Abstract
Infectious bronchitis virus (IBV) infections in poultry cause great economic losses to the poultry industry worldwide. The emergence of viral variants complicates disease control. The IBV strains in Taiwan were clustered into two groups, Taiwan group I and Taiwan group II, based on the S1 gene. A variant was previously identified and showed a distinct S1 gene homology with other local strains. This study investigated the 3′ 7.3 kb genome of eight Taiwan strains isolated from 1992 to 2007. The genes of interest were directly sequenced. Sequence analyses were performed to detect any recombination event among IBVs. The results demonstrated that all of the examined viruses maintained the typical IBV genome organization as 5′-S-3a-3b-E-M-5a-5b-N-UTR-3′. In the phylogenetic analyses, various genes from one strain were clustered into separate groups. Moreover, frequent recombination events were identified in the Simplot analyses among the Taiwan and China CK/CH/LDL/97I-type strains. Putative crossover sites were located in the S1, S2, 3b, M genes and the intergenic region between the M and 5a genes. All of the recombinants showed chimeric IBV genome arrangements originated from Taiwan and China-like parental strains. Field IBVs in Taiwan undergo genetic recombination and evolution.
Keywords: China, Infectious bronchitis virus, Recombination, Sequence analysis, Taiwan
1. Introduction
Avian infectious bronchitis virus (IBV), with a large infectious RNA genome (27.6 kb), belongs to the family Coronaviridae, the group 3 Coronavirus. The RNA molecule of the IBV is linear, single-stranded, positive sense and possesses a 5′ cap and a 3′ poly-A tail. IBVs can replicate in respiratory, alimentary and urogenital tracts in chickens of all ages, resulting in great economic losses to the poultry industry worldwide (Cavanagh and Naqi, 2003). Clinical signs associated with respiratory and enteric tracts and renal damage might be found in infected chickens (Cavanagh, 2007). During infection, IBV possesses a unique discontinuous transcription system, i.e. a nested set of 3′ co-terminal subgenomic mRNAs sharing a common leader sequence in the 5′ end are transcribed in the presence of negative-stranded RNA intermediates. The viral polymerase “jumping” possibly contributes to the high RNA recombination frequency in coronaviruses (Lai, 1992, Lai and Holmes, 2001).
The IBV genome encodes four structural proteins: spike glycoprotein (S), envelope protein (E; also known as sM), membrane glycoprotein (M), and nucleocapsid protein (N) (Cavanagh and Naqi, 2003). The spike glycoprotein is post-translationally cleaved into S1 and S2 subunits. The S1 subunit anchors onto the viral outer membrane using the S2 subunit to form a club-shaped projection on the mature virion (Cavanagh, 1983). The S1 glycoprotein is involved in cell attachment and carries epitopes for serotype-specific hemagglutination-inhibition and virus-neutralization antibodies (Cavanagh and Davis, 1986, Hodgson et al., 2004, Ignjatovic and Sapats, 2005, Koch et al., 1990). The S2 glycoprotein, in which two antigenic determinants were identified (Koch et al., 1990), may possess different secondary structures that affect the S1 specific antibody binding (Callison et al., 1999). The membrane glycoprotein is associated with virus assembling and budding (Lai and Holmes, 2001). The nucleocapsid protein interacts with genomic RNA to form the viral nucleocapsid, playing a role in viral RNA synthesis and cell immunity (Lai and Holmes, 2001). In addition, four non-structural proteins of unknown function are expressed by the polycistronic genes, gene 3 and 5 (Britton et al., 2006). Virus mutants carrying truncated 3b genes demonstrate increased virulence and growth advantages in vitro and in ovo (Shen et al., 2003). The open reading frames (ORFs) of 3a, 3b, 5a, and 5b encode accessory proteins not essential for IBV replication (Casais et al., 2005, Hodgson et al., 2006, Youn et al., 2005).
As a signature of avian coronaviruses, IBV strains are continuously evolving through point mutations and recombination of their genomes. Those variants have better adaptation or increased virulence advantageous to IB outbreaks. To date, a large number of IBV sero- or genotypes have been identified worldwide (Cavanagh, 2007). Most molecular epidemiologic studies have focused on the spike glycoprotein gene. (Bochkov et al., 2006, Dolz et al., 2006, Dolz et al., 2008, Jackwood et al., 2007). It was reported that slight sequence differences in the S1 gene probably lead to poor cross-protection (Cavanagh et al., 1997). Viruses of different types can co-circulate within a region (Capua et al., 1999, Liu et al., 2006), raising the inter-strain RNA recombination frequency (Bochkov et al., 2007, Jia et al., 1995, Lee and Jackwood, 2000). The wide use of live virus vaccine may also critically contribute to the genetic evolution of IBVs by acting as a heterologous RNA donor template (Kusters et al., 1990, Wang et al., 1993). The emergence of viral variants has complicated disease control requiring persistent IBV molecular surveys.
