental setup in mechanism-based assays is specifically designed to identify molecules able to interfere with a particular process previously identified as essential for productive virus replication or involved in viral pathogenesis. Assays based on a specific mechanism, that is, inhibition of a transcription factor, protection from apoptosis by interfering with a specific activating mechanism, or inhibition of virus entry, are the most popular approaches in DR screenings. Cell-free systems and in vitro assays with purified proteins can be used; however, cellular assays with engineered cell lines, recombinant viruses and/or replicon systems, or pseudoviruses, are preferred as information on target modulation is obtained directly in a physiological environment. Compounds identified by mechanism-based assays in DR screenings against ZIKV, EBOV, and DNA viruses are described below. Although DR screenings based on mechanism-based assays could be more convenient (because the target is known), sometimes false positives can be identified. Other limits are the possible toxicity of the hits due to off-target effects and missing antiviral compounds acting by different mechanisms because the antiviral activity is restricted to a single process established a priori.
In the following sections we describe the most recent advances of repurposing of existing and candidate drugs to treat infections caused by both RNA and DNA viruses (Table 3 ). Priority has been given to drugs that showed efficacy in clinical trials, or in animal models, and to investigational drugs exhibiting effective concentrations compatible with clinically achievable plasma levels in humans.
Table 3.
Approved and Candidate Drugs with Repurposing Potential as Antiviral Agents
| Compound | Status/indication | Virus | Experimental modela | Target | Refs |
|---|---|---|---|---|---|
| Mycophenolic acid | Approved/immunomodulator | ZIKV | Infected cells in vitro | NDb | [8] |
| Daptomycin | Approved/antibacterial | ZIKV | Infected cells in vitro | ND | [8] |
| Niclosamide | Approved/antiparasitic | ZIKV | Infected cells in vitro | ND and NS2B/NS3 protease |
9, 12 |
| Azithromycin | Approved/antibacterial | ZIKV | Infected cells in vitro | ND | [10] |
| Novobiocin | Approved/antibacterial | ZIKV | Infected cell lines in vitro, mouse model | NS2B/NS3 protease | [11] |
| Nanchangmycin | Investigational | ZIKV | Infected cells in vitro, mouse neuron–glia ex vivo cultures | Virus entry | [13] |
| Hippeastrine hydrobromide | Investigational | ZIKV | Infected cells in vitro, organoids, mouse model | ND | [14] |
| Sofosbuvir | Approved/antiviral | ZIKV | Infected cells in vitro, mouse model | NS5 RNA polymerase | 15, 19, 20 |
| Ribavirin | Approved/antiviral | ZIKV | Infected cells in vitro, mouse model | NS5 RNA polymerase | [17] |
| Chloroquine | Approved/antimalarial | ZIKV | Infected cells in vitro, mouse model of vertical transmission | ND | 22, 23 |
| MERS- and SARS-CoV | Infected cells in vitro | ND | [46,47] | ||
| Memantine | Approved/treatment of Alzheimer’s disease | ZIKV | Primary neurons in vitro, mouse model | ND | [24] |
| Prochlorperazine | Approved/antiemetic | DENV | Infected cells in vitro, mouse model | Entry | [28] |
| Chlorcyclizine | Approved/antihistamine | HCV | Chimeric mouse model | Entry? | [29] |
| Manidipine | Approved/antihypertensive | JEV ZIKV |
Infected cells in vitro, mouse model | NS4B | [25] |
| HCMV | Infected cells in vitro | IE2 | [68] | ||
| Favipiravir | Approved/antiviral | EBOV | Phase 2 clinical trial | RNA polymerase L | [33] |
| GS-5734 | Investigational/antiviral | MERS- and SARS-CoV | Nonhuman primates | RNA polymerase | 34, 35 |
| Imatinib | Approved/anticancer | MERS- and SARS-CoV | Infected cells in vitro | Viral fusion | [3] |
| Chlorpromazine | Approved/antipsychotic | MERS- and SARS-CoV | Infected cells in vitro | ND | 46, 47 |
| Chlarithromycin/Naproxen + Oseltamivir | Approved/antibacterial, anti-inflammatory (+antiviral) | Influenza | Phase 2b/3 clinical trials | ND | [56] |
| Nitazoxanide | Approved/antiparasitic | Influenza | Phase 3 clinical trials | Maturation of hemagglutinin | [60] |
| Rotavirus | Phase 2 clinical trials | Viral morphogenesis | www.clinicaltrials.gov/ct2/show/NCT01328925 | ||
| Norovirus | Phase 2 clinical trials | ND | www.clinicaltrials.gov/ct2/show/NCT03395405 | ||
| Raltegravir | Approved/antiviral | Herpesvirus | Infected cells in vitro | Terminase | [70] |
| Lopinavir/ritonavir + interferon β-1b | Approved/antiviral | MERS-CoV | Nonhuman primates, phase 2/3 clinical trial | Protease | 50, 51 |
| Lopinavir/ritonavir | HPV | Proof-of-concept clinical trial | Overexpression RNAse L and? | [72] |
The most advanced phase of drug development is reported.
