Highlights
-
•
Diarrhea samples from 65 foals were collected in Europe and screened for rotavirus.
-
•
From 26 qPCR positive stool samples, 11 could be (partially) genotyped.
-
•
In addition to the common G3/G14P[12] strains, the rare P[18] genotype was detected.
-
•
A vaccine based on an inactivated G3P[12] genotype is still relevant in Europe.
Keywords: Rotavirus, Vaccine, VP7, VP4, G3, G14, P[12], P[18]
Abstract
Equine group A rotavirus (RVAs) mainly cause disease in foals under the age of 3 months. Only sporadic data are available on the circulation of RVAs in equine populations in Europe. In this study, 65 diarrheic samples from foals under 4 months of age were collected in Belgium (n = 32), Germany (n = 17), Slovenia (n = 5), Sweden (n = 4), Hungary (n = 3), Italy (n = 2), France (n = 1) and The Netherlands (n = 1). Forty percent of these samples (n = 26) were found to be RVA positive by a quantitative RT-PCR assay. The viral load in 11 of these samples was sufficiently high to be (partially) genotyped. G3, G14 and P[12] were the main genotypes detected, and phylogenetic analyses revealed that they were closely related to contemporary equine RVA strains detected in Europe as well as in Brazil and South Africa. Regional variation was observed with only G14 and P[12] being detected in Germany, whereas mainly G3P[12] was encountered in Belgium. Surprisingly the only G14P[12] RVA strain detected in Belgium was also found to possess the very rare P[18] genotype, which has been described only once from equine RVA strain L338 detected in the UK in 1991. Despite the identification of this uncommon P[18] genotype, G3P[12] and G14P[12] RVA strains remained the most important genotypes in Europe during the study period. Based on this finding and the knowledge that G3P[12] and G14P[12] serotypes are partially cross-reactive it can be assumed that a vaccine based on an inactivated virus of the G3P[12] genotype is still relevant in the current European epidemiological situation, although the addition of a G14 strain would most likely be beneficial.
1. Introduction
Group A rotaviruses (RVAs) have been detected in horse populations around the world (Papp et al., 2013). Nowadays, classification of RVAs is performed almost exclusively by sequencing analyses of the gene segments encoding the VP7 and VP4 outer capsid proteins, and 27 G- and 37 P-genotypes have been recognized, respectively (Matthijnssens et al., 2011, Trojnar et al., 2013). To date, only genotypes G3P[12] and G14P[12] are considered to be of epidemiological importance in horses (Papp et al., 2013). Equine G3 RVA strains have been further subdivided into subtypes G3A and G3B based on their reactivity with mAbs (Browning et al., 1992). Interestingly, the subtype G3B seems to be restricted to Japan, whereas G3A seems to be prevalent in the rest of the world (Bailey et al., 2013).
Equine RVAs mainly cause disease in animals below the age of 3 months, resulting in damage to the small intestine leading to severe watery diarrhea, which may lead to death (Imagawa et al., 1991, Sellon and Long, 2014). They cause significant economic losses because infected foals need to be quarantined from other foals and pregnant mares to avoid the further spread of the virus, which is a labor-intensive and costly event for horse-breeders (Ntafis et al., 2010, Papp et al., 2013). Besides good farm management, vaccination is currently the most widely used method to reduce the risk of disease caused by RVA in horse farms (Sellon and Long, 2014). Currently 3 inactivated vaccines are licensed globally and administered to pregnant mares. Vaccines based on the inactivated H-2 strain (G3AP[12]) are licensed in the US, New Zealand, Australia and several European countries (Zoetis Inc.). The vaccine licensed in Japan uses the HO-5 strain (G3BP[12]) (Nisseiken Co., Ltd). The third vaccine is licensed in Argentina and contains the equine RVA strain H-2, a simian G3P[2] RVA strain and the bovine G6P[1] RVA strain (Biogénesis Bagó).
The aim of this study was to investigate which equine RVAs genotypes circulated in 2013 in Europe and, hence, the relevance of the vaccine licensed in the European Union which contains an inactivated equine RVA strain of the G3AP[12] genotype.
2. Materials and methods
2.1. Sample collection
Local veterinarians in Belgium, France, Germany, Hungary, Italy, Slovenia, Sweden and The Netherlands were offered the opportunity to send diarrheic samples from sick foals under three months of age to a Belgian laboratory (Dierengezondheidszorg, Torhout) for RVA testing in 2013. Samples tested positive were subsequently sent to a second lab (Laboratory of Clinical and Epidemiological Virology, Rega Institute for Medical Research, KULeuven) for genotyping.