IBV strains in Taiwan were previously clustered into two groups, Taiwan group I (TW-I) and Taiwan group II (TW-II), on the basis of the S1 gene (Wang and Tsai, 1996). However, a variant isolated in 2002 showed an unusually high S1 gene homology with China strains, but not in the N gene, suggesting an inter-strain recombination event (Huang et al., 2004). In this study, the 3′ 7.3 kb genomes from Taiwan IBV strains were investigated to elucidate the genetic diversity of viruses.
2. Materials and methods
2.1. Viruses
Eight IBVs isolated in Taiwan from 1992 to 2007 (Huang et al., 2004) and the vaccine strain H120 (ABIC Biological Laboratories Teva Ltd., Israel) were recovered for this study. The case histories of local strains are listed in Table 1 . Viruses were propagated in the allantoic cavity of 9–11-day-old specific pathogen free embryonated eggs (Animal Health Research Institute, Council of Agriculture, Tamsui, Taiwan). Each egg received 0.1–0.2 ml inoculation. After 48–72 h incubation, allantoic fluid was collected and frozen at −80 °C until use.
Table 1.
Eight IBVs isolated in Taiwan from 1992 to 2007.
| Strain | Year of isolation | Chicken type | Age (weeks)a | Location | Pathogenicity | Genotypeb |
|---|---|---|---|---|---|---|
| 1171/92 | 1992 | Broiler | 3 | Taoyuan | Nephropathogenic | TW-I |
| 2296/95 | 1995 | Broiler | 2 | Taoyuan | Nephropathogenic | TW-II |
| 2575/98 | 1998 | Broiler | 4 | Changhua | Nephropathogenic | TW-I |
| 2992/02 | 2002 | Broiler | 4 | Yilan | Nephropathogenic | Variant |
| 3071/03 | 2003 | Broiler | 5.5 | Yilan | Nephropathogenic | TW-I |
| 3263/04 | 2004 | Broiler | 5.2 | Yilan | Nephropathogenic | TW-II |
| 3374/05 | 2005 | Broiler | NAc | Changhua | NA | Variant |
| 3468/07 | 2007 | Taiwan country chicken | 12 | NAc | NA | TW-I |
Age of chickens at the time of virus isolation.
Genotype is determined based on the S1 gene. TW-I, Taiwan group I; TW-II, Taiwan group II; Variant, neither TW-I nor TW-II.
NA: Not available.
2.2. Viral RNA extraction, RT-PCR and DNA sequencing
Viral RNA was extracted from 200 μL of virus-infected allantoic fluid using a viral nucleic acid extraction kit (Geneaid Biotech Ltd., Taipei, Taiwan) following the manufacturer's protocol. Previously published primers (Huang and Wang, 2007) were employed in this study to amplify the gene fragments. For sequencing the 3′ un-translated region (UTR) of H120, one additional forward primer was designed as 5′-GGAACAATGCACAGCTGGAA-3′ from strain H120-GD (GenBank accession no. AY028296).
Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed with one step in a reaction volume of 50 μL containing 0.5 μL of 5 U/μL RealTaq DNA polymerase (Real Biotech, Taipei, Taiwan), 5 μL of 10× buffer (Real Biotech), 12 μL of 2.5 mM dNTPs (GeneTeks BioScience, Taipei, Taiwan), 0.5 μL of 50 pmol/μL upstream primer (Mission Biotech, Taipei, Taiwan), 0.5 μL of 50 pmol/μL downstream primer (Mission Biotech), 0.4 μL of 40 U/μL Recombinant RNasin ribonuclease inhibitor (Promega, Madison, WI), 0.1 μL of 200 U/μL M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA), 10 μL of viral RNA and 21 μL of diethyl pyrocarbonate-treated water. Reverse transcription was carried out at 40 °C for 30 min, and followed by initial DNA polymerase activation at 94 °C (3 min). PCR was then performed for 35 cycles of denaturation at 94 °C (30 s), annealing at 53 °C (30 s), and polymerization at 72 °C for (1 min 40 s). The cycling reaction was completed with the final polymerization step at 72 °C for 10 min. The amplified products were analyzed in 1.5% agarose gel electrophoresis, stained with ethidium bromide and visualized under UV light.
DNA sequencing in both strands from separate RT-PCR products was conducted by a commercial service (Mission Biotech). Each nucleotide was determined from at least four identical results.
2.3. Sequence analysis
The obtained nucleotide sequences were compiled and the amino acid sequences were deduced using DNAStar software (DNAStar, Madison, WI). The IBV reference strains were retrieved from the GenBank database with the accession number listed in Table 2 . The AlignX program of the Vector NTI Suite 8 software (InforMax, North Bethesda, ML) was used to generate multiple sequence alignments and determine the nucleotide identity. Phylogenetic analyses were performed with the neighbor-joining method using MEGA version 4 (Tamura et al., 2007). The bootstrap values were determined from 1000 replicates of the original data.