ND: not determined.
Drug Repurposing for RNA Virus Infections
ZIKV and Other Flaviviruses
ZIKV is an arbovirus that recently caused a large outbreak in Latin America. Generally, ZIKV causes a self-limiting disease; however, ZIKV infection has been also associated with neurologic disorders (such as Guillain–Barré syndrome and others) and with severe congenital defects in newborns (microcephaly and ophthalmological alterations) when infection occurs during pregnancy. As the virus is capable of human-to-human spread through both vertical (transplacental) and sexual transmission, possible ZIKV pandemics are of particular concern for global public health [7]. Neither a specific antiviral treatment nor a vaccine to counteract ZIKV diffusion is available to date. DR campaigns were reported shortly after the WHO’s declaration of ZIKV as a public health emergency in early 2016. In some cases, collections of FDA-approved drugs and bioactive molecules were randomly screened, while in others a specific class of compounds was considered based on previous activity against other flaviviruses or in molecular pathways relevant for pathogenesis, in particular for ZIKV-induced neuropathogenesis.
In the first DR study, among the identified inhibitors of ZIKV replication, the immunosuppressant drug mycophenolic acid and the antibiotic daptomycin were the most promising, because of their wide use during pregnancy to treat other diseases and their ability to cross the placenta [8]. Importantly, for both drugs, the effective anti-ZIKV concentration falls in a range achievable in humans, thus they represent promising candidates for clinical trials. Successive DR screenings identified the FDA-approved antihelminthic drug niclosamide and the macrolide azithromycin, approved as an antibacterial agent, as potent inhibitors of ZIKV replication in different neural cell lines targeted by ZIKV fetal infection 9, 10. Niclosamide and azithromycin are widely used drugs that can also be administered during pregnancy (category B) and their active concentration against ZIKV is clinically achievable. Thus, although further studies are needed to evaluate their antiviral efficacy in vivo, they also represent promising candidates for anti-ZIKV repurposing. Finally, structure-based in silico screenings, followed by in vitro and in vivo experimental studies, led to the identification of another antibiotic, novobiocin, and other FDA-approved drugs (including niclosamide and temoporfin), as novel inhibitors of ZIKV that act by targeting the NS3/NS2B protease 11, 12. In another study, nanchangmycin, a polyether of bacterial origin, blocked ZIKV infection in different cell lines as well as in ex vivo midbrain neuron–glia mixed cultures from embryonic mice [13]. Importantly, nanchangmycin showed antiviral activity also against other viruses that share the same clathrin-dependent entry mechanism. Another natural compound, the alkaloid hippeastrine hydrobromide (HH), recently emerged as a promising inhibitor of ZIKV replication and microcephaly-related effects [14]. The anti-ZIKV effect of HH in human fetal-like forebrain organoids was reported, as well as the rescue of organoid cells from virus-induced death. Importantly, an 18-day treatment of ZIKV-infected organoids with HH resulted in size and thickness comparable to those of uninfected organoids, thus suggesting a potential inhibitory effect of this compound on virus-induced microcephaly during fetal infection. Finally, HH was also effective in a mouse model of ZIKV infection even when added 5 days postinfection, thus highlighting its impressive therapeutic potential.
Studies reporting the repurposing potential against ZIKV of drugs approved for the treatment HCV infections, that is, ribavirin and sofosbuvir, are worth mentioning, although conflicting results on the efficacy of the latter have been reported 15, 16, 17, 18, 19, 20. Discrepancies may be due to the different cell lines or animal models used and should be explained at a molecular level; moreover, deeper investigation on the safety of sofosbuvir during pregnancy is needed before considering this anti-HCV drug as a candidate for the treatment of ZIKV fetal infections. Moreover, several antimalarial drugs showed anti-ZIKV activity 21, 22. In particular, chloroquine was effective in reducing ZIKV vertical transmission in infected pregnant mice [23].