2.2. RVA detection (qPCR)
Viral RNA was extracted from the fecal samples using the QIAamp Viral RNA mini kit (Qiagen/Westburg, Leusden, The Netherlands) according to the manufacturer's instructions. The extracted RNA was stored at −60 °C or below prior and after analysis. To confirm presence or absence of RVA, the commercial kit TaqVet™Triplex Ruminant Rotavirus & Coronavirus (Laboratoire Service International, Lissieu, France) which is suitable to detect equine RVAs was used. Based on the manufacturer declaration, the test is able to detect all subtypes by detecting the conserved NSP5 gene on genome segment 11, encoding a non-structural protein of RVAs. It has a limit of detection of 7 RVA RNA copies and was shown to be more sensitive than an antigen ELISA in a comparative study (Ramos et al., 2009).
2.3. RVA genotyping
The extracted RNA was denatured at 95 °C for 2 min. Reverse transcriptase PCR (RT-PCR) was carried out using the Qiagen OneStep RT-PCR Kit (Qiagen/Westburg) using VP7 primers Beg9 (5′-GGCTTTAAAAGAGAGAATTTCCGTCTGG-3′) and End9 (5′-GGTCACATCATACAATTCTAATCTAAG-3′), and the VP4 primers GEN_VP4_P12_10F (5′-TGGCTTCTCTTATTTACAGACAG-3′) and GEN_VP4_P12_2360R (5′-TCACATCCTTCAGAAGCTACTC-3′). For sample EQ44, also primers GEN_P[18]_44F (5′-ATTCTTACGCAGTAGACTTGTCAG- 3′) and GEN_P[18]_2362R (5′-GGTCACATCCTGCATAAGCTAC-3′) were used. The reaction was carried out with an initial reverse transcription step at 50 °C for 30 min, followed by PCR activation at 95 °C for 15 min, 35 cycles of amplification (30 s at 94 °C, 30 s at 50 °C for VP7 and 1 min at 50 °C for VP4, 1 min at 72 °C), and a final extension of 10 min at 72 °C in a Thermocycler Biometra T3000 (Biometra, Westburg BV, The Netherlands). PCR products (1062 bp, 2351 bp and 2319 bp, respectively) were run on a polyacrylamide gel, stained with ethidium bromide and visualized under UV-light. The PCR amplicons were purified using ExoSAP-IT (Affymetrix USB, Santa Clara, USA), and sequenced using the dideoxynucleotide chain termination method with the ABI PRISM® BigDye Terminator Cycle Sequencing Reaction kit (Perkin-Elmer Applied Biosystems, Foster City, CA) on an automated sequencer (ABI PRISMTM 3130). The primers Beg9, VP4-P12-10F and GEN_P[18]_44F described above were used as sequencing primers. The chromatogram sequencing files were inspected using the computer application Chromas 2.3 (Technelysium, Helensvale, Australia), and samples were genotyped using BLAST.
2.4. Phylogenetic analyses
Sequences were aligned using Muscle and the model test in MEGA determined T92 + G (VP7) and T92 + I (VP4) as the appropriate substitution models. Phylogenetic analyses for VP7 and VP4 sequences obtained in this study and references strains from GenBank were performed using the Maximum Likelihood algorithm with 1000 bootstrap replicates.
2.5. Accession numbers
Sequences obtained in this study were deposited in GenBank under the accession numbers: KM515863–KM515873 (VP4) and KM515874–KM515883 (VP7).
3. Results
3.1. Samples
Sixty-four (64) diarrheic samples were received from foals less than 3 months of age and 1 sample was obtained from a foal that was 112 days old at the day of sampling. Thirty-two (32) samples were received from Belgium, 1 from France, 17 from Germany, 3 from Hungary, 2 from Italy, 5 from Slovenia, 4 from Sweden and 1 from The Netherlands (Table 1 ). From each location (stud farm or private owner) a maximum of 3 samples was collected, with the exception of 1 stud farm in Germany from which 5 samples were received. The breed, gender, date of birth of and clinical signs observed in the foals are reported in Supplementary Data 1, and Table 2 .
Table 1.