Table 2.
IBV reference strains included in this study.
| Strain | Country | Accession numbers |
|---|---|---|
| Armidale | Australia | DQ490205 (S1-N)a |
| Vic | Australia | DQ490221 (S1-N) |
| BJ | China | AY319651 (S1-N) |
| CK/CH/LDL/97I | China | EF030996 (S1), EF602445 (S2-N) |
| CK/CH/LDL/98I | China | DQ167132 (S1), EF602446 (S2-N) |
| J2 | China | AF286303 (S1) |
| LX4 | China | AY338732 (S1-N) |
| SAIBK | China | DQ288927 (S1-N) |
| T3 | China | AF227438 (S1) |
| Q1 | China | AF286302 (S1) |
| QXIBV | China | AF193423 (S1), AF288146 (S2), AF221667 (M-ORF 5), AF199412 (N) |
| 1171/92 | Taiwan | DQ646406 (S1-N) |
| 2296/95 | Taiwan | DQ646404 (S1-N) |
| 2575/98 | Taiwan | DQ646405 (S1-N) |
| Beaudette | USA | NC_001451 (S1-N) |
| Cal99 | USA | AY514485 (S1-N) |
| CU-T2 | USA | U49858 (S1-N) |
| DE072 | USA | AF274435 (S1), AY024337 (S2), AF202998 (ORF 3), AF202999 (M), AF203000 (ORF 5), AF203001 (N) |
| Gray | USA | L18989 (S1), AF394180 (S2), AF318282 (E), AF286180 (M), AF469011 (ORF 5), M85245 (N) |
| M41 | USA | AY851295 (S1-N) |
Sequences of the gene fragments used within the parenthesis.
2.4. Simplot analysis
The consecutive IBV nucleotide sequences from the S to N genes (6.8 kb) based on the multiple alignment results were introduced into similarity plots with SimPlot version 3.5.1 (Lole et al., 1999). The nucleotide identity was calculated using the Kimura 2-parameter method with a transition-transversion ratio of 2 in each window of 500 bp. The window was successively furthered along the alignment using a 20-bp increment. At least four sequences were required to initiate an analysis.
2.5. GenBank accession numbers
The IBV sequences resulting from this study were submitted to the GenBank database. The accession numbers are EU822336 (strain 3468/07), EU822337 (strain 3374/05), EU822338 (strain 3263/04), EU822339 (strain 3071/03), EU822340 (strain 2992/02), and EU822341 (strain H120).
3. Results
3.1. Sequence comparisons
Sequences that covered the 3′ 7.3 kb genome were determined from the Taiwan IBVs and strain H120. All of the examined viruses were found to maintain the typical IBV genome organization as 5′-S-3a-3b-E-M-5a-5b-N-3′ (data not shown). The S gene size ranged from a minimum of 3471 nucleotides (strain 3071/03) to a maximum of 3501 nucleotides (strains 2992/02 and 3374/05). Compared with other local strains, single base mutations in the 3′ end of the S and 3b genes from the strain 3071/03 changed the genetic code from GAA to TAA (Glutamine → stop codon), resulting in 27- and 3-base truncated ORFs, respectively. Similarly, in the 3374/05, a single base T-insertion in the 3b gene created an early stop codon, leading to a 48-base truncation. However, the 3a, 5b, and N gene ORF sizes were conserved among IBVs. All of the virus genomes carried an intergenic (IG) region located between the M and 5a genes with a size of 351–362 nucleotides. The IG region of the strain H120 was 55 bases longer than that of the strain Beaudette. The 3′ UTR, downstream of the stop codon in the N gene, a region of 475 and 500 nucleotides was sequenced from local strains and H120, respectively. All of the local strains shared a high sequence identity (96–100%) in this region. However, only 52% and 73% identity were observed between H120 and its closely related M41 and Beaudette strains, respectively.
3.2. Phylogenetic analyses
Phylogenetic analyses were performed based on the nucleotide sequence alignment using each ORF from the S to N genes among eight Taiwan and reference strains (Fig. 1 ). In the S1 gene, the Taiwan strains showed the highest identity (84%) with the Beaudette reference strain and the lowest (60%) with the DE072. The Taiwan strains (except for 2992/02 and 3374/05) could be clustered into two groups, TW-I and TW-II, based on the S1 gene. Strains 2992/02 and 3374/05 were closely related (>95% identity) to the China genotype VII strains (CK/CH/LDL/97I, CK/CH/LDL/98I, Q1, J2, and T3) (Liu et al., 2006). In the S2 gene, all of the local strains were grouped with CK/CH/LDL/97I and CK/CH/LDL/98I except for the two viruses isolated before 1995 (strains 1171/92 and 2296/95). Strains 2575/98 and 2992/02 were classified with CK/CH/LDL/97I and CK/CH/LDL/98I in the analyses of the 3a and 3b genes. The E gene analysis segregated the 2992/02 into the China group. Strains 2992/02 and 3468/07 were distributed with the China strains in the M gene analysis. The 5a, 5b and N genes analyses revealed that all of the local strains were in the same group. Nucleotide sequences from the three ORFs were conserved among the Taiwan strains with >92% homology.