Neuroprotection, when coupled with drugs able to block, directly, virus replication during fetal infection, could be able to attenuate the neurodegenerative effects of ZIKV in the brain and ophthalmological damage derived from virus infection of the optic nerve and ocular tissues. N-methyl-d-aspartate receptor antagonists (in particular memantine, approved for the treatment of Alzheimer’s disease) were the most promising neuroprotective drugs in both human primary neurons and mice infected with ZIKV [24].
Many of the drugs with a repurposing potential against ZIKV are also active against other flaviviruses, suggesting that conserved pathways could be explored for the development of broad- spectrum anti-flavivirus agents. This is the case for mycophenolic acid, niclosamide, nanchangmycin, cloroquine, and other antimalarial drugs. On the other hand, independent DR screenings against Japanese encephalitis virus (JEV), HCV, and dengue virus (DENV) led to the identification of antiviral activity of different FDA-approved drugs 25, 26, 27. In particular, the dopamine D2 receptor antagonist prochlorperazine was identified as a potent inhibitor of HCV and DENV that acts by interfering with virus attachment and entry 26, 28. Another drug with anti-HCV repurposing potential is the antihistamine chlorcyclizine, which showed antiviral activity in both an infectious assay and a chimeric mouse model [29]. To date, however, none of these compounds has been yet evaluated in clinical trials.
Ebola Virus
EBOV is a filovirus responsible for several outbreaks since its discovery in the late 1970s, but the last one, in 2014–2016, was the most alarming due to its size and spread. Indeed, it caused an international health emergency since accidentally imported EBOV cases were recorded in nonendemic geographical areas such as the USA and Europe. EBOV causes a lethal disease characterized by acute hemorrhagic fever and a very high fatality rate (90%). Despite this, and the ease of transmission, no specific therapeutics are available yet. This is worsened by the high level of biocontainment (BSL-4) necessary for the manipulation of this virus that considerably hampers the development of antiviral drugs and vaccines. In this context, DR studies were prompted by the last outbreak (extensively reviewed in 30, 31, 32). EBOV provides a unique example of a DR approach applied directly in clinical trials, as during the last outbreak several drugs were evaluated in infected patients (under both clinical trials and compassionate use) to test their ability to protect from lethal EBOV infection. These included the viral RNA polymerase inhibitors favipiravir (approved in Japan for the treatment of influenza A virus) 31, 33, GS-5734 (an adenosine analog active against highly pathogenic coronaviruses) 34, 35, and amodiaquine (an antimalarial drug widely used in Africa). Treatment of EBOV-infected patients with these drugs has been associated with a decrease in fatality rate compared to control patients [30]; however, their efficacy remains to be confirmed in randomized clinical studies. Preclinical DR studies were also performed with both live viruses and pseudoviruses and led to the identification of a series of approved drugs able to confer protection from lethal EBOV infection in animal models. However, these drugs have not been tested in humans yet. Most of these drugs exert antiviral activity by blocking EBOV at the entry step, and this is biased by the fact that the use of pseudoviruses allows the identification of compounds only inhibiting viral entry. Promising anti-EBOV activity was determined for selective estrogen receptor modulators (cloremiphene and toremiphene 36, 37), chloroquine and related antimalarial drugs [38], sertraline and bepridil [39], teicoplanin [40], cationic amphiphilic drugs such as amiodarone [41], and the broad-spectrum RNA polymerase inhibitor BCX-4430 [42]. Finally, the frontier of DR to directly test a combination of drugs that have shown antiviral activity (although at concentrations not clinically reachable in humans with the approved regimen) gave successful results [43]. Indeed, a targeted drug combination approach resulted in the identification of two sets of three-drug cocktails (i.e., toremifene–mefloquine–posaconazole and toremifene–clarithromycin–posaconazole, all previously identified by DR) that act synergistically in an EBOV entry-inhibition assay and at concentrations achievable in humans [44].
