Abstract
The aim of this study was to perform a current molecular characterization of bovine pathogenic Escherichia coli strains isolated from random samplings in Argentinean dairy farms. Rectal swabs were obtained from 395 (63.7 %) healthy and 225 (36.3 %) diarrheic calves, belonging to 45 dairy farms in Cordoba Province, Argentina. E. coli isolates were examined for virulence genes (f5, f41, f17, sta, stb, lt, eae, vt) using PCR and the prevalence of E. coli virulence profiles was spatially described in terms of spatial distribution. A total of 30.1 % isolates were found to be positive for at least one of the virulence genes. Depending on the different gene combinations present, 11 virulence profiles were found. Most of the isolates analyzed had a single gene, and no combination of fimbrial and enterotoxin gene was predominant. There was no association between the frequency and distribution of E. coli virulence genes and calf health status. Most of the virulence profiles were compatible with ETEC strains and showed a homogeneous distribution over the sampled area. A clustering pattern for E. coli virulence profiles could not be recognized. This work provides updated information on the molecular characterization of pathogenic E. coli strains from dairy herds in Cordoba, Argentina. These findings would be important to formulate prevention programs and effective therapies for diarrhea in calves caused by E. coli.
Keywords: Escherichia coli, Calves, Diarrhea, PCR, Virulence genes, Argentina
Resumen
El objetivo de este trabajo fue realizar una caracterización molecular actualizada de cepas patógenas bovinas de Escherichia coli aisladas de un muestreo aleatorio en tambos de una de las principales zonas lecheras de Argentina. Se obtuvieron hisopados rectales de 395 terneros neonatos sanos (63,7 %) y 225 diarreicos (36,3 %) pertenecientes a 45 tambos de la provincia de Córdoba, Argentina. Los genes de virulencia f5, f41, f17, sta, stb, lt, eae y vt se analizaron mediante PCR y se investigó la prevalencia de los perfiles de virulencia en función de la distribución geográfica. La prevalencia de aislamientos de E. coli patogénicos con al menos un gen de virulencia fue del 30,1 %. Once perfiles de virulencia fueron identificados, dependiendo de la combinación de genes presentes. La mayor parte de las muestras presentó un solo gen de virulencia, y no predominó ninguna combinación de genes de fimbrias y toxinas. No hubo asociación entre la frecuencia y la distribución de los genes de virulencia y el estado de salud de los terneros. La mayoría de los perfiles de virulencia fueron compatibles con cepas ECET y se distribuyeron cubriendo toda el área geográfica muestreada. No se reconoció ningún patrón de agrupamiento espacial para dichos perfiles. Este trabajo provee información actualizada sobre la caracterización molecular de E. coli patógena en rodeos lecheros de Córdoba, Argentina. Estos resultados serían importantes para formular programas preventivos y terapias eficaces contra la diarrea bovina causada por E. coli.
Palabras clave: Escherichia coli, Terneros, Diarrea, PCR, Genes de virulencia, Argentina
Introduction
Neonatal calf diarrhea is an important cause of morbidity and mortality worldwide in newborn calves10. This multifactorial disease involves the calf immune status, environmental factors and farm management practices (housing, feeding and hygienic conditions)3, 25 as well as the interaction of different pathogens such as bacteria, viruses and protozoa.
Escherichia coli is the predominant aerobic organism in the normal intestinal microbiota of mammals; here it plays an important role in host metabolism, immunology and nutrition42. However, a reduced number of highly adapted pathogenic strains are capable of causing intestinal or extraintestinal diseases and great economic losses22, 40.
Pathogenic E. coli strains have different virulence factors that allow them to colonize the host's small intestine, avoiding the immune response and stimulating the deleterious inflammatory response to produce diarrhea11, 45. These virulence factors include the antigens of colonization or adhesion (F2–F6, F17, F18, F41 fimbriae and intimin) and exotoxins (heat-labile enterotoxin [LT], heat-stable enterotoxins [STa and STb] and verotoxins [VT])26. Only the most successful combinations of virulence factors have persisted to become specific E. coli pathotypes that are capable of causing disease in healthy individuals22.
Farm animal and human diarrhea are frequently due to infection by one or several of E. coli pathotypes: enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), Vero toxin-producing/Shiga toxin-producing E. coli (VTEC/STEC) which include its well-known subgroup enterohemorrhagic E. coli (EHEC), enteroaggregative E. coli (EAggEC) and enteroadherent E. coli (EAdEC)28. ETEC infection is the most common type of colibacillosis in young animals especially in calves and piglets45.