Fig. 1.
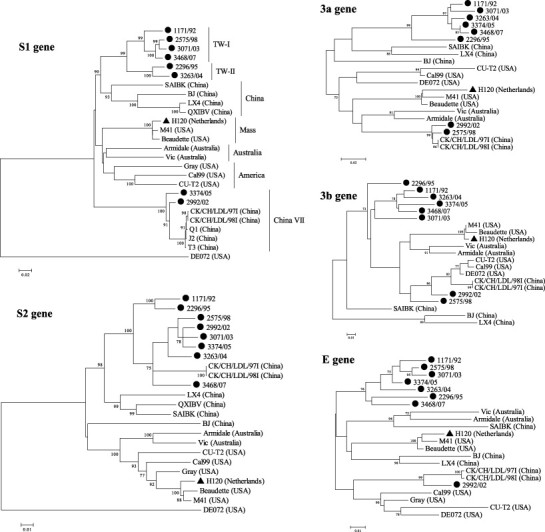
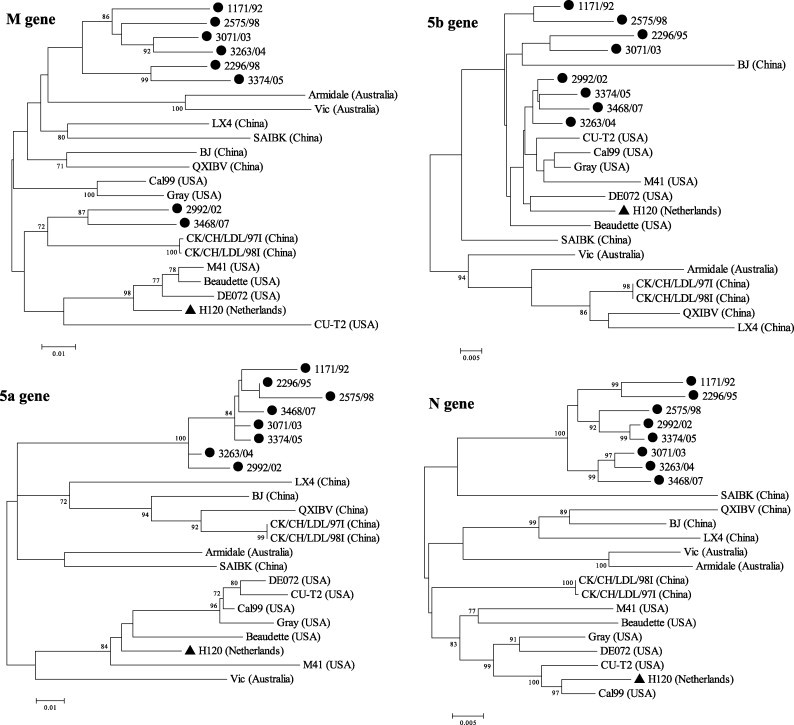
Phylogenetic analyses of the Taiwan strains (●), H120 (▴) and reference strains for structural and non-structural protein genes of IBVs. The phylogenetic trees were constructed using the MEGA version 4 by the neighbor-joining method (bootstrapping for 1000 replicates with its value >70%).
3.3. Inter-strain recombinants identification
The 3′ 6.8 kb genome sequence (S–N gene) of six Taiwan IBVs were queried in the Simplot analysis. Strains 1171/92, H120 and the China strain CK/CH/LDL/97I were used as putative parental strains when strains 2575/98, 2992/02, 3071/03, 3374/05, 3468/07 (TW-I) were queried. The parental strain 1171/92 was replaced with 2296/95 when strain 3263/04 (TW-II) was queried. The similarity plot displays the consecutive nucleotide identity (%) among the queried strain and parental strains. Strains were considered as recombinants if any crossover event took place between two putative parental strains. The breakpoint in which the parental strains have equal identity to the query strain is the predicted recombination site. As Fig. 2 demonstrates, crossover events between parental 1171/92 or 2296/95 and CK/CH/LDL/97I were detected in each plot. The recombination sites were located in the S1, S2, 3b, M genes, and the IG region between the M and 5a genes. Each putative recombinant was schematically assembled using Taiwan and China-like sequence fragments. The genomic positions of those crossover sites were indicated within the plot (numbers in red).
Fig. 2.