Number of equine stool samples collected in different European countries, positivity rate by qRT-PCR targeting the NSP5 gene, and number of samples that were genotyped.
| qRT-PCR |
RT-PCR/Sequence Positive |
||||||
|---|---|---|---|---|---|---|---|
| Country | Total Number | POS |
NEG |
VP7 | VP4 | ||
| Number | % | Number | % | ||||
| Belgium | 32 | 15 | 46.9 | 17 | 53.1 | 4 | 4 |
| Germany | 17 | 7 | 41.2 | 10 | 58.8 | 4 | 5 |
| France | 1 | 0 | 0.0 | 1 | 100.0 | 0 | 0 |
| Hungary | 3 | 0 | 0.0 | 3 | 100.0 | 0 | 0 |
| Italy | 2 | 1 | 50.0 | 1 | 50.0 | 1 | 1 |
| The Netherlands | 1 | 0 | 0.0 | 1 | 100.0 | 0 | 0 |
| Sweden | 4 | 2 | 50.0 | 2 | 50.0 | 0 | 0 |
| Slovenia | 5 | 1 | 20.0 | 4 | 80.0 | 1 | 0 |
| Total | 65 | 26 | 40.0 | 39 | 60.0 | 10 | 10 |
Table 2.
Clinical information of the 11 foals from which the (partially) genotyped RVA strains were detected.
| Date of birth | Sample collection | Country | Sex | Vaccinated? | Bottle fed | Profuse watery diarrhea | Dehydration | Anorexia | Abdominal pain | Depression | CtVP6 qRT-PCR | VP7 sequencing | VP4 sequencing | RVA strain name |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25-Feb-13 | 4-Mar-13 | BE | M | NO | NO | YES | NO | YES | NO | NO | 33,62 | G3 | P[12] | RVA/Horse-wt/BEL/EQ12/2013/G3P[12] |
| 18-Feb-13 | 9-Mar-13 | IT | M | NO | NO | YES | YES | YES | NO | YES | 27,73 | G3 | P[12] | RVA/Horse-wt/ITA/EQ16/2013/G3P[12] |
| 24-Mar-13 | 2-Apr-13 | BE | F | NO | NO | YES | YES | YES | NO | YES | 32,01 | G3 | P[12] | RVA/Horse-wt/BEL/EQ20/2013/G3P[12] |
| 27-Mar-13 | 8-Apr-13 | DE | F | NO | NO | YES | NO | YES | YES | YES | 26,18 | G14 | P[12] | RVA/Horse-wt/GER/EQ24/2013/G14P[12] |
| 21-Mar-13 | 8-Apr-13 | DE | M | NO | NO | YES | NO | YES | NO | YES | 27,55 | G14 | P[12] | RVA/Horse-wt/GER/EQ25/2013/G14P[12] |
| 20-Feb-13 | 8-Apr-13 | DE | F | NO | NO | YES | NO | YES | NO | YES | 32,05 | NEG | P[12] | RVA/Horse-wt/GER/EQ26/2013/GxP[12] |
| 8-May-13 | 14-May-13 | BE | F | NO | YES | YES | YES | NO | NO | NO | 33,36 | G14 | P[12]+P[18] | RVA/Horse-wt/BEL/EQ44/2013/G14P[12+18] |
| 4-May-13 | 15-May-13 | SL | NO | NO | YES | NO | NO | NO | NO | 35,56 | G14 | NEG | RVA/Horse-wt/SLO/EQ46/2013/G14P[x] | |
| 28-Feb-13 | 28-May-13 | DE | F | NO | NO | YES | NO | NO | NO | NO | 30,23 | G14 | P[12] | RVA/Horse-wt/GER/EQ50/2013/G14P[12] |
| 18-Apr-13 | 13-Jun-13 | BE | M | NO | NO | YES | NO | NO | NO | NO | 26,79 | G3 | P[12] | RVA/Horse-wt/BEL/EQ62/2013/G3P[12] |
| 14-May-13 | 19-Jun-13 | DE | F | NO | NO | YES | NO | NO | NO | NO | 36,01 | G14 | P[12] | RVA/Horse-wt/GER/EQ68/2013/G14P[12] |
3.2. RVA positivity rate
From a total of 65 samples collected overall, 26 were found RVA positive (40.0%) by a diagnostic qRT-PCR targeting the RVA NSP5 gene, showing a wide range in the Ct values (26.18-38.88; Supplementary Data 1). The positivity rate for samples from Belgium and Germany (the two only countries with more than 5 RVA positive samples) was very similar with 46.9% and 41.2%, respectively, whereas it ranged between 0 and 50% for the remaining countries with 2 RVA positive samples or less (France, Hungary, Italy, Slovenia, Sweden and The Netherlands) (Table 1).