In Argentina, neonatal calf diarrhea is a severe and common disease, affecting both beef and dairy herds with a morbidity above 60 %31. Previous reports2, 30 have shown the critical role of pathogenic E. coli in calf diarrhea. However, these studies are not recent and have generally been restricted to few animals or herds.
The aim of this study was to perform a current molecular characterization of pathogenic E. coli strains isolated from a random sampling of neonatal calves.
Materials and methods
Calves and data collection
Forty-five farms from Villa María (Córdoba) dairy area, with a herd size of 100–250 cows were randomly selected from a roster provided by the producers’ association. Such herd size strata represent 80 % of the dairy operations in Villa María and 10 % of Argentina's dairies41. A cross-sectional study involving Holstein calves less than 10 days of age, both healthy and diarrheic, was carried out between February and October 2008. The identification, age and gender of each animal were recorded. A clinical examination of each calf was performed, and the clinical parameters related to diarrhea were registered.
Bacterial isolates
Individual rectal swabs (n = 620) were collected and transported in Stuart's transport medium (Britania®, Argentina) to the Bacteriology laboratory. The swabs were plated in MacConkey agar plates (Oxoid, UK) and Gram staining was performed on isolated lactose-positive colonies. Gram-negative bacteria were biochemically characterized as belonging to the Enterobacteriaceae family, Escherichia genus, according to the Bergey's Manual19. Colonies compatible with E. coli were transferred to tubes containing minimal agar medium24.
Molecular typing by PCR
E. coli-positive strains were randomly selected and cultured on MacConkey agar plates.
Following overnight incubation, a loop was taken from the bacterial confluent growth zone, suspended in 300 μl of distilled water, boiled for 5 min, and centrifuged at 11,000 rpm for 2 min. The supernatant was used for PCR. From each PCR-positive sample, 10 colonies were streaked on MacConkey agar plates, pooled and processed by PCR. If a positive result was obtained, individual colonies were tested by PCR to identify positive isolates.
PCR technique was performed as described by Siqueira et al.39, with minor modifications. Briefly, the amplification reaction was carried out with 7 μl of template, 6 μl of reaction buffer (5× Green Go Taq®, Promega) containing MgCl2 (1.5 mM), 0.6 μl of dNTP (10 mM, Promega), 0.5 μl of primer (300 ng/μl, Ruralex Fago) and 0.2 μl of Taq DNA polymerase (5 U/μ, Go Taq® DNA Polimerasa, Promega). Volumes were adjusted to 30 μl with sterile distilled water. Templates were screened for the presence of E. coli virulence factors coding gene by conventional PCR, using the primers described in Table 1 . PCR cycling conditions consisted of initial denaturation at 94 °C for 2 min, followed by 30 cycles of amplification (denaturation at 93 °C for 1 min, annealing for 1 min, extension at 72 °C for 1 min) and final elongation at 72 °C for 7 min. All PCR reactions were performed in an MJ Research Thermocycler Bio-Rad model PTC-220. The amplified PCR products were separated by electrophoresis on a 2 % agarose gel with Tris–acetate–EDTA (TAE) buffer (Sigma–Aldrich®). A 100-bp DNA ladder (CienMarker, Biodynamics) was used as molecular weight marker. Control E. coli strains for each gene (Table 2 ) and distilled water were used as positive and negative controls respectively. The gel was stained with ethidium bromide (Sigma–Aldrich®) and visualized under a UV-transilluminator at 302 nm.
Table 1.
Characteristics of the oligonucleotide primers used for the PCR reaction.
| Target gene | Primer | Primer sequence (5′–3′) | Amplicon size (bp) | Annealing temperature | Reference |
|---|---|---|---|---|---|
| vt | VT VT-F VT VT-R |
GAGCGAAATAATTTATATGT CGAAATCCCCTCTGTATTTGCC |
322 | 48 °C | 36 |
| lt | LT LT-B LT LT-FN |
CCGAATTCTGTTATATATGTC GGCGACAGATTATACCGTGC |
696 | 55 °C | 6 |
| stb | STb STb-F STb STb-R |
ATCGCATTTCTTCTTGCATC GGGCGCCAAAGCATGCTCC |
175 | 55 °C | 6 |
| sta | STa STa-A STa STa-B |
ATTTTTATTTCTGTATTGTCTTT GGATTACAACACAGTTCACAGCAGT |
176 | 48 °C | 14 |
| eae | EaeA-F EaeA-R |
AGGCTTCGTCACAGTTG CCATCGTCACCAGAGGA |
570 | 55 °C | 8 |
| f17 | F17 F17-F F17 F17-R |
GGGCTGACAGAGGAGGTGGGGC CCCGGCGACAACTTCATCACCGG |
411 | 60 °C | 29 |
| f5 | K99 K99-A K99 K99-B |
CCAGCGCCCGGCAGTAATGACTGC CCACCATTAGACGGAGCGCGG |
278 | 60 °C | 14 |
| f41 | F41 F41-A F41 F41-R |
GGCTATGGAAGACTGGAGAGGG GGGGTGACTGAGGTCATCCC |
551 | 55 °C | 14 |
Table 2.
