Abstract
To expand the epidemiological understanding of feline calicivirus (FCV) in dogs, genotypic and phenotypic characterizations of a FCV strain isolated from a puppy showing enteritis were performed. After isolation in cell culture, the novel isolate was analysed by RT-PCR and the amplicons obtained were sequenced. In order to characterize the growth properties of the isolate, the size of the plaques, the temperature of inactivation and the kinetics of growth were evaluated. Moreover, the novel strain was used to perform a serological study on 86 canine serum samples and 81 feline sera by virus-neutralization assay. The comparative analysis of nucleotide and amino acid sequences of the isolate, named FCV-Te/10/07, revealed the highest identity to strain FCV-F65. The growth kinetic revealed that strain Te/10/07 grew more rapidly than F9 strain. By virus-neutralization assay, dogs from the same region of the isolate showed antibodies against the FCV-F9 vaccinal strain in 63.9% (55/86) of sera, while antibodies against the Te/10/07 were found in seven sera (8.13%). In cats neutralizing antibodies against Te/10/07 strain were recovered in 50.62% (41/81) of samples tested, even if 38 sera were positives for F9 strain with similar titres or higher. In three cats neutralization to Te/10/07 alone was seen.
Our results confirmed the interspecific circulation of FCV strains among different animal species, but new investigations are needed to establish whether FCV is pathogenic in the dog and the role of interspecies circulation in pathogen spreading.
Keywords: Dog, Feline calicivirus, Isolation, Characterization, Serology
1. Introduction
Feline calicivirus (FCV), belonging to the family Caliciviridae genera Vesivirus, is a small non-enveloped virus with a diameter of 35–38 nm. The virus has a single-stranded, positive sense RNA genome approximately 7.7 kb in length and contains three open reading frames (ORFs): ORF1 (Sosnovtsev et al., 2002), ORF2 (Carter et al., 1992) and ORF3 (Sosnovtsev et al., 2005).
Although FCV isolates are closely related and constitute one serotype, there are several strains which differ in pathogenicity and antigenicity able to adapt in many heterogeneous conditions (Knowles et al., 1990).
In cats, FCV infection is usually inapparent or associated with mild upper respiratory disease and/or signs of oral ulceration. Less commonly some isolates can cause an acute febrile lameness syndrome (Pedersen et al., 1983, Dawson et al., 1994) as well as abortion (Ellis, 1981) and severe pneumonia. Moreover, during the last decade, highly virulent strains of FCV (VSD-FCV: virulent systemic disease-FCV) have emerged that are associated with outbreaks of disease with high mortality (about 50%) (Pedersen et al., 2000, Hurley et al., 2004, Pesavento et al., 2004, Coyne et al., 2006) and a new range of clinical features characterized by high persistent fever, severe edema of face and limbs, pancreatitis or pneumonia (Schorr-Evans et al., 2003).
FCV is a common pathogen of domestic cats, but has been isolated from several large felids in captivity and in the wild; for this reason it was suggested that all members of the Felidae are susceptible and there are no alternative hosts (Radford et al., 2007).
Occasionally some caliciviruses have also been found in dogs. Four apparently distinct types of calicivirus in dogs have been described: calicivirus serologically related to FCV (FCV-like) associated to glossitis or enteric infection (Evermann et al., 1981, Evermann et al., 1985, Gabriel et al., 1996, Pratelli et al., 2000a, Martella et al., 2002); canine calicivirus (CaCV), isolated from the feces of diarrheal dogs and which differed genetically and antigenically from FCV (Schaffer et al., 1985, Mochizuki et al., 1993); Norovirus GIV.2 detected from a puppy with a severe gastroenteritis (Martella et al., 2008); and calicivirus isolated from dogs with genital disease (Crandell, 1988).
A seroprevalence study (Pratelli et al., 2000b) has been shown that dogs FCV are widespread in the dogs population, as well as in another report (Binns et al., 2000) has been observed an association between the presence of dogs and FCV infection in cats. However, to date, the role of these viruses in the epidemiology of FCV in cats and dogs is uncertain (Radford et al., 2007).
We report the results of a study based on genotypic and phenotypic characterization of a FCV strain isolated from a dog with enteritis and preliminary serological survey performed on a canine and feline population.