3.3. RVA genotyping
The 26 qRT-PCR positive samples were subjected to VP7 and VP4 RT-PCR and sequence analyses in order to determine the G- and P-genotypes, respectively. The G and P-genotype could be determined unambiguously for only 8 samples (3 from Belgium [EQ12, EQ20, and EQ62], 4 from Germany [EQ24, EQ25, EQ50 and EQ68] and 1 from Italy [EQ16]). Furthermore, for 1 sample from Germany, only the P-genotype could be determined [EQ26], for 1 sample from Slovenia only the G-genotype could be determined [EQ46], and from 1 sample from Belgium [EQ44] a mixed P-genotype was found (Table 2 and supplementary data 1). In the sequence chromatogram of latter sample the first short part (≈ 230 bp, high sequence peaks intensity) belonged to genotype P[18], whereas the second longer part of the chromatogram (≈ 680 bp, low sequence peaks identity) belonged to genotype P[12]. Since P[18] is a very rare equine P-genotype, we developed P[18] specific primers, not cross reactive with the P[12] genotype, which were used for RT-PCR and sequence analyses of sample EQ44. This analyses confirmed the presence of the P[18] sequence in this sample. As the only currently identified P[18] strain (L338) was found in combination with the unusual G13 genotype, we also performed a novel RT-PCR with G13 specific primers, but no RT-PCR product was detected (data not shown).
Fig. 1 shows the genotype distribution of all (n = 26) investigated samples (referred to as “Europe”), and the genotype distribution for Belgium and Germany in particular. The data for Europe suggest that both the G3P[12] and the G14P[12] genotypes are more or less equally present in the samples analyzed. However, both genotype combinations do not seem to be equally distributed geographically, as only the G14 genotype was identified in German samples, whereas mainly G3 was identified in Belgian samples. Furthermore, a single G3P[12] strain was detected in an Italian sample, and a G14P[x] strain in a Slovenian sample.
Fig. 1.
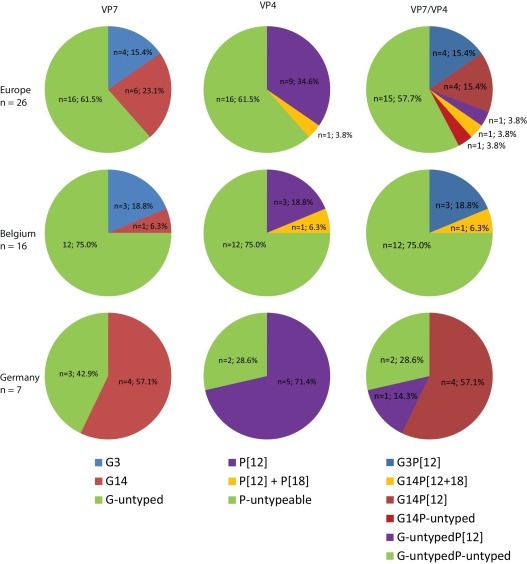
G- and P-genotype distribution of equine RVA strains. G- and P-genotype distribution of equine RVA strains over all the study sites (referred to as “Europe”), and the two sites, Belgium and Germany, with more than 5 samples tested.
3.4. RVA phylogenetic analyses
Fig. 2 shows the phylogenetic analyses of the obtained VP7 and VP4 sequences together with representative strains from GenBank. The 6 G14 RVA strains isolated from Belgian, German and Slovenian samples are very closely related to each other (showing nucleotide identities ranging from 98.9–100%), and to contemporary equine G14 strains detected in Italy, Brazil, South Africa and Ireland. The Italian and the 3 Belgian G3 RVA strains fall into two clusters (nucleotide identities ranging from 96.1-100%), closely related to G3A RVA strains from Ireland, Argentina, Germany and Australia (Fig. 2A). In the VP4 phylogenetic tree the same strains cluster together, as in the VP7 tree. The P[12] sequences of the G14 carrying strains are nearly identical (99.4-100%), and cluster with P[12] strains from South Africa and Ireland. These P[12] sequences are located in a different cluster as compared to the G3 carrying P[12] strains (nucleotide identities ranging from 94.8–95.9%). The former P[12] strains are closely related with equine P[12] RVA strains from Ireland and Greece, and show a genetic diversity as low as 98.6% (Fig. 2B). The P[18] VP4 sequence obtained from sample EQ44 was shown to cluster with reference strain L338 and showed a 96.0% nucleotide similarity.