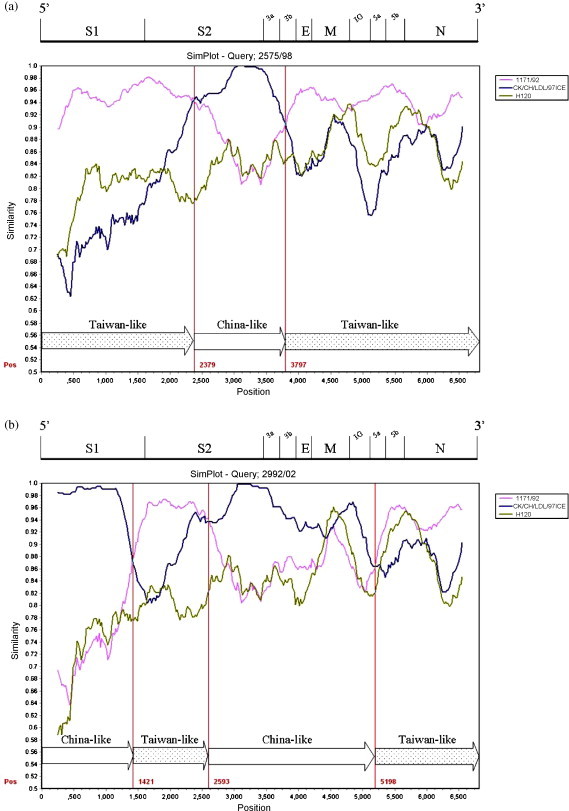
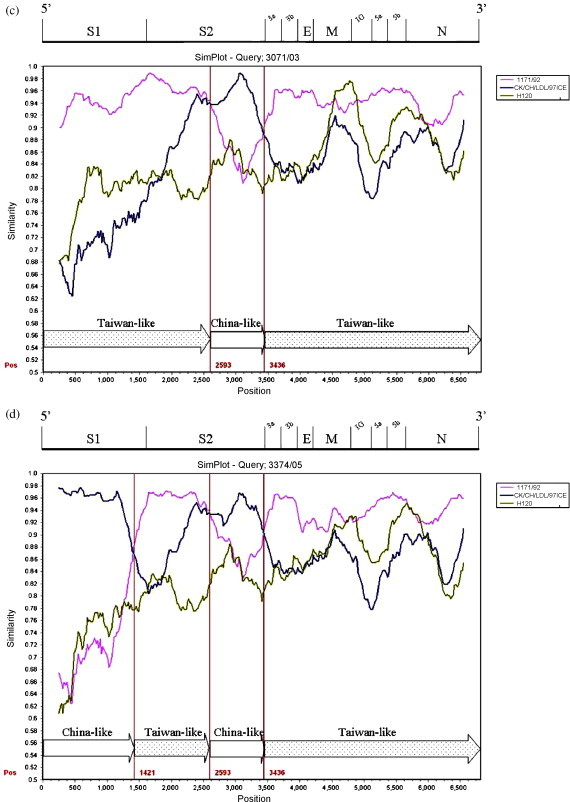
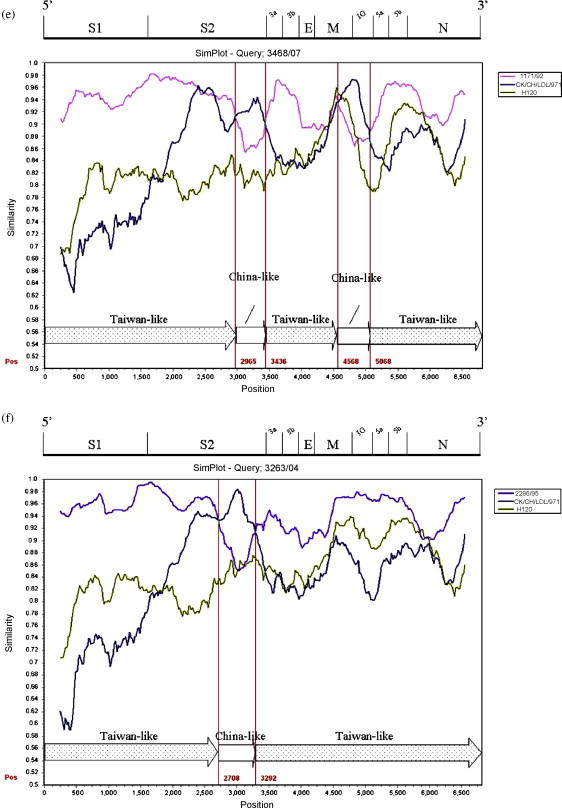
Simplot analyses of the Taiwan IBVs. Strains 1171/92 (pink), H120 (green), and the China strain CK/CH/LDL/97I (deep blue) were used as putative parental strains when strains 2575/98 (a), 2992/02 (b), 3071/03 (c), 3374/05 (d), 3468/07 (e) were queried. The parental strain 1171/92 was replaced with 2296/95 (light blue) when the strain 3263/04 (f) was queried. Each graph displays the consecutive nucleotide identity (%) from the S to N genes among the queried strain and parental strains. The breakpoint where the parental strains have equal identity to the query strain is the predicted recombination site. Each putative recombinant was schematically assembled using Taiwan and China-like sequence regions. The genomic positions of the crossover sites were indicated by numbers in red. The genomic scale was given at the top of the plot. IG: Intergenic region. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
4. Discussion
In Taiwan, field outbreaks are frequently reported despite routine vaccine use. There were originally two IBV genotypes (TW-I and TW-II) circulating in the field (Liu et al., 2003, Wang and Tsai, 1996) until genetic variants 2992/02 (Huang et al., 2004) and 3374/05 emerged. Both variants showed a high S1 gene homology with the proventriculus pathogenic strains J1, Q2, T3, CK/CH/LDL/97I and CK/CH/LDL/98I, belonging to the China genotype VII (Liu et al., 2006, Yu et al., 2001). The 3′ 6.8 kb gene of CK/CH/LDL/97I was used as a putative parental strain. Mass type viruses have been widely used as vaccine strains in Taiwan. Our primary concern was the possible viral recombination resulting from the co-circulation of heterologous vaccine strains in flocks. The consecutive sequence of 3′ 7.3 kb genome in strain H120 was obtained first in this study and served as another putative parental strain. To our surprise, six local strains experienced crossover events with the strain CK/CH/LDL/97I instead of H120. Thus, inter-strain recombination events had occurred between the IBVs from Taiwan and China. Taiwan and China are geographically separate. Neither live poultry nor processed poultry products from China have been allowed to import into Taiwan for years. How those recombinants arose remains unknown. A recently identified IBV isolate in Korea (Kr/D64/05) also revealed a close relationship to the China CK/CH/LDL/97I-type strains (Lee et al., 2008). Furthermore, the appearance of the China-like strains in Taiwan is reminiscent of the spread of the China QXIBV strain in European countries (Beato et al., 2005, Domanska-Blicharz et al., 2006). In this case, it could be speculated that migrating birds provide the genetic sources of IBV variants in Taiwan. A chicken-nephropathogenic IBV strain was identified from a non-diseased teal (Anas sp.) in China (Liu et al., 2005). Thus, the transport of IBVs over long distances by other avian species appears to be possible (Cavanagh, 2005). However, we still cannot overlook illegal trafficking or unapproved vaccine use in the field.