2. Materials and methods
2.1. Virologic investigation
One faecal sample collected from a 3-month old dog with clinical signs of gastroenteritis was forwarded to our laboratories for virologic examination. Faecal sample was homogenised (10%, w/v) in Dulbecco's Modified minimal essential medium (D-MEM) plus antibiotics (penicillin 5000 IU/ml, streptomycin 2500 μg/ml, amphotericin B 10 μg/ml), filtered and inoculated onto confluent Madin–Darby canine Kidney (MDCK), feline Crandell Kidney (CrFK) and dog adenocarcinoma (A72) cell line. After 3 days of incubation at 37 °C, the inoculated cells were tested for FCV, canine coronavirus (CCoV) and canine adenovirus (CAV) antigens by indirect immunofluorescence (IF) assay using a cat serum positive for FCV vaccinal strain F9, a dog serum positive for CCoV and a dog serum positive for CAV antibodies. Faecal sample was passaged three times considered to be negative for, CCoV and CAV. The supernatant of the faecal homogenate and the inoculated monolayers were also subjected to hemoagglutination test (HA) using a suspension containing 1% pig erythrocytes and 1% foetal calf serum (FCS) in a 96-well V-plate to screen against canine parvovirus type 2 (CPV-2). Results were read after 4 h at +4 °C and expressed as the reciprocal of the highest sample dilution able to produce HA.
Faecal sample was also tested for type A rotavirus antigens by an enzyme-linked immunosorbent assay (ELISA) (Biox Diagnostics, Jemelle, Belgium).
2.2. Molecular identification
Viral RNA from the original faecal sample and infected cells was extracted using the QIAamp RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions. Template RNAs were eluted in 50 μl of RNase-free water and stored at −70 °C until their use. DNA was extracted from faecal homogenate by boiling for 10 min and chilling on ice, whereas nucleic acid purification from infected cells was achieved using the DNeasy Tissue Kit (Qiagen GmbH, Hilden, Germany), followed by final elution of DNA with 200 μl of AE buffer and storage at −70 °C.
Viral RNA was analysed by reverse transcription (RT)-PCR using the following primer sets:
-
-
Cali1–Cali2 (Marsilio et al., 2005) targeted to a region of the ORF2 gene of FCV.
-
-
CaCV1–CaCV2 (Roerink et al., 1999) to amplify the complete ORF2 gene of CaCV.
-
-
p289–p290 (Jiang et al., 1999), a broadly reactive primer pair, targeted to highly conserved motives of the RdRp region of the polymerase complex of Vesivirus, Lagovirus, Norovirus and Sapovirus genera.
-
-
CCV1, CCV2 and CCV3 (Pratelli et al., 2001) to amplify a segment of the gene encoding for transmembrane protein M of CCoV.
Viral DNA was analysed by PCR using primers 555/For–555/Rev (Buonavoglia et al., 2001) that amplify a 583 bp fragment of the capsid protein-encoding gene of CPV-2. RT-PCR and PCR assays were performed as previously reported (Jiang et al., 1999, Roerink et al., 1999, Buonavoglia et al., 2001, Pratelli et al., 2001, Marsilio et al., 2005). The PCR products were analysed on agarose gel electrophoresis, stained with ethidium bromide and visualized using UV light.
2.3. Sequence analysis and phylogeny
PCR products obtained were purified from agarose gel using the commercial kit Qiaex Gel Extraction (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. The DNA was then used as template for direct sequencing (Eurofins MWG Operon, Ebersberg, Germany) using primers p289–p290 (Jiang et al., 1999), Cali1–Cali2 (Marsilio et al., 2005) and specific primers designed according to an overlapping strategy. The sequences were assembled and analysed using the BioEdit software package version 2.1 (Hall, 1999) and the NCBI's (http://www.ncbi.nlm.nih.gov) and EMBL's (http://www.ebi.ac.uk) analysis tools. Phylogenetic and molecular analyses were conducted using Mega 3 program (Kumar et al., 2004). The nucleotide sequence data of ORF1 and ORF2 genes have been deposited in the EMBL/GenBank database under accession nos. EU980609 and EU980610, respectively.
2.4. Phenotypic characterization
Phenotypic characterization was performed following the method described by Ossiboff et al. (2007). In particular, the growth properties of the novel isolate were analysed by plaque assay, temperature inactivation experiments (for 30 min at 41.8, 46.2 and 52.2 °C) and by generation of single and multiple cycle growth curves [4,8, 12, 24 h post-infection (p.i.) at multiplicities of infection (MOI) of 10 and 0.02 respectively]. Characterization was done comparing the growth properties of the isolate with those of the F9 vaccine strain.