Fig. 2.
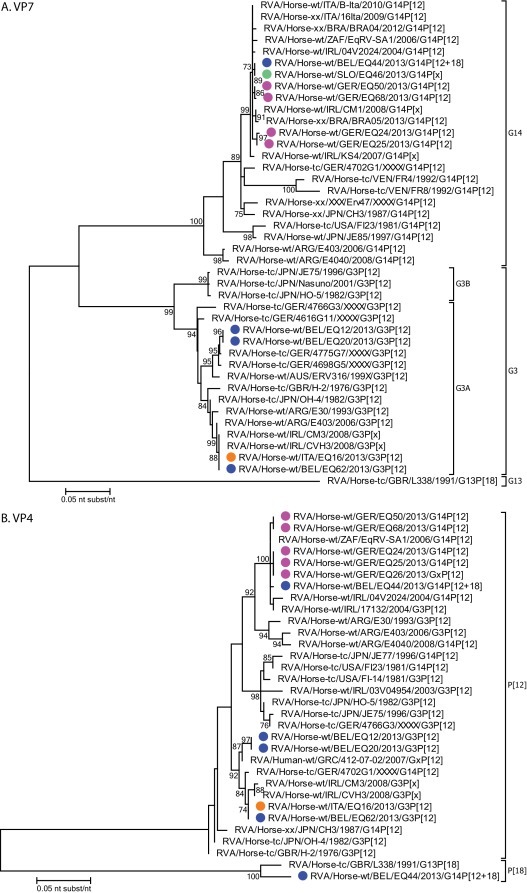
Phylogenetic analyses. Phylogenetic analyses of the VP7 (Figure 2A) and VP4 (Figure 2B) gene segments obtained in this study, together with relevant strains available in GenBank. Strains from this study are indicated with a circle. Blue, magenta, orange and green colors represent samples from Belgium, Germany, Italy and Slovenia, respectively.
4. Discussion and conclusions
Vaccination of infants or pregnant mothers has been shown one of the most effective measurements to prevent RVA gastroenteritis in humans as well as in a variety of animal species (Dhama et al., 2009). For the prevention of gastroenteritis caused by RVA in horses, the approach of using inactivated RVA vaccines to administer to pregnant mares has been selected. A number of studies have been conducted assessing the vaccine efficacy of equine RVA vaccines. It was shown in several field studies that following vaccination of mares, antibodies can be detected in blood, colostrum and their foals without adverse effects (Barrandeguy et al., 1998, Imagawa et al., 2005, Powell et al., 1997). Follow-up of clinical parameters associated with RVA infections in these studies demonstrated variable vaccine efficacy, which suggests that vaccination provides reasonable but not complete protection. However, the interpretation of field data is in general difficult. As the overall amount of RVA is reduced in the stable or herd when horses are vaccinated, this reduced infection pressure benefits also unvaccinated horses in the study which makes the assessment of vaccine efficacy difficult. Although little is known about cross protection between G3 and G14 serotypes, a recent study in Japan suggests that a vaccine based on a G3BP[12] RVA strain also generated cross protective virus-neutralizing antibodies against G14P[12] RVA strains, albeit at a slightly lower level (Nemoto et al., 2012). This confirms previous studies, which demonstrated that G3 and G14 serotypes partly cross-react with each other (Imagawa et al., 2005, Imagawa et al., 2002). This cross-reaction in combination with the fact that G3 and G14 are also genetically related G types suggests that a vaccine based on one of these serotypes should offer a certain degree of (cross-)protection against both serotypes. A previous study demonstrated that diarrhea caused by a serotype G14 rotavirus infection was comparatively milder in the group of horses vaccinated with an inactivated G3 serotype RVA vaccine as compared to the non-vaccinated group (Imagawa et al., 2005). This raises questions about the benefit of using a G3/G14 multivalent vaccine as suggested by some researchers (Bailey et al., 2013). Nonetheless, there are a few observational studies, which have suggested that vaccination may have caused a shift in the genotype distribution of circulating equine RVA strains in Ireland (Collins et al., 2008) and Argentina (Garaicoechea et al., 2011). Our investigation was an additional attempt to increase the knowledge on the epidemiological situation of equine RVAs in Europe and predict the usefulness of currently available commercial vaccines.