Coronaviruses
Coronaviruses (CoVs) are RNA viruses responsible for gastrointestinal, respiratory, and neurologic diseases in animals, and for zoonotic infections in humans. Of particular importance is their established potential for cross-species transmission with domesticated animals, which act as intermediate hosts and are responsible for infections in humans. The major concerns about CoV infections are their significant morbidity and mortality, emergence of new viruses competent for human-to-human transmission, the potential of domestic animal adaptation, and the difficulty in identifying the intermediate host. Severe acute respiratory syndrome CoV (SARS-CoV), a highly pathogenic CoV, emerged in China in 2002/2003 and was responsible for a pandemic with over 8098 infected people and 10% mortality (www.cdc.gov). MERS-CoV emerged in 2012, has spread to 27 countries so far, and is characterized by a 35% mortality rate [45]. Thus, CoVs represent a huge threat to public health; in this scenario, the search for effective therapeutic strategies is a priority for global public health management. Prompted by the MERS-CoV outbreak, three independent studies reporting an approach by DR for anti-CoV drug discovery were published 46, 47, 48. Approved drugs were tested against MERS- and SARS-CoV, and two molecules were identified in two independent screenings, that is, the dopamine receptor antagonist chlorpromazine and the antimalarial drug chloroquine (both reported to also have antiflavivirus activity). These two drugs exhibited antiviral activity against both viruses, and in a range clinically achievable in humans. However, they did not progress further in preclinical studies. The DR approach also allowed identification of the ABL tyrosine kinase oncogene pathway as essential for the entry of CoVs 3, 46. Indeed, the ABL kinase inhibitor imatinib, approved as an oral anticancer agent, inhibited the replication of both MERS- and SARS-CoV by preventing the fusion of the virus with the endosomal membrane. This is an important example of how DR can be determinant in the identification of host factors required for pathogen replication and in finding novel targets for antiviral intervention. Other host-targeting anti-CoV drugs are cyclophylin A inhibitors such as cyclosporin A and alispovir [49]. This latter drug had antiviral activity against both MERS- and SARS-CoV in cell culture, but failed to show a protective effect in a mouse model of SARS-CoV infection. Nonetheless, cyclophylin A inhibitors deserve further investigation for their pan anti-CoV activity. HIV protease inhibitors were also identified by DR screenings and showed efficacy in combination with interferon β-1b against MERS-CoV in infected cells and in a nonhuman primate model [50]. Currently, a combination of lopinavir/ritonavir and interferon β-1b for the treatment of MERS-CoV syndrome is under clinical evaluation [51].
Influenza Virus
Influenza virus belongs to the family Orthomyxoviridae and is a human pathogen of global public health importance as it causes seasonal, pandemic, and zoonotic influenza disease outbreaks. Pandemic and zoonotic influenza strains are of particular concern because of their high potential for transmission, given the absence of pre-existing immunity in human populations. As opposed to other RNA viruses described so far in this review, there are some current antiviral treatment options for influenza infections. However, both adamantanes (inhibitors of viral ion channel M2) and neuraminidase inhibitors suffer from the rapid emergence of drug-resistant viral strains and side effects, thus limiting their therapeutic efficacy. Therefore, new anti-influenza drugs are being developed [52]. DR campaigns identified some drugs already approved or under clinical evaluation that showed anti-influenza properties, such as BAY 81-8781 (approved as intravenous Aspirin and exerting antiviral activity by blocking NF-kB pathway activation [53]), dapivirine (a non-nucleoside inhibitor of HIV-1 retrotranscriptase [54]), naproxen (which targets influenza nucleoprotein [55]), and the antibiotic clarithromycin. Clarithromycin and naproxen − along with oseltamivir in a three-drug combination – have been evaluated in a phase 2b/3 clinical trial that showed efficacy in the treatment of severe influenza [56]. Also, nalidixic acid and dorzolamide were identified by an in silico screening specifically targeting mutant viral neuraminidase and showed efficacy against oseltamivir-resistant influenza, but not against wild-type viruses [57]. Perhaps the most advanced example of DR is the case of the antiparasitic drug nitazoxanide, which is a safe drug used for the treatment of Giardia and Cryptosporidium spp. infections currently being repurposed for the treatment of influenza 58, 59. Nitazoxanide is a promising anti-influenza drug candidate that interferes with the maturation of viral hemagglutinin at a post-translational level and showed efficacy in different late-stage clinical trials [60]. Moreover, the broad-spectrum antiviral activity demonstrated by this drug (able to block the replication of more than 20 different viruses) raises hopes that it has other possible future indications as an antiviral.