All six recombinants defined in this study were inter-genotypic recombination and the “China-like” sequence substitutions took place in multiple genes. In particular, partial S gene replacement was observed in every recombinant. Since coronaviruses possess different host range or cell tropism through the variance in the S gene (Casais et al., 2003, Kuo et al., 2000), alterations in the antigenic characteristic in those variants could be expected. In this study, however, nearly all of the strains showed nephropathogenicity (lesions of kidney) in chickens, rather than the proventricular lesions caused by the CK/CH/LDL/97I-type strains. How a variant with a chimeric genome arrangement from heterologous strains demonstrates its tissue tropism or pathogenicity in a host is not clear. In addition, it was found that chicks challenged with virulent CK/CH/LDL/97I were incompletely protected by commercial vaccines and other heterologous strains (Liu et al., 2007). To effectively control the IB disease in Taiwan, the protective effect of vaccines against challenges from those recombinants awaits to be investigated.
To our knowledge, this is the first use of Simplot for genetic analyses of IBV strains. The similarity plot can depict the genetic distance among the aligned sequences in a graphical window. In this study IBV recombinants could be defined directly from the Simplot analyses, and the crossover events and corresponding genome positions were readily observed. In phylogenetic analyses, strains were deduced as recombinants if different genes from the genomes were clustered into separate phylogenetic groups. Parallel results were obtained from both analyses.
The emergence of IBV variants through RNA recombination was previously described (Brooks et al., 2004, Jia et al., 1995, Lee and Jackwood, 2000, Mondal and Cardona, 2007, Wang et al., 1993). Recombination events occurred in multiple genes. The consensus IG sequences CT(T/G)AACAA or the conserved regions around were assumed as the recombination “hot spots” in IBVs (Lee and Jackwood, 2000). In addition, the CTTTTG sequence was observed adjacent to the putative recombinant junction (Brooks et al., 2004, Wang et al., 1993). In this study, T-rich motifs (A/T/G)TTTTG, consensus among a recombinant and its parental strains, were located upstream of several crossover sites. For instance, the motifs were found in the S2 gene of the 3263/04, the 3b gene of the 2575/98, and the IG region of the 2992/02 and 3468/07. All of the recombinants experienced crossover events in the 3′ end of the S gene, indicating a possible region for template switching in viral RNA synthesis. Similar results were obtained from the IBV strain CU-T2 and Cal99, whose recombination sites were deduced in the 3′ 751 and 700 nucleotide of the S gene, respectively (Jia et al., 1995, Mondal and Cardona, 2007).