Characteristics of E. coli strains used as positive control for virulence genes in PCR reaction.
| Identification | Origin | Virulence profiles |
|---|---|---|
| FV-10185 | OVINE | vt1, vt2 |
| FV-10186 | PORCINE | eae |
| FV-10187 | PORCINE | vt2, sta, stb, f18 |
| FV-10188 | PORCINE | lt, stb, f4 |
| FV-10189 | PORCINE | lt, sta, stb, f18 |
| FV-10190 | PORCINE | sta, f6 |
| FV-10191 | BOVINE | sta, f41, f5 |
| FV-10192 | OVINE | f17 |
Note: These control E. coli strains were kindly provided by the Animal Health Group (EEA INTA Balcarce) and come from the strain collection of Dr. Jorge Blanco (E. coli Reference Laboratory, University of Santiago de Compostela, Lugo, Spain).
The E. coli strains were classified into different pathotypes, based on their specific virulence genes26.
Data analysis and mapping
All data analyses were performed using the commercially available statistical software, SPSS for Windows (Version 10.0, Chicago, IL, USA). Differences were considered significant at the level of p < 0.05. Herd geographic coordinates were recorded using a Global Positioning System (GPS). The prevalence of E. coli pathotypes was spatially described using a dot map displayed in Arcview 3.2 (ESRI, Redlands, CA, USA).
A spatial scan statistic was used to identify and locate significant spatial clusters of E. coli virulence profiles within the study area. The null hypothesis to test was that herds within a particular window have the same prevalence level as herds outside the window. All analyses were performed by using SaTScan® software (Version 9.0, http://www.satscan.org).
Results
Diarrhea prevalence
Overall, 620 calves from 45 dairy farms were sampled. The average number of calves sampled per herd was 14 ± 7.95 (media ± standard deviation). At the time of sampling, 36.3 % of calves (n = 225) presented diarrhea. Diarrhea prevalence found in dairy farms markedly varied from 0 % to 71.4 % with a median of 35.7 % (percentile 25: 20.0 % and percentile 75: 50.0 %). There were no cases of diarrhea in 2 farms, while prevalence of diarrhea was higher than 50 % in 8 farms (Fig. 1 ).
Figure 1.
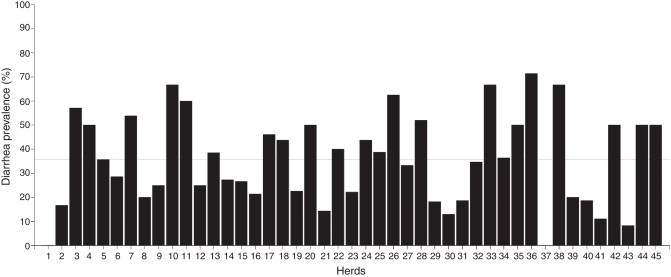
Herd prevalence of diarrhea in dairy calves from Córdoba, Argentina. The X axis represents all the analyzed herds (n = 45). The dashed line indicates diarrhea median prevalence (35.7 %).
Molecular characterization of E. coli isolates
A total of 156 samples from 363 E. coli-positive isolates were analyzed by PCR to detect 8 virulence genes. A total of 109 samples (69.9 %) were negative for all genes investigated (probably non-pathogenic strains) and 47 (30.1 %) were found to be positive for at least one of the virulence genes. A total of 39 isolates were obtained from the PCR-positive samples, Depending on the different gene combinations present, 11 different virulence profiles were found (Table 3 ). Most of the isolates analyzed had a single gene. The prevalent virulence gene was f17, while the only enterotoxin gene observed was sta; lt and stb genes were not detected, while vt and f41 genes were only found in combination with other genes. No combination for both fimbriae and toxins was prevalent.
Table 3.