Drug Repurposing for DNA Virus Infections
DR has also been explored in the search for new antiviral strategies directed against DNA viruses not considered ‘emerging’ viral pathogens, but responsible for latent, life-long infections that can cause high morbidity and be life-threatening for at-risk populations. Human cytomegalovirus (HCMV) is a β-herpesvirus that establishes persistent infection and can reactivate under conditions of immunosuppression [61]. HCMV is one of the best characterized examples of host adaptation and of the ability of viruses to subvert, completely, cellular physiological processes in the infected cell. In this context, different DR campaigns identified several approved or investigational drugs with an anti-HCMV mechanism different from that of the currently available drugs 62, 63. This is the case for statins [64], cardiac glycosides [65], the antiparasitic drugs emetine and nitazoxanide 62, 66, kinase inhibitors [67], and the antihypertensive drug manidipine [68]. All of these drugs pharmacologically modulate host proteins; thus, although a specific viral target cannot be excluded, the anti-HCMV activity most likely relies on the interference with host pathways that are intercepted by the virus. Further studies are needed to identify the precise molecular mechanism.
A DR campaign was also performed to identify inhibitors of hepatitis B virus (HBV). Calcium-channel blockers, dopamine receptor antagonist, and the antifungal terbinafine emerged as promising inhibitors of HBV RNA transcription and DNA synthesis [69].
Virus-targeting drugs also showed repurposing potential. This is the case for HIV integrase inhibitors that demonstrated broad-spectrum activity against different herpesviruses by inhibiting the viral terminase 70, 71, and the HIV protease inhibitors lopinavir/ritonavir which were clinically evaluated as a potential treatment for human papilloma virus (HPV)-related preinvasive cervical malignancies [72].
Concluding Remarks and Future Perspectives
The lack of specific therapy for most viral infectious diseases remains a major medical need. The development of effective therapeutics is subordinated to the understanding of molecular mechanisms underlying virus replication and pathogenesis, as well as virus–host interactions, which remain puzzling for several known viral pathogens and also represent the main challenge against future emerging viruses. In this context, evaluating the repurposing potential of existing or candidate drugs can represent an effective strategy for the development of ready-to-use antiviral agents as well as for the identification of new pathways and targets for intervention. Active drugs identified by DR could, in principle, skip preclinical and phase 1 safety/tolerability clinical trials and directly enter phase 2 clinical trials for testing efficacy. Even though this strategy already demonstrated feasibility in the development of new anticancer drugs (such as the antifungal drug itraconazole and its ‘second life’ as an anticancer drug) [73], in antiviral drug discovery there are still only a few successful examples (against influenza [60], EBOV [33], and MERS-CoV [51]) (see Outstanding Questions). Important issues related to DR (summarized in Table 1) still remain to be addressed, in particular the translational aspects of this approach as well as the possible obstacles in pharmaceutical development of drugs showing repurposing potential. The main obstacle to successful repurposing relies perhaps on the effective concentrations required for antiviral activity, which are often higher than those clinically achievable with the approved regimens. Reconsidering the administration protocols in order to reach higher plasma levels is not convenient (because it would mean losing one of the advantages of DR) and could not be feasible due to toxicity issues. The most effective solution to this issue seems to be the development of combination therapies by exploiting the synergism of two or three drugs with different mechanisms of action 43, 74. Promising results in this direction have been accomplished with EBOV [44] and influenza [56], and could be obtained for other viruses. Besides the lower toxicity that can be obtained by decreasing the doses of the combined drugs, combination therapy has also the advantage of reducing resistance issues. There is still a caveat: to act synergistically at a lower dose, drugs need to not interfere with each other and to have different mechanisms of action. Antiviral combination therapies should be most effective when drugs inhibit, as early as possible, different viral functions or host pathways involved in virus replication. This is the reason why the search for new antiviral targets for therapeutic interventions by different approaches still remains mandatory and can be addressed by DR campaigns.
Outstanding Questions.
Will combination therapy overcome the issue of low potency of drugs identified by drug repurposing?
Will the gap left by the lack of effective animal models that recapitulate human viral diseases and virus-associated pathogenesis be filled?
Will a DR approach in antiviral drug discovery translate into the evaluation of repurposed drugs directly to phase II clinical trials similar to what occurs for other diseases such as cancer and neurodegenerative disorders?
Will DR benefit from productive public/private partnerships aimed at overcoming intellectual property, program restriction, and business issues?
Are we prepared for the clinical evaluation of potentially repurposed drugs during a future outbreak of highly pathogenic/pandemic viruses such as EBOV, ZIKV, or other respiratory viruses?
Could all the information and experience acquired in DR campaigns be useful and determinant for the containment of newly emerging viruses in the future?
Acknowledgments
The research in our laboratory was supported by the University of Padua (Progetto di Ricerca di Ateneo 2014, grant PDA141311 to A.L.; STARS-CoG ‘FINDER’ to B.M.) and Associazione Italiana per la Ricerca sul Cancro (AIRC, grant no. IG 18855).