In addition to viral recombination, point mutation is a way of virus evolution. The single base nonsense mutations resulted in truncations of the S and 3b genes in the Taiwan strains 3071/03 and 3374/05. The naturally occurring IBV strains with mutations or complete deletions in non-structural ORFs 3a, 3b, 5a, and 5b revealed lower virus growth rate or titer in ovo (Liu et al., 2008) and in vivo (Mardani et al., 2008).
This study describes the dual viral genomes incorporation in the separated areas having no known interactions between poultry. The molecular investigation in the 3′ 7.3 kb of the IBV genome demonstrates that inter-strain recombination events contribute to the genetic diversity of the Taiwan strains. Field IBVs in Taiwan undergo genetic recombination and evolution, which might lead to disease control difficulty. The biological and antigenic characteristics of the IBV variants await further studies.
Acknowledgment
The financial support of the National Science Council and Council of Agriculture, Executive Yuan, Taiwan, is greatly appreciated.
References
- Beato M.S., De Battisti C., Terregino C., Drago A., Capua I., Ortali G. Evidence of circulation of a Chinese strain of infectious bronchitis virus (QXIBV) in Italy. Vet. Rec. 2005;156:720. doi: 10.1136/vr.156.22.720. [DOI] [PubMed] [Google Scholar]
- Bochkov Y.A., Batchenko G.V., Shcherbakova L.O., Borisov A.V., Drygin V.V. Molecular epizootiology of avian infectious bronchitis in Russia. Avian Pathol. 2006;35:379–393. doi: 10.1080/03079450600921008. [DOI] [PubMed] [Google Scholar]
- Bochkov Y.A., Tosi G., Massi P., Drygin V.V. Phylogenetic analysis of partial S1 and N gene sequences of infectious bronchitis virus isolates from Italy revealed genetic diversity and recombination. Virus Genes. 2007;35:65–71. doi: 10.1007/s11262-006-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton P., Casais R., Hodgson T., Davis M., Cavanagh D. Genes 3 and 5 of infectious bronchitis virus are accessory protein genes. Adv. Exp. Med. Biol. 2006;581:363–368. doi: 10.1007/978-0-387-33012-9_64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J.E., Rainer A.C., Parr R.L., Woolcock P., Hoerr F., Collisson E.W. Comparisons of envelope through 5B sequences of infectious bronchitis coronaviruses indicates recombination occurs in the envelope and membrane genes. Virus Res. 2004;100:191–198. doi: 10.1016/j.virusres.2003.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callison S.A., Jackwood M.W., Hilt D.A. Infectious bronchitis virus S2 gene sequence variability may affect S1 subunit specific antibody binding. Virus Genes. 1999;19:143–151. doi: 10.1023/A:1008179208217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capua I., Minta Z., Karpinska E., Mawditt K., Britton P., Cavanagh D., Gough R.E. Co-circulation of four types of infectious bronchitis virus (793/B, 624/I, B1648 and Massachusetts) Avian Pathol. 1999;28:587–592. doi: 10.1080/03079459994380. [DOI] [PubMed] [Google Scholar]
- Casais R., Davies M., Cavanagh D., Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005;79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Dove B., Cavanagh D., Britton P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J. Virol. 2003;77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus IBV: structural characterization of the spike protein. J. Gen. Virol. 1983;64(Pt 12):2577–2583. doi: 10.1099/0022-1317-64-12-2577. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67(Pt 7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Elus M.M., Cook J.K. Relationship between sequence variation in the S1 spike protein of infectious bronchitis virus and the extent of cross-protection in vivo. Avian Pathol. 1997;26:63–74. doi: 10.1080/03079459708419194. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Naqi S.A. Infectious bronchitis. In: Saif A.M., Fadly Y.M., McDougald L.R., Swayne D.E., editors. Diseases of Poultry. 11th ed. Iowa State University Press; Ames, IA: 2003. pp. 101–119. [Google Scholar]
- Dolz R., Pujols J., Ordonez G., Porta R., Majo N. Antigenic and molecular characterization of isolates of the Italy 02 infectious bronchitis virus genotype. Avian Pathol. 2006;35:77–85. doi: 10.1080/03079450600597295. [DOI] [PubMed] [Google Scholar]
- Dolz R., Pujols J., Ordonez G., Porta R., Majo N. Molecular epidemiology and evolution of avian infectious bronchitis virus in Spain over a fourteen-year period. Virology. 2008;374:50–59. doi: 10.1016/j.virol.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanska-Blicharz K., Minta Z., Smietanka K., Porwan T. New variant of IBV in Poland. Vet. Rec. 2006;158:808. doi: 10.1136/vr.158.23.808-c. [DOI] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Casais R., Dove B., Britton P., Cavanagh D. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J. Virol. 2004;78:13804–13811. doi: 10.1128/JVI.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.P., Lee H.C., Cheng M.C., Wang C.H. S1 and N gene analysis of avian infectious bronchitis viruses in Taiwan. Avian Dis. 2004;48:581–589. doi: 10.1637/7186-033004R. [DOI] [PubMed] [Google Scholar]