Virulence profiles detected in the E. coli isolates (n = 39).
| Number of genes | Virulence profiles | Number of isolates | Frequencies (%) | Dairy farms (Calf health status)a |
|---|---|---|---|---|
| 1 | f17 | 16 | 41 | 11(H), 12(D), 18(D), 29(H), 32(H), 41(H), 42(D), 43(H), 75(D), 72(H), 76(H), 80(D), 85(H), 86(D), 93(H), 632(H) |
| 1 | eae | 5 | 12.7 | 2(D), 25(D), 57(D), 63(H), 632(H) |
| 1 | sta | 3 | 7.7 | 10(D), 90(H), 632(D) |
| 1 | f5 | 1 | 2.6 | 11(D) |
| 2 | f17 vt | 3 | 7.7 | 11(H), 93(H), 43(H) |
| 2 | eae vt | 3 | 7.7 | 2(D), 57(D), 86(D) |
| 2 | f17 sta | 3 | 7.7 | 25(D), 90(H), 92(H) |
| 2 | f17 f5 | 2 | 5.1 | 1(H), 15(H) |
| 2 | sta eae | 1 | 2.6 | 632(D) |
| 3 | sta f5 f41 | 1 | 2.6 | 42(D) |
| 3 | sta f5 f17 | 1 | 2.6 | 25(D) |
D, diarrheic calf; H, healthy calf.
Regarding the frequency and distribution of E. coli virulence genes, no significant differences (p = 0.3008, Chi Square) were observed between healthy and diarrheic calves (Table 3).
Identification and spatial distribution of E. coli genetic profiles
The map with the locations of all the farms visited and the spatial distribution of E. coli virulence profiles is farm is shown in Fig. 2 . Most of the virulence profiles were compatible with ETEC strains and showed a homogeneous distribution over the sampled area. A clustering pattern for E. coli virulence profiles could not be recognized.
Figure 2.
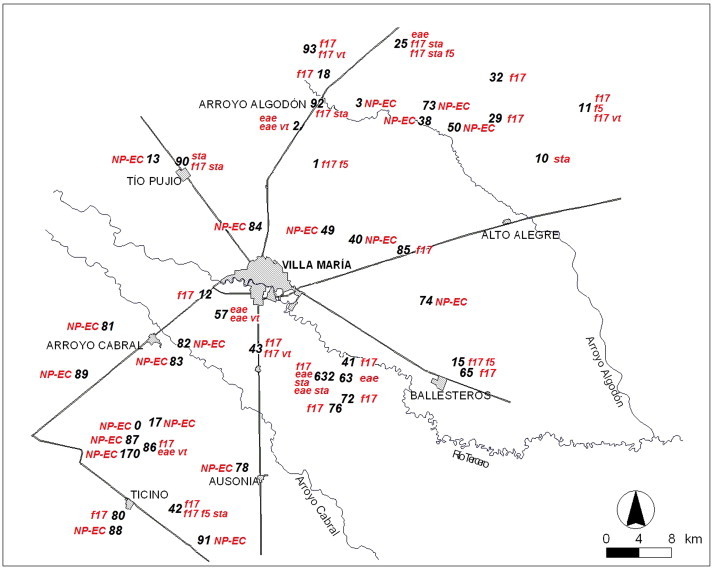
Map with the locations of all the dairy farms visited (n = 45), showing the spatial distribution of Escherichia coli virulence profiles in each farm. Córdoba, Argentina. NP-EC, non-pathogenic E. coli.
Discussion
Diarrhea is a frequent and growing concern in young calves, especially in the first week of age. Regarding the main enteropathogens causing diarrhea, ETEC, Rotavirus, Coronavirus and Cryptosporidium parvum have been reported in 75–95 % of cases of intestinal infections in young calves3, 13, 15. In general, pathogenic E. coli appears to be less important compared with other diarrheagenic pathogens, since it is shed during a short period of time, resulting in a low prevalence. However, this bacterium plays an important role in the occurrence of enterotoxic and septicemic colibacillosis, and its incidence and impact on morbidity/mortality of newborn calves might be relatively high3, 5, 27, 45. In Argentina, the role of pathogenic E. coli isolated from healthy and diarrheic calves has been previously investigated. However, these investigations have been limited to the study of few animals or herds and were conducted over thirty years ago. Odeón30 and Barrandeguy et al.2, demonstrated that ETEC strains were commonly found in beef and dairy herds. Conversely, Bellinzoni et al.4, reported that pathogenic E. coli was not detected in any calf. In this study, pathogenic E. coli prevalence was 30.1 %. The differences in these findings could be due to regional variations, management and hygienic conditions, the age of the animals, or to the fact that the analysis of F5 fimbriae in the previous reports was determined only by serology, without searching for virulence genes with molecular methods. The present work involved a great number of dairy farms and calves, and was focused on the molecular characterization of pathogenic E. coli strains among dairy farms from Cordoba province, Argentina. Previous studies in the same dairy farms of Cordoba province investigated the role of Cryptosporidium spp. and Giardia spp. producing calf diarrhea43 and the participation of E. coli causing subclinical mastitis12. However, investigations about the molecular characterization of pathogenic E. coli causing diarrhea in neonatal calves have not been conducted. The results of the present study provide new and complementary information about one of the pathogens causing diarrhea in neonatal calves in this dairy area.