2.5. Virus-neutralization (VN) assay
Eighty-six canine serum samples and 81 feline sera collected during years 2006–2008, were tested for neutralizing antibodies against the new isolate and the FCV-F9 vaccine strain. The neutralization assays were performed as previously described (Povey, 1974). Briefly, 50 μl of serial twofold diluted serum and 50 μl of infectious culture medium containing 100 TCID50 of the FCV-F9 strain were mixed and incubated for 1 h at 37 °C. One hundred microliters (1 × 105 cell/ml) of CrFK cell suspension was then added to each well. The cultures were then incubated for 3 days at 37 °C, 5% with CO2. The assay was performed three times for each serum.
3. Results
Typical FCV cythopatic effect was evident from the first passage on CrFK cell cultures. No growth was observed in cultured cells of canine origin. Results were negative for CPV-2, CCoV, CAV and type A rotavirus antigens. The isolate was identified as FCV by IF test using a cat serum anti-FCV (Fig. 1 ).
Fig. 1.
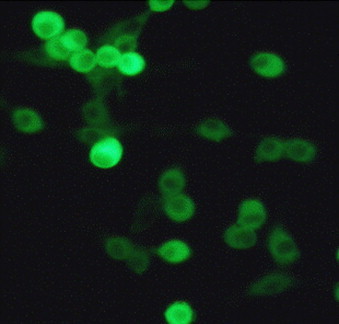
Indirect immunofluorescence analysis on CRFK cells. The intracytoplasmatic fluorescence was revealed by using a cat polyclonal antiserum raised against FCV-F9 vaccinal strain. Magnification, 400×.
Furthermore, the faecal sample and the infected cells at the passage one and three, were screened for Caliciviridae family, FCV, CaCV, CCoV and CPV-2 by conventional RT-PCR or PCR. Results were negative for CCoV, CaCV and CPV-2. Caliciviruses were identified by using primers p289–p290 (Jiang et al., 1999), whereas the virus was characterized as FCV by using primers Cali1–Cali2 (Marsilio et al., 2005) specific for the FCV capsid gene (Fig. 2 ).
Fig. 2.
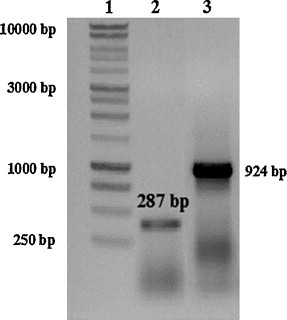
RT-PCR with 289/290 and Cali1/Cali2 primer sets. Line 1: marker (Promega 1 kb DNA ladder); line 2: PCR product obtained using generic calicivirus primers (289/290) of 287 bp in size; line 3: amplicon obtained by specific FCV (Cali1/Cali2) primer set of 924 bp in size.
Nucleotide analysis of the PCR product (924 bp in length) obtained by using specific primers for FCV (Cali1–Cali2), confirmed that the virus was closely related to FCV. The novel isolate was designated FCV-Te/10/07.
A sequence of 1557-nucleotides (nt) of ORF1 gene, encoding partial proteinase–polymerase region (pro–pol) (corresponding at nt positions 3176–4733 of FCV-F9 vaccinal strain: GenBank accession no. M86379) and 1834-nt of ORF2 gene encoding A–E regions of the VP1 capsid protein (corresponding at nt 5371–7260 of F9 strain), were determined. BLAST and FASTA analyses of the ORF1 sequence, revealed nucleotide identity of Te/10/07 to the FCV-F9 of 78.4%, while identity to CaCV/48 was equal to 52.6%. The highest sequence match was found to the F65 strain (UK/1990) (81% nt identity) isolated from cat with lameness and oral disease (Glenn et al., 1999). Alignment of ORF2 sequences showed nucleotide identity of Te/10/07 to FCVs sequence available on GenBank database, ranged from about 71% to 80%, while the identity to the F9 strain was 75%. The phylogenetic tree based on the capsid amino acid sequence confirmed that the novel isolate clustered with the FCV group (Fig. 3 ).
Fig. 3.