Glossary
- Arbovirus
(ARthropod BOrne VIRUS) virus that can replicate in arthropods and is transmitted to vertebrates (including humans) by hematophagous vectors (mainly mosquitoes, flies, and ticks). Humans, animals, and arthropods can therefore be reservoirs.
- Gene editing by CRISPR/Cas9 system
a type of genetic engineering that is based on the use of the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system, which acts as molecular scissors to delete or insert genetic material in a precise location within the genome of a single cell or a living organism.
- Microcephaly
a medical condition due to a congenital defect where the circumference of the newborn’s head is abnormally smaller than expected. As a consequence, smaller brains in microcephalic babies may have not developed properly or have stopped growing. Microcephaly can be caused by viral infections, toxin exposure, and drugs or alcohol abuse during pregnancy.
- Organoids
three-dimensional (3D) cell cultures that are a simplified yet representative version of an organ, resembling its microanatomy and cell-type composition, and are able to recapitulate some of the represented organ’s functions. These culture systems are obtained in vitro from one or a few cells derived from a tissue, from stem cells, or induced pluripotent stem cells and are able to self-renew and differentiate in multiple, organ-specific cell types. Several 3D organ structures have been obtained to date and represent essential physiological models for the study of biological and pathogenic processes, including viral infections.
- Polypharmacology
describes the ability of an approved drug or a pharmaceutical molecule to act on multiple targets. It can be related to toxic or side-effects of the molecule, or it represents an important opportunity for drug repurposing.
- Pseudovirus
noninfectious, recombinant virus-like particle that resembles the envelope or the capsid of a specific virus, being constituted by the same components. Pseudoviruses may or may not contain genetic material, but they do not have the ability to replicate.
- Synergism
joint action of two or more drugs that produces a combined effectiveness greater than the sum of the efficacy of each individual drug in monotherapy.
- Zoonotic infection
infectious disease that is transmitted from animals to humans. Zoonotic infections may occur through direct contact with infected animals or infected material of animal origin, or may be vector-, air-, water-, or food-borne.
Footnotes
Supplemental information associated with this article can be found, in the online version, at https://doi.org/10.1016/j.tim.2018.04.005.
Supplemental Information
The following are Supplemental information to this article:
References
- 1.Jones L.H., Bunnage M.E. Applications of chemogenomic library screening in drug discovery. Nat. Rev. Drug Discov. 2017;16:285–296. doi: 10.1038/nrd.2016.244. [DOI] [PubMed] [Google Scholar]
- 2.Strittmatter S.M. Overcoming drug development bottlenecks with repurposing: old drugs learn new tricks. Nat. Med. 2014;20:590–591. doi: 10.1038/nm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman C.M. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol. 2016;90:8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colson P., Raoult D. Fighting viruses with antibiotics: an overlooked path. Int. J. Antimicrob. Agents. 2016;48:349–352. doi: 10.1016/j.ijantimicag.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strating J.R.P.M. Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 2015;10:600–615. doi: 10.1016/j.celrep.2014.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loregian A., Palù G. How academic labs can approach the drug discovery process as a way to synergize with big pharma. Trends Microbiol. 2013;21:261–264. doi: 10.1016/j.tim.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Wikan N., Smith D.R. Zika virus: history of a newly emerging arbovirus. Lancet Infect. Dis. 2016;16:e119–e126. doi: 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- 8.Barrows N.J. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu M. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Retallack H. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. U. S. A. 2016;113:14408–14413. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan S. Structure-based discovery of clinically approved drugs as Zika virus NS2B-NS3 protease inhibitors that potently inhibit Zika virus infection invitro and invivo. Antiviral Res. 2017;145:33–43. doi: 10.1016/j.antiviral.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Li Z. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res. 2017;27:1046–1064. doi: 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch K. Screening bioactives reveals nanchangmycin as a broad spectrum antiviral active against Zika virus. Cell Rep. 2017;18:804–815. doi: 10.1016/j.celrep.2016.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell. 2017;21:274–283. doi: 10.1016/j.stem.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullard-Feibelman K.M. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res. 2017;137:134–140. doi: 10.1016/j.antiviral.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adcock R.S. Evaluation of anti-Zika virus activities of broad-spectrum antivirals and NIH clinical collection compounds using a cell-based, high-throughput screen assay. Antiviral Res. 2017;138:47–56. doi: 10.1016/j.antiviral.