- Huang Y.P., Wang C.H. Sequence changes of infectious bronchitis virus isolates in the 3′ 7.3 kb of the genome after attenuating passage in embryonated eggs. Avian Pathol. 2007;36:59–67. doi: 10.1080/03079450601110015. [DOI] [PubMed] [Google Scholar]
- Ignjatovic J., Sapats S. Identification of previously unknown antigenic epitopes on the S and N proteins of avian infectious bronchitis virus. Arch. Virol. 2005;150:1813–1831. doi: 10.1007/s00705-005-0541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hilt D.A., Williams S.M., Woolcock P., Cardona C., O’Connor R. Molecular and serologic characterization, pathogenicity, and protection studies with infectious bronchitis virus field isolates from California. Avian Dis. 2007;51:527–533. doi: 10.1637/0005-2086(2007)51[527:MASCPA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Jia W., Karaca K., Parrish C.R., Naqi S.A. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 1995;140:259–271. doi: 10.1007/BF01309861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch G., Hartog L., Kant A., van Roozelaar D.J. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J. Gen. Virol. 1990;71(Pt 9):1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodoma: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters J.G., Jager E.J., Niesters H.G., van der Zeijst B.A. Sequence evidence for RNA recombination in field isolates of avian coronavirus infectious bronchitis virus. Vaccine. 1990;8:605–608. doi: 10.1016/0264-410X(90)90018-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Holmes K.V. Coronaviridae: the virus and their replication. In: Knipe D.M, Howley P.M., editors. Field Virology. 4th ed. Linppincott Williams & Wilkins Publisher; Philadelphia: 2001. pp. 1163–1185. [Google Scholar]
- Lee C.W., Jackwood M.W. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 2000;145:2135–2148. doi: 10.1007/s007050070044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.K., Jeon W.J., Lee Y.J., Jeong O.M., Choi J.G., Kwon J.H., Choi K.S. Genetic diversity of avian infectious bronchitis virus isolates in Korea between 2003 and 2006. Avian Dis. 2008;52:332–337. doi: 10.1637/8117-092707-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Liu H.J., Lee L.H., Shih W.L., Lin M.Y., Liao M.H. Detection of infectious bronchitis virus by multiplex polymerase chain reaction and sequence analysis. J. Virol. Methods. 2003;109:31–37. doi: 10.1016/s0166-0934(03)00041-7. [DOI] [PubMed] [Google Scholar]
- Liu S., Chen J., Chen J., Kong X., Shao Y., Han Z., Feng L., Cai X., Gu S., Liu M. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) J. Gen. Virol. 2005;86:719–725. doi: 10.1099/vir.0.80546-0. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang Q., Chen J., Han Z., Shao Y., Kong X., Tong G. Identification of the avian infectious bronchitis coronaviruses with mutations in gene 3. Gene. 2008;412:12–25. doi: 10.1016/j.gene.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang X., Wang Y., Li C., Liu Q., Han Z., Zhang Q., Kong X., Tong G. Evaluation of the protection conferred by commercial vaccines and attenuated heterologous isolates in China against the CK/CH/LDL/97I strain of infectious bronchitis coronavirus. Vet. J. 2007;179:130–136. doi: 10.1016/j.tvjl.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.W., Zhang Q.X., Chen J.D., Han Z.X., Liu X., Feng L., Shao Y.H., Rong J.G., Kong X.G., Tong G.Z. Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch. Virol. 2006;151:1133–1148. doi: 10.1007/s00705-005-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardani K., Noormohammadi A.H., Hooper P., Ignjatovic J., Browning G.F. Infectious bronchitis viruses with a novel genomic organization. J. Virol. 2008;82:2013–2024. doi: 10.1128/JVI.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S.P., Cardona C.J. Genotypic and phenotypic characterization of the California 99 (Cal99) variant of infectious bronchitis virus. Virus Genes. 2007;34:327–341. doi: 10.1007/s11262-006-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Wen Z.L., Liu D.X. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology. 2003;311:16–27. doi: 10.1016/S0042-6822(03)00117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wang C.H., Tsai C.T. Genetic grouping for the isolates of avian infectious bronchitis virus in Taiwan. Arch. Virol. 1996;141:1677–1688. doi: 10.1007/BF01718291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Junker D., Collisson E.W. Evidence of natural recombination within the S1 gene of infectious bronchitis virus. Virology. 1993;192:710–716. doi: 10.1006/viro.1993.1093. [DOI] [PubMed] [Google Scholar]
- Youn S., Leibowitz J.L., Collisson E.W. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology. 2005;332:206–215. doi: 10.1016/j.virol.2004.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Jiang Y., Low S., Wang Z., Nam S.J., Liu W., Kwangac J. Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis. 2001;45:416–424. [PubMed] [Google Scholar]