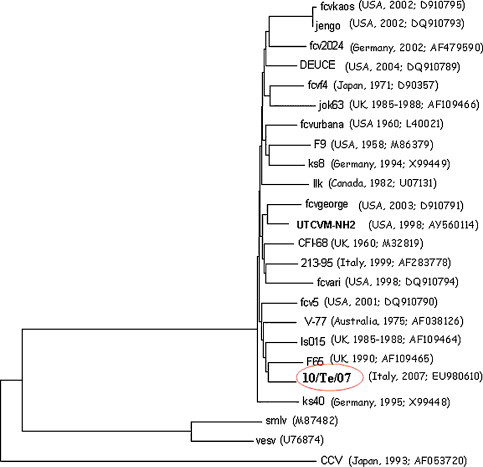
Phylogenetic analysis of the capsid amino acid sequence of the Te/10/07. Tree was generated by using the neighbour-joining distance method. For each strain, the country and year of isolation and the GenBank accession number are indicated.
Comparative analysis of the deduced amino acids (aa) sequences of the hypervariable region E (426–520 aa), revealed the highest similarity to strain LS015 (92%) (80% nt identity) (UK, 1985–1988) associated with chronic stomatitis (Glenn et al., 1999) and to strain F65 (91.5%) (79% nt identity) (Glenn et al., 1999); similarity to FCV-F9 was about 87% (75% nt identity), whereas to the canine strain 213/95 (Italy, 2000) (Pratelli et al., 2000a, Martella et al., 2002) was 84% (73% of nucleotide identity).
Investigation on the growth properties showed that the Te/10/07 strain tended to produce larger plaques (Fig. 4 ) than FCV-F9 in CrFK cells. Moreover, consistent with their larger plaque size, we found that the growth kinetic of the novel isolate was faster than F9 strain. As described in Table 1 , during the single-cycle growth kinetic experiment (MOI 10) we observed that the peak yield of FCV-Te/10/07 strain was produced between T1 (4 h p.i.) and T2 (8 h p.i.), while FCV F9 strain peak yield occurred at T3 (12 h p.i.) and T4 (24 h p.i.). In the multiple-cycle growth kinetic (MOI 0.02) we found that the titres of Te/10/07 increased exponentially by 4 h p.i. In contrast the F9 vaccine strain showed an exponential growth by 8–12 h p.i.
Fig. 4.
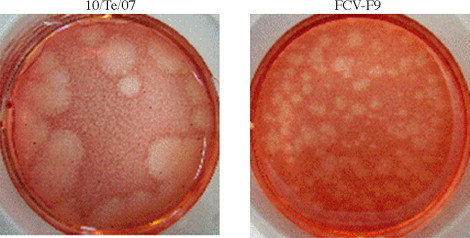
Plaque assay of Te/10/07 and FCV-F9. After incubation at 37 °C for 48 h in humidified 5% CO2, the overlay was removed and CrFK cells were stained with 1% (w/v) Neutral Red solution.
Table 1.
Single- (MOI 10) and multiple-cycle (0.02) growth kinetics of FCV-F9 and Te/10/07.
| MOI 10 |
MOI 0.02 |
|||||||
|---|---|---|---|---|---|---|---|---|
| 4 h (T1) | 8 h (T2) | 12 h (T3) | 24 h (T4) | 4 h (T1) | 8 h (T2) | 12 h (T3) | 24 h (T4) | |
| FCV-F9 | 1.2 × 107 | 1.6 × 107 | 4 × 107 | 3 × 106 | 1.4 × 106 | 1.2 × 107 | 1.1 × 108 | 2 × 107 |
| FCV-Te/10/07 | 1 × 106 | 1.1 × 108 | 8 × 107 | 1.2 × 107 | 1.9 × 108 | 2 × 107 | 1 × 107 | 1.2 × 107 |
In thermal inactivation experiments we observed that the loss of infectivity was similar for both strains analysed following 30 min incubation at 46.2 °C and full inactivation at 52.2 °C (data not shown).
By serological investigation, neutralizing antibodies were present against the strain F9 in 63.9% (55/86) of dog sera with titres from 1:2 to 1:16; whereas antibodies anti-FCV-Te/10/07 were found in only seven sera (8.13%) all of which had concurrent neutralization antibodies to F9. A serum of these showed neutralizing antibodies titres against Te/10/07 higher than F9. However, none of the samples tested resulted positive only for the dog strain (Table 2 ).
Table 2.