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Kamiyama N. Ribavirin inhibits Zika virus (ZIKV) replication in vitro and suppresses viremia in ZIKV-infected STAT1-deficient mice. Antiviral Res. 2017;146:1–11. doi: 10.1016/j.antiviral.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mumtaz N. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antiviral Res. 2017;146:161–163. doi: 10.1016/j.antiviral.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Mesci P. Blocking Zika virus vertical transmission. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-19526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacramento C.Q. The clinically approved antiviral drug sofosbuvir inhibits Zika virus replication. Sci. Rep. 2017;7 doi: 10.1038/srep40920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balasubramanian A. Antiviral activities of selected antimalarials against dengue virus type 2 and Zika virus. Antiviral Res. 2017;137:141–150. doi: 10.1016/j.antiviral.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Delvecchio R. Chloroquine, an endocytosis blocking agent, inhibits Zika virus infection in different cell models. Viruses. 2016;8:322. doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiryaev S.A. Repurposing of the anti-malaria drug chloroquine for Zika virus treatment and prophylaxis. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-15467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa V.V. N-Methyl-d-aspartate (NMDA) receptor blockade prevents neuronal death induced by Zika virus infection. mBio. 2017;8 doi: 10.1128/mBio.00350-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S. Screening of FDA-approved drugs for inhibitors of Japanese encephalitis virus infection. J. Virol. 2017;91 doi: 10.1128/JVI.01055-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gastaminza P. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. U. S. A. 2010;107:291–296. doi: 10.1073/pnas.0912966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai J.-H. Pharmacological intervention for dengue virus infection. Biochem. Pharmacol. 2017;129:14–25. doi: 10.1016/j.bcp.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Simanjuntak Y. Repurposing of prochlorperazine for use against dengue virus infection. J. Infect. Dis. 2015;211:394–404. doi: 10.1093/infdis/jiu377. [DOI] [PubMed] [Google Scholar]
- 29.He S. Repurposing of the antihistamine chlorcyclizine and related compounds for treatment of hepatitis C virus infection. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.3010286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweiti H. Repurposed therapeutic agents targeting the Ebola virus: a systematic review. Curr. Ther. Res. Clin. Exp. 2017;84:10–21. doi: 10.1016/j.curtheres.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G. Clinical evaluation of Ebola virus disease therapeutics. Trends Mol. Med. 2017;23:820–830. doi: 10.1016/j.molmed.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bixler S.L. Discovering drugs for the treatment of Ebola virus. Curr. Treat. Options Infect. Dis. 2017;9:299–317. doi: 10.1007/s40506-017-0130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sissoko D. Experimental treatment with favipiravir for Ebola virus disease (the JIKI trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren T.K. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheahan T.P. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansen L.M. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kouznetsova J. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg. Microbes Infect. 2014;3:e84. doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madrid P.B. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansen L.M. A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aaa5597. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y. Teicoplanin inhibits Ebola pseudovirus infection in cell culture. Antiviral Res. 2016;125:1–7. doi: 10.1016/j.antiviral.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salata C. Amiodarone and metabolite MDEA inhibit Ebola virus infection by interfering with the viral entry process. Pathog. Dis. 2015;71:280–281. doi: 10.1093/femspd/ftv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor R. BCX4430 – A broad-spectrum antiviral adenosine nucleoside analog under development for the treatment of Ebola virus disease. J. Infect. Public Health. 2016;9:220–226. doi: 10.1016/j.jiph.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng W. Drug repurposing screens and synergistic drug-combinations for infectious diseases. Br. J. Pharmacol. 2018;175:181–191. doi: 10.1111/bph.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun W. Synergistic drug combination effectively blocks Ebola virus infection. Antiviral Res. 2017;137:165–172. doi: 10.1016/j.antiviral.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dyall J. Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs. 2017;77:1935–1966. doi: 10.1007/s40265-017-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dyall J. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Wilde A.H. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan J.F.W. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Wilde A.H. Alisporivir inhibits MERS- and SARS-coronavirus replication in cell culture, but not SARS-coronavirus infection in a mouse model. Virus Res. 2017;228:7–13. doi: 10.1016/j.virusres.