In order to categorize E. coli pathotypes, the presence of virulence characteristics needs to be identified. The use of PCR-based technology to identify virulence genes has become widely adopted to distinguish pathogenic E. coli strains from normal gut flora. In this study, ETEC-compatible strains were the most prevalent strains determined by the PCR technique. Nagy and Fekete28 reported that diarrhea caused by ETEC is considered to be the major infectious disease of newborn calves during the pre-weaning and weaning periods. Infected animals are important reservoirs of ETEC and their feces are the major source of environmental contamination with the bacteria, which are acquired by newborn calves soon after birth. Although bovine ETEC strains do not represent a real hazard to man and cannot be regarded as zoonotic agents44, the high circulation of such strains affects the productive capacity and efficiency of calves in artificial breeding.
ETEC bacteria are known to adhere to the small intestinal epithelium without inducing significant morphological changes and to secrete enterotoxins that alter the functions of enterocytes by increasing secretion and reducing absorption. The main colonization factors or fimbriae described in bovine pathogenic E. coli are F5 (K99), F17 and F41. F5 fimbria has been associated with most ETEC strains and is responsible for most cases of ETEC infection in newborn calves13. In this study, the prevalence of ETEC F5+ was 12.9 %. Similar results were obtained by Younis et al.45, in Egypt (10.4 %), Ok et al.32, in Turkey (13.4 %) and Pourtaghi et al.35, in Iran (14.1 %). A higher prevalence of ETEC F5+ strains was reported in Egypt and Israel34 (23 %), Mozambique1 (25 %) and Brazil23 (35 %). On the contrary, a lower prevalence of E. coli F5+ was found in the United States9, the Netherlands3, Brazil33 and Turkey21, with rates of 1.8 %, 2.6 %, 5.8 % and 9.4 % respectively.
F17 fimbriae have been found on some bovine ETEC strains and mediate attachment of these bacteria to the intestinal epithelium. We found that the most prevalent virulence factor in both diarrheic and healthy calves was individual F17 fimbria (41 %). The observation that a high number of E. coli strains carried genes for fimbriae but not toxin genes has been reported in others studies. Güler et al.18, found similar results (44 %) for F17 fimbriae in E. coli isolates from calves. Nguyen et al.29, showed a low prevalence (16 %) of F17 fimbriae among E. coli strains isolated from diarrheic calves. It is important to take into account that E. coli F17+ strains have also been associated with extraintestinal disease and usually show other virulence factors associated with the ability to cause septicemia29. In Iran, Ghanbarpour and Oswald16, 17 found strains of E. coli F17+ isolated from septicemic calves (29 %) and from bovine mastitis (20.4 %). Thus, in our study, an additional PCR screening of extraintestinal virulence factors would be necessary to confirm the likely presence of non-enterotoxigenic F17+ strains.
After adherence to the intestinal mucosa, ETEC strains produce one or both enterotoxins, LT and ST, which are responsible for hypersecretion of electrolytes and water into the small intestine. In this study, we found that sta was the only enterotoxin gene detected, while lt and stb genes were not detected in any of the examined strains. These results agree with Nagy and Fekete28, who reported that the STa enterotoxin is produced in porcine and bovine ETEC strains, while the LT and STb enterotoxins are predominantly produced by human and porcine ETEC strains. Conversely, there are some reports describing the isolation of LT and STb enterotoxins from cows and calves. In Brazil, Salvadori et al.38, recorded 3.9 % ETEC possessing st and lt genes from diarrheic calves and Rigobelo et al.37, reported a prevalence rate of E. coli isolated from diarrheic calves carrying st (25.4 %) and lt (13.2 %) genes. In China, Huasai et al.20, reported that 15.5 % of ETEC strains isolates from healthy cows were positive for lt and st genes. In Italy, it has been found that all the ETEC strains isolated from diarrheic water buffalo calves had the lt gene but lacked st gene7. Finally, Acha et al.1, reported that the sta gene was not detected in any of the E. coli F5 strains isolated from diarrheic calves in Mozambique. Since ETEC enterotoxins are plasmid-encoded secreted proteins, the variation in the prevalence of these toxins could be explained by the existence of conjugative plasmids widely distributed among bovine ETEC strains.
Regarding the distribution of E. coli virulence profiles, most of them were compatible with ETEC strains and were widely distributed throughout the examined geographical area. This pattern of spatial distribution was similar to the one reported for Giardia spp. in the same dairy farms, but disagrees with the primary cluster shown by Cryptosporidium spp43. The ETEC virulence gene distribution on the farms showed different combinations but no profile was prevalent. There was no association between the virulence profiles and dairy farm location.
In conclusion, this study showed a high frequency of pathogenic E. coli strains that were widely distributed either among diarrheic or healthy calves in the examined farms. Mainly, F17 and F5 fimbriae were found to be the most common virulence factors of E. coli strains isolated from dairy calves. This work provides updated information on the molecular characterization of pathogenic E. coli strains obtained from a random sampling of dairy herds in Cordoba, Argentina. These findings would be important to formulate prevention programs and effective therapies for calf diarrhea caused by pathogenic E. coli.
Ethical disclosures
Protection of human and animal subjects
The authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of data
The authors declare that no patient data appear in this article.
Right to privacy and informed consent
The authors declare that no patient data appear in this article.
Conflict of interest
The authors declare that they have no conflicts of interest.
Acknowledgements
The authors thank Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). This work was supported by grants from Secretaría de Ciencia y Técnica de la Universidad Nacional de Río Cuarto (SECYT-UNRC) and Agencia Nacional de Promoción Científica y Tecnológica (FONCYT).
References
- 1.Achá S., Kühn I., Jonsson P., Mbazima G., Katouli M., Möllby R. Studies on calf diarrhea in Mozambique: prevalence of bacterial pathogens. Acta Vet Scand. 2004;45:27–36. doi: 10.1186/1751-0147-45-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrandeguy M., Cornaglia E., Gottschalk M., Fijtman N., Pasini M., Yafal A., Parraud J., Schudel A. Rotavirus, enterotoxigenic Escherichia coli and other agents in the feces of dairy calves with and without diarrhea. Rev Latinoam Microbiol. 1988;30:239–245. [Google Scholar]
- 3.Bartels C., Holzhauer M., Jorritsma R., Swart W., Lam T. Prevalence, prediction and risk factors of enteropathogens in normal and non-normal faeces of young Dutch dairy calves. Prev Vet Med. 2010;93:162–169. doi: 10.1016/j.prevetmed.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellinzoni R., Blackhall J., Terzolo H., Moreira A., Auza N., Mattion N., Micheo G., La Torre J., Scodeller E. Microbiology of diarrhea in beef and dairy calves in Argentina. Rev Argent Microbiol. 1990;22:130–137. [PubMed] [Google Scholar]
- 5.Bendali F., Bichet H., Schelcher F., Sanaa M. Pattern of diarrhoea in newborn beef calves in south-west France. Vet Res. 1999;30:61–74. [PubMed] [Google Scholar]
- 6.Blanco M., Blanco J.E., González E., Mora A., Jansen W., Gómes T., Zerbini F., Yano T., Pestana de Castro A., Blanco J. Genes coding for enterotoxins and verotoxins in porcine Escherichia coli strains belonging to different O:K:H serotypes: relationship with toxic phenotypes. J Clin Microbiol. 1997;35:2958–2963. doi: 10.1128/jcm.35.11.2958-2963.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borriello G., Lucibelli M., De Carlo E., Auriemma C., Cozza D., Ascione G., Scognamiglio F., Iovane G., Galiero G. Characterization of enterotoxigenic E. coli (ETEC), Shiga-toxin producing E. coli (STEC) and necrotoxigenic E. coli (NTEC) isolated from diarrhoeic Mediterranean water buffalo calves. Res Vet Sci. 2012;93:18–22. doi: 10.1016/j.rvsc.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.China B., Pirson V., Mainil J. Typing of bovine attaching and effacing Escherichia coli by multiplex in vitro amplification of virulence-associated genes. Appl Environ Microbiol. 