Antibody titres determined by VN assay: the number of dog sera with each titre against FCV-F9 and Te/10/07.
| STRAINS | Positives |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1:2a | 1:4 | 1:8 | 1:16 | 1:32 | 1:64 | 1:128 | 1:256 | |
| FCV-F9 | 7 | 5 | 18 | 15 | – | – | – | – |
| Te/10/07 | – | 1 | 2 | 4 | – | – | – | – |
Serum dilutions.
As reported in Table 3 , the prevalence of FCV-F9-antibodies in cats was 76.54% (62/81). Neutralizing antibodies against Te/10/07 strain were recovered in 50.62% (41/81) of samples tested, and 38 sera were positives for F9 strain too, with similar titres or higher. In three cats antibodies were found only against the dog strain.
Table 3.
Antibody titres determined by VN assay: the number of cat sera with each titre against FCV-F9 and Te/10/07.
| Strains | Positives |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1:2a | 1:4 | 1:8 | 1:16 | 1:32 | 1:64 | 1:128 | 1 ≥ 256 | |
| FCV-F9 | 6 | 3 | 2 | 35 | 1 | 1 | 2 | 12 |
| Te/10/07 | 5 | 7 | 14 | 10 | 2 | 1 | 2 | – |
Serum dilutions.
4. Discussion
The high degree of genomic plasticity that characterizes FCV has contributed to the emergence of several variants (Radford et al., 2007), some of which highly transmissible and associated with severe clinical disease (Dawson et al., 1994, Geissler et al., 1997, Pedersen et al., 2000). A number of studies investigated the properties of FCV infection in cats, in contrast little is known about the occurrence and the epidemiology of this virus in dogs, probably because only a few strains have been isolated in cell culture (Evermann et al., 1981, Evermann et al., 1985, Gabriel et al., 1996, Pratelli et al., 2000a).
In the present report, an isolate of calicivirus from a 3-month old puppy (Te/10/07) showing enteritis, submitted to genotypic and phenotypic characterization.
Sequence analysis of ORF1 and ORF2 genes showed that FCV-Te/10/07, is a member of genogroup I (Geissler et al., 1997, Glenn et al., 1999, Sato et al., 2002). As previously demonstrated for other FCVs (Ossiboff et al., 2007, Radford et al., 2007), our strain not form a distinct group based on geographic origin, host range or clinical signs. Comparison of hypervariable region E to the available sequences revealed no consistent differences among Te/10/07 strain and those associated with the most characteristic disease. By BLAST and FASTA analysis of ORF1 and ORF2 genes, our isolate appeared to be most genetically related to the F65 strain (UK, 1990), that has previously been demonstrated experimentally to induce a febrile lameness syndrome (Dawson et al., 1994, Glenn et al., 1999). This observation allowed us to hypothesize that Te/10/07 does not arose from a recombinant event between two different FCV strains or between a FCV strain and a closely related calicivirus.
Investigation on the in vitro growth properties of the Te/10/07 strain revealed that our isolate had rapid growth kinetic compared to F9. As previously hypothesized (Ossiboff et al., 2007) it may represent a marker of increased virulence of FCVs in vivo.
In accordance with another study performed in Italy (Pratelli et al., 2000b), the serological investigation showed that FCV-F9 is widely recognized in dog populations with a prevalence of 63.9%. In contrast, the percentage of dogs with antibodies to Te/10/07 was very low (8.13%) and none of the samples tested were positive solely for Te/10/07. Similarly in cats, despite of the high prevalence of antibodies against FCV-F9 vaccine strain, only three samples were positive against the Te/10/07. These results suggest that FCV-Te/10/07 is less related antigenically than other isolates to the commonly used vaccine strain F9.
In conclusion, this study confirms the interspecific circulation of FCVs among dogs and cats. The presence of this novel virus into dog population raises some concerns about the role of FCVs in puppy gastroenteritis. Further investigations are needed to determine if FCV-Te/10/07 may be an important cause of diarrhea in dog and if domestic dogs may represent a potential source of infection for cats.
Acknowledgement
The authors wish to thank Mr. Palucci for his technical assistance.