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan J.F.W. Treatment with lopinavir/ritonavir or interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arabi Y.M. Treatment of Middle East respiratory syndrome with a combination of lopinavir-ritonavir and interferon-β1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials. 2018;19:81. doi: 10.1186/s13063-017-2427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loregian A. Antiviral strategies against influenza virus: towards new therapeutic approaches. Cell. Mol. Life Sci. 2014;71:3659–3683. doi: 10.1007/s00018-014-1615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Droebner K. Pharmacodynamics, pharmacokinetics, and antiviral activity of BAY 81-8781, a novel NF-kappaB inhibiting anti-influenza drug. Front. Microbiol. 2017;8:2130. doi: 10.3389/fmicb.2017.02130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Y. Discovery of dapivirine, a nonnucleoside HIV-1 reverse transcriptase inhibitor, as a broad-spectrum antiviral against both influenza A and B viruses. Antiviral Res. 2017;145:103–113. doi: 10.1016/j.antiviral.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lejal N. Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob. Agents Chemother. 2013;57:2231–2242. doi: 10.1128/AAC.02335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hung I.F.N. Efficacy of clarithromycin-naproxen-oseltamivir combination in the treatment of patients hospitalized for influenza A(H3N2) infection: an open-label randomized, controlled, phase IIb/III trial. Chest. 2017;151:1069–1080. doi: 10.1016/j.chest.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 57.Bao J. Drug repurposing identifies inhibitors of oseltamivir-resistant influenza viruses. Angew. Chem. Int. Ed. Engl. 2016;55:3438–3441. doi: 10.1002/anie.201511361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossignol J.F. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral Res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haffizulla J. Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial. Lancet Infect. Dis. 2014;14:609–618. doi: 10.1016/S1473-3099(14)70717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKimm-Breschkin J.L. Prevention and treatment of respiratory viral infections: Presentations on antivirals, traditional therapies and host-directed interventions at the 5th ISIRV Antiviral Group conference. Antiviral Res. 2018;149:118–142. doi: 10.1016/j.antiviral.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mercorelli B. Early inhibitors of human cytomegalovirus: state-of-art and therapeutic perspectives. Pharmacol. Ther. 2011;131:309–329. doi: 10.1016/j.pharmthera.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mercorelli B. Drug repurposing approach identifies inhibitors of the prototypic viral transcription factor IE2 that block human cytomegalovirus replication. Cell Chem. Biol. 2016;23:340–351. doi: 10.1016/j.chembiol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 63.Gardner T.J. Development of a high-content screen for the identification of inhibitors directed against the early steps of the cytomegalovirus infectious cycle. Antiviral Res. 2015;113:49–61. doi: 10.1016/j.antiviral.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ponroy N. Statins demonstrate a broad anti-cytomegalovirus activity in vitro in ganciclovir-susceptible and resistant strains. J. Med. Virol. 2015;87:141–153. doi: 10.1002/jmv.23998. [DOI] [PubMed] [Google Scholar]
- 65.Kapoor A. Human cytomegalovirus inhibition by cardiac glycosides: evidence for involvement of the HERG gene. Antimicrob. Agents Chemother. 2012;56:4891–4899. doi: 10.1128/AAC.00898-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukhopadhyay R. Efficacy and mechanism of action of low dose emetine against human cytomegalovirus. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arend K.C. Kinome profiling identifies druggable targets for novel human cytomegalovirus (HCMV) antivirals. Mol. Cell. Proteom. 2017;16:S263–S276. doi: 10.1074/mcp.M116.065375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mercorelli B. Repurposing the clinically approved calcium antagonist manidipine dihydrochloride as a new early inhibitor of human cytomegalovirus targeting the Immediate-Early 2 (IE2) protein. Antiviral Res. 2017;150:130–136. doi: 10.1016/j.antiviral.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 69.van de Klundert M.A.A. Identification of FDA-approved drugs that target hepatitis B virus transcription. J. Viral Hepat. 2016;23:191–201. doi: 10.1111/jvh.12479. [DOI] [PubMed] [Google Scholar]
- 70.Yan Z. HIV integrase inhibitors block replication of alpha-, beta-, and gammaherpesviruses. mBio. 2014;5 doi: 10.1128/mBio.01318-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nadal M. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16078–16083. doi: 10.1073/pnas.1007144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hampson L. A single-arm, proof-of-concept trial of lopimune (lopinavir/ritonavir) as a treatment for HPV-related pre-invasive cervical disease. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coussens N.P. Small-molecule screens: a gateway to cancer therapeutic agents with case studies of Food and Drug Administration-approved drugs. Pharmacol. Rev. 2017;69:479–496. doi: 10.1124/pr.117.013755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun W. Drug combination therapy increases successful drug repositioning. Drug Discov. Today. 2016;21:1189–1195. doi: 10.1016/j.drudis.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang R. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.