1996;62:3462–3465. doi: 10.1128/aem.62.9.3462-3465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho Y., Han J., Wang C., Cooper V., Schwartz K., Engelken T., Yoon K. Case–control study of microbiological etiology associated with calf diarrhea. Vet Microbiol. 2013;166:375–385. doi: 10.1016/j.vetmic.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Constable P. Antimicrobial use in the treatment of calf diarrhea. J Vet Intern Med. 2004;18:8–17. doi: 10.1111/j.1939-1676.2004.tb00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croxen M., Finlay B. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 12.Dieser S., Vissio C., Lasagno M., Bogni C., Larriestra A., Odierno L. Prevalence of pathogens causing subclinical mastitis in Argentinean dairy herds. Pak Vet J. 2014;34:124–126. [Google Scholar]
- 13.Foster D., Smith G. Pathophysiology of diarrhea in calves. Vet Clin North Am Food Anim Pract. 2009;25:13–36. doi: 10.1016/j.cvfa.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franck S., Bosworth B., Moon H. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin producing Escherichia coli strains from calves. J Clin Microbiol. 1998;36:1795–1797. doi: 10.1128/jcm.36.6.1795-1797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García A., Ruiz-Santa-Quiteria J., Orden J., Cid D., Sanz R., Gómez-Bautista M., De la Fuente F. Rotavirus and concurrent infections with other enteropathogens in neonatal diarrheic dairy calves in Spain. Comp Immunol Microbiol Infect Dis. 2000;23:175–183. doi: 10.1016/S0147-9571(99)00071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghanbarpour R., Oswald E. Characteristics and virulence genes of Escherichia coli isolated from septicemic calves in southeast of Iran. Trop Anim Health Prod. 2009;1:1091–1099. doi: 10.1007/s11250-008-9289-0. [DOI] [PubMed] [Google Scholar]
- 17.Ghanbarpour R., Oswald E. Phylogenetic distribution of virulence genes in Escherichia coli isolated from bovine mastitis in Iran. Res Vet Sci. 2010;88:6–10. doi: 10.1016/j.rvsc.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Güler L., Gündüz K., Ok Ü. Virulence factors and antimicrobial susceptibility of E. coli. Zoonoses Public Health. 2008;55:249–257. doi: 10.1111/j.1863-2378.2008.01121.x. [DOI] [PubMed] [Google Scholar]
- 19.Holt J., Krieg J., Sneath P., Staley P., Williams S. Genus Escherichia. In: Holt J., Krieg J., Sneath P., Staley P., Williams S., editors. Bergey's Manual of Determinative Bacteriology. 9th ed. Williamn & Wilkins; Baltimore: 1994. pp. 179–180. [Google Scholar]
- 20.Huasai S., Chen A., Wang C., Li Y., Tongrige B. Occurrence and characteristics of virulence genes of Escherichia coli strains isolated from healthy dairy cows in Inner Mongolia, China. Braz J Microbiol. 2012;43:528–534. doi: 10.1590/S1517-83822012000200013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Içen H., Arserim N., Işik N., Özkan C., Kaya A. Prevalence of four enteropathogens with immunochromatographic rapid test in the feces of diarrheic calves in east and southeast of Turkey. Pak Vet J. 2013;33:496–499. [Google Scholar]
- 22.Kaper J., Nataro J., Mobley H. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 23.Langoni H., Linhares A., Avila F., Da Silva A., Elias A. Contribution to the study of diarrhea etiology in neonate dairy calves in Sao Paulo state, Brazil. Braz J Vet Res Anim Sci. 2004;41:313–319. [Google Scholar]
- 24.Lazo Pérez L., Dahbi G., Blanco M., Blanco J.E., Blanco J., LLorens F. Aplicación de técnicas moleculares en la caracterización de aislados de Escherichia coli procedentes de cerdos con síndrome diarreico en la Provincia Villa Clara. Rev Salud Anim. 2009;31:93–104. [Google Scholar]
- 25.Lorenz I. Diarrhoea of the young calf: an update. Proceedings of the XXIV World Buiatrics Congress; Nice, France; 2006. pp. 130–138. [Google Scholar]
- 26.Mainil J. Escherichia coli virulence factors. Vet Immunol Immunopathol. 2013;152:2–12. doi: 10.1016/j.vetimm.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 27.Morrell E., Moore D., Odeón A., Poso M., Odriozola E., Cantón G., Paolicchi F., Malena R., Leunda M., Morsella C., Campero C. Retrospective study of bovine neonatal mortality: cases reported from INTA Balcarce, Argentina. Rev Argent Microbiol. 2008;40:151–157. [PubMed] [Google Scholar]