References
- Binns S.H., Dawson S., Speakman A.J., Cuevas L.E., Hart C.A., Gaskell C.J., Morgan K.L., Gaskell R.M. A study of feline upper respiratory tract disease with reference to prevalence and risk factors for infection with feline calicivirus and feline herpesvirus. J. Feline Med. Surg. 2000;2:123–133. doi: 10.1053/jfms.2000.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonavoglia C., Martella V., Pratelli A., Tempesta M., Cavalli A., Buonavoglia D., Bozzo G., Elia G., Decaro N., Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 2001;82:1555–1560. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- Carter M.J., Milton I.D., Turner P.C., Meanger J., Bennett M., Gaskell R.M. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch. Virol. 1992;122:223–235. doi: 10.1007/BF01317185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne K.P., Reed F.C., Porter C.J., Dawson S., Gaskell R.M., Radford A.D. Recombination of feline calicivirus within an endemically infected cat colony. J. Gen. Virol. 2006;87:921–926. doi: 10.1099/vir.0.81537-0. [DOI] [PubMed] [Google Scholar]
- Crandell R.A. Isolation and characterization of caliciviruses from dogs with vesicular genital diseases. Arch. Virol. 1988;98:65–71. doi: 10.1007/BF01321006. [DOI] [PubMed] [Google Scholar]
- Dawson S., Bennett D., Carter S.D., Bennett M., Meanger J., Turner P.C., Carter M.J., Milton I., Gaskell R.M. Acute arthritis of cats associated with feline calicivirus infection. Res. Vet. Sci. 1994;56:133–143. doi: 10.1016/0034-5288(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Ellis T.M. Jaundice in a Siamese cat with in utero feline calcivirus infection. Aust. Vet. J. 1981;57:383–385. doi: 10.1111/j.1751-0813.1981.tb00527.x. [DOI] [PubMed] [Google Scholar]
- Evermann J.F., Bryan G.M., Mckeirnan A. Isolation of a calicivirus from a case of canine glossitis. Canine Pract. 1981;8:36–39. [Google Scholar]
- Evermann J.F., Mckeirnan A.J., Smith A.W., Skilling D.E., Ott R.L. Isolation and identification of caliciviruses from dogs with enteric infections. Am. J. Vet. Res. 1985;46:218–220. [PubMed] [Google Scholar]
- Gabriel S., Tohya Y., Mochizuki M. Isolation of a calicivirus antigenically related to feline caliciviruses from feces of a dog with diarrhoea. J. Vet. Med. Sci. 1996;58:1041–1043. doi: 10.1292/jvms.58.10_1041. [DOI] [PubMed] [Google Scholar]
- Geissler K., Schneider K., Platzer G., Truyen B., Kaaden O.R., Truyen U. Genetic and antigenic heterogeneity among feline calicivirus isolates from distinct disease manifestation. Virus Res. 1997;48:193–206. doi: 10.1016/s0168-1702(97)01440-8. [DOI] [PubMed] [Google Scholar]
- Glenn M., Radford A.D., Turner P.C., Carter M., Lowery D., Desilver D.A., Meanger J., Baulch Brown C., Bennet M., Gaskell R.M. Nucleotide sequence of UK and Australian isolates of feline calicivirus (FCV) and phylogenetic analysis of FCV strains. Vet. Microbiol. 1999;67:175–193. doi: 10.1016/s0378-1135(99)00043-7. [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hurley K.E., Pesavento P.A., Pederesen N.C., Poland A.M., Wilson E., Foley J.E. An outbreak of virulent systemic feline calicivirus disease. JAVMA. 2004;224:241–249. doi: 10.2460/javma.2004.224.241. [DOI] [PubMed] [Google Scholar]
- Jiang X., Huang P.W., Zhong W.M., Farkas T., Cubitt D.W., Matson D.O. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods. 1999;83:145–154. doi: 10.1016/s0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- Knowles J.O., Dawson S., Gaskell R.M., Gaskell C.Y., Harvey C.E. Neutralisation patterns among recent British and North American feline calicivirus isolates from different clinical origins. Vet. Rec. 1990;127:125–127. [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:153–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Marsilio F., Di Martino B., Decaro N., Buonavoglia C. A novel nested PCR for the diagnosis of calicivirus infections in the cat. Vet. Microbiol. 2005;105:1–7. doi: 10.1016/j.vetmic.