Our study indicated that RVA RNA could be detected in 40% of the 65 fecal samples from diarrheic foals all over Europe, which was similar to the prevalence seen in countries with more than 5 submitted positive samples such as Belgium (46.9%) and Germany (41.2%). The prevalence observed in this study is in line with published data. Bailey and colleagues (2013) reported that the frequency of detection of RVA in clinical cases varies from 20 to 77%. They appear to be endemic in most, if not all, horse populations (Browning et al., 1991b, Conner and Darlington, 1980, Higgins et al., 1990, Monini et al., 2011, Netherwood et al., 1996, Ohta et al., 1990).
The Ct values obtained by the diagnostic qRT-PCR indicated a wide range in the amount of viral particles found in stool samples (Table 2). The low amount of RVA particles in certain stool samples could be explained by the fact that those samples were taken after the peak of RVA shedding, that a fraction of the virus particles was degraded during sample transportation and storage, or that the RVA infection was only very mild, which raises the question as to whether RVA or another pathogen was the primary cause or only a contributing factor of the observed diarrhea.
These differences in the viral load present in stool samples were also reflected in the number of samples that could be G and/or P-genotyped. All samples with a Ct value lower than 32.03 (n = 6) could be fully typed. None of the samples with a Ct value above 36.10 (n = 8) resulted in PCR products needed for sequencing. The genotyping results for samples with a Ct between 32.03 and 36.10 were variable ranging from fully typed (n = 3), over partially typed (n = 2) to untyped (n = 6). These results indicate that the diagnostic qRT-PCR has a higher sensitivity as compared to the VP4 and VP7 PCRs.
Our genotyping results indicate that the G3P[12] and G14P[12] genotype combinations are the most prevalent, which confirms global data on equine RVAs (Papp et al., 2013). Our data also suggest that regional differences in the genotype distribution exist, as can be seen in the data from Belgium and Germany, where G3 and G14, respectively, were most prevalent, although this was based on rather small numbers. In this context, an interesting observation was the finding of the P[18] genotype in the Belgian RVA strain EQ44, in a mixed infection with G14 and P[12]. L338 is thus far the only P[18] RVA strain detected worldwide, isolated from a foal with diarrhea in the UK in 1991 (Browning et al., 1991a). Complete genome analyses of this strain revealed that the 11 gene segments were unique, each belonging to their own genotype (Matthijnssens et al., 2012). The single isolation of this particular RVA strain has always raised questions, whether or not L338 was a genuine equine RVA, or may have been the result of an interspecies transmission from a yet unidentified host species. The detection of a novel P[18] RVA strain in this study, 22 years after the detection of L338, lends support to the former hypothesis of a genuine equine RVA strain. This hypothesis is further supported by the fact that the obtained P[18] sequence showed 4.0% differences on the nucleotide level compared to L338, suggesting an accumulation of mutations has occurred over 2 decades. However, it cannot be excluded that this evolution has occurred in a different host species, and that this virus was transmitted to horses on two different occasions. L338 was detected in combination with G13, whereas we could not detect G13 in the sample. This might be explained by (a) the P[18] from EQ44 was combined with G14, (b) P[18] was combined with a genotype that was not detected by our VP7 primers or (c) G13 has accumulated mutations in the G13 primer binding sites, and was therefore not detected. Intensified RVA surveillance programs in horses are needed to address these questions.
Phylogenetically all G14P[12] RVA strains identified (Belgium, Slovenia and Germany) were very closely related to each other and to other contemporary equine RVA strains, probably reflecting the swift migration of viruses together with their hosts. The identified G3P[12] RVA strains (Belgium and Italy) fell into two distinct lineages inside the phylogenetic clusters containing RVA strains belonging to the G3A lineage, as could be expected for strains detected outside Japan (Bailey et al., 2013).
In conclusion, although the uncommon P[18] genotype was found on one occasion in this study, G3P[12] and G14P[12] were demonstrated to be the most important genotypes in Europe. Based on this finding and the knowledge that G3P[12] and G14P[12] serotypes are partially cross-reactive it can be assumed that a vaccine based on an inactivated G3P[12] virus is still relevant in the current European epidemiological situation, although the addition of a G14 strain would most likely be beneficial.
Conflict of interests
The study design, sample collection, analyses and interpretation of data