- 28.Nagy B., Fekete P. Enterotoxigenic Escherichia coli in veterinary medicine. Int J Med Microbiol. 2005;295:443–454. doi: 10.1016/j.ijmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen T., Vo T., Vu-Khac H. Virulence factors in Escherichia coli isolated from calves with diarrhea in Vietnam. J Vet Sci. 2011;12:159–164. doi: 10.4142/jvs.2011.12.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odeón A. Diarrea neonatal de los terneros en el sudeste de la Provincia de Buenos Aires (Argentina), aspectos epidemiológicos, etiológicos, inmunológicos y patológicos. Rev Arg Prod Anim. 1981;1:51. [Google Scholar]
- 31.Odeón A. La diarrea neonatal: una enfermedad que puede afectar la eficiencia productiva de los rodeos de cría. Visión Rural. 2004;11:17–20. [Google Scholar]
- 32.Ok M., Güler L., Turgut K., Ok U., Şen I., Gündüz I., Birdane M., Güzelbekteş H. The studies on the aetiology of diarrhoea in neonatal calves and determination of virulence gene markers of Escherichia coli strains by multiplex PCR. Zoonoses Public Health. 2009;56:94–101. doi: 10.1111/j.1863-2378.2008.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira-Filho J., Silva D., Pacheco M., Mascarini L., Ribeiro M., Alfieri A., Alfieri A., Stipp D., Barros B., Borges A. Diarrhea in Nelore calves: clinical and etiologic study. Pesq Vet Bras. 2007;27:419–424. [Google Scholar]
- 34.Perk K., Moussa A., Tromp A., Reda I., Refai M., Friedman A., Farid A., Gallily O., Salah S., Saif L. Neonatal diarrheal disease of dairy cattle in Egypt and Israel. Israel J Vet Med. 2000;55:13–18. [Google Scholar]
- 35.Pourtaghi H., Dahpahlavan V., Momtaz H. Virulence genes in Escherichia coli isolated from calves with diarrhoea in Iran. Comp Clin Pathol. 2013;22:513–515. [Google Scholar]
- 36.Read S., Clarke R., Martin A., De Grnadis S., Hii J., McEwen S., Gyles C. Polymerase chain reaction for detection of verotoxigenic Escherichia coli isolated from animal and food sources. Mol Cell Probes. 1992;6:153–161. doi: 10.1016/0890-8508(92)90060-b. [DOI] [PubMed] [Google Scholar]
- 37.Rigobelo E., Gamez H., Marin J., Macedo C., Ambrosin J., Avila F. Virulence factors of Escherichia coli isolated from diarrhoeic calves. Arq Bras Med Vet Zootec. 2006;58:305–310. [Google Scholar]
- 38.Salvadori M., Valadares G., Leite D., Blanco J., Yano T. Virulence factors of Escherichia coli isolated from calves with diarrhea in Brazil. Braz J Microbiol. 2003;34:230–235. [Google Scholar]
- 39.Siqueira A., Ribeiro M., Leite D., Tiba M., De Moura C., Lopes M., Prestes N., Salerno T., Da Silva A. Virulence factors in Escherichia coli strains isolated from urinary tract infection and pyometra cases and from feces of healthy dogs. Res Vet Sci. 2009;86:206–210. doi: 10.1016/j.rvsc.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 40.Sousa C.P. Escherichia coli as a specialized bacterial pathogen. Rev Biol Ciên Terra. 2006;6:341–352. [Google Scholar]
- 41.Taverna M., Fariña S. FunPEL; 2013. La producción de leche en Argentina. Anuario de la Lechería Argentina; pp. 7–14. http://www.vet.unicen.edu.ar/html/Areas/Prod_Animal/Documentos/2014/Capitulo1_LaProducciondeLecheenArgentina.pdf. [Google Scholar]
- 42.Tenaillon O., Skurnik D., Picard B., Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 43.Tiranti K., Larriestra A., Vissio C., Picco N., Alustiza F., Degioanni A., Vivas A. Prevalence of Cryptosporidium spp. and Giardia spp., spatial clustering and patterns of shedding in dairy calves from Córdoba, Argentina. Rev Bras Parasitol Vet. 2011;20:65–72. doi: 10.1590/s1984-29612011000200009. [DOI] [PubMed] [Google Scholar]
- 44.Wasteson Y. Zoonotic Escherichia coli. Acta Vet Scand. 2001;95:79–84. [PubMed] [Google Scholar]
- 45.Younis E., Ahmed A., El-Khodery S., Osman S., El-Naker Y. Molecular screening and risk factors of enterotoxigenic Escherichia coli and Salmonella spp. in diarrheic neonatal calves in Egypt. Res Vet Sci. 2009;87:373–379. doi: 10.1016/j.rvsc.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