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Martella V., Lorusso E., Decaro N., Elia G., Radogna A., D’abramo M., Desario C., Cavalli A., Corrente M., Camero M., Germinario C.A., Bányai K., Di Martino B., Marsilio F., Carmichael L.E., Buonavoglia C. Detection and molecular characterization of a canine Norovirus. Emerg. Infect. Dis. 2008;14:1306–1308. doi: 10.3201/eid1408.080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Pratelli A., Gentile M., Buonavoglia D., Decaro N., Fiorante P., Buonavoglia C. Analysis of the capsid protein gene of a feline-like calicivirus isolated from a dog. Vet. Microbiol. 2002;85:315–322. doi: 10.1016/s0378-1135(01)00521-1. [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Kawanishi A., Sakamoto H., Tashiro S., Fujimoto R., Ohwaki M. A calicivirus isolated from a dog with fatal diarrhoea. Vet. Rec. 1993;132:221–222. doi: 10.1136/vr.132.9.221. [DOI] [PubMed] [Google Scholar]
- Ossiboff R.J., Sheh A., Shotton J., Pesavento P.A., Parker J.S. Feline calicivirus (FCVs) isolated from cats with virulent systemic disease possess in vitro phenotypes distinct from those of other FCV isolates. J. Gen. Virol. 2007;88:506–517. doi: 10.1099/vir.0.82488-0. [DOI] [PubMed] [Google Scholar]
- Pedersen N.C., Elliott J.B., Glasgow A., Poland A., Keel K. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Vet. Microbiol. 2000;73:281–300. doi: 10.1016/S0378-1135(00)00183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Laliberte L., Ekman S. A transient febrile limping syndrome of kittens caused by two different strains of feline calicivirus. Feline Pract. 1983;13:26–35. [Google Scholar]
- Pesavento P.A., Maclachlan N.J., Dillard-Telm L., Grant C.K., Hurley K.F. Pathologic, immunohistochemical, and electron microscopic findings in naturally occurring virulent systemic feline calicivirus infection in cats. Vet. Pathol. 2004;41:257–263. doi: 10.1354/vp.41-3-257. [DOI] [PubMed] [Google Scholar]
- Povey R.C. Serological relationship among feline caliciviruses. Infect. Immun. 1974;10:1307–1314. doi: 10.1128/iai.10.6.1307-1314.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Greco G., Camero M., Normanno G., Buonavoglia C. Isolation and identification of a calicivirus from a dog with diarrhoea. New Microbiol. 2000;23:257–260. [PubMed] [Google Scholar]
- Pratelli A., Martella V., Elia G., Terio E., Lavazza A., Buonavoglia C. Characterization of a calicivirus strain isolated from a pup with enteritis. In: Brocchi E., Lavazza A., editors. Proceedings Fifth International Congress of ESVV. 2000. pp. 196–197. [Google Scholar]
- Pratelli A., Tempesta M., Greco G., Martella V., Buonavoglia C. Development of a nested PCR assay for the detection of canine coronavirus. J. Virol. Methods. 2001;84:91–94. doi: 10.1016/S0166-0934(99)00017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford A.D., Coyne K.P., Dawson S., Porter C.J., Gaskell R.M. Feline calicivirus. Vet. Res. 2007;38:319–335. doi: 10.1051/vetres:2006056. [DOI] [PubMed] [Google Scholar]
- Roerink F., Hashimoto M., Tohya Y., Mochizuki M. Organization of the canine calicivirus genome from the RNA polymerase gene to the Poly(A) tail. J. Gen. Virol. 1999;80:929–935. doi: 10.1099/0022-1317-80-4-929. [DOI] [PubMed] [Google Scholar]
- Sato Y., Ohe K., Murakami M., Fukuyama M., Furuhata K., Kishikawa S., Suzuki Y., Kiuchi A., Hara M., Ishikawa Y., Taneno A. Phylogenetic analysis of field isolates of feline calicivirus (FCV) in Japan by sequencing part of its capsid gene. Vet. Res. Commun. 2002;26:205–219. doi: 10.1023/a:1015253621079. [DOI] [PubMed] [Google Scholar]
- Schaffer F.L., Soergel M.E., Black J.W., Skilling D.E., Smith A.W., Cubitt W.D. Characterization of a new calicivirus isolated from feces of a dog. Arch. Virol. 1985;84:181–195. doi: 10.1007/BF01378971. [DOI] [PubMed] [Google Scholar]
- Schorr-Evans E.M., Poland A., Johnson W.E., Pedersen N.C. An epizootic of highly virulent feline calicivirus disease in a hospital setting in New England. J. Feline Med. Surg. 2003;5:217–226. doi: 10.1016/S1098-612X(03)00008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.O., Onwudiwe O., Green K.Y. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Garfied M., Green K.Y. Processing map and essential cleavage sites of the non-structural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 2002;76:7060–7072. doi: 10.1128/JVI.76.14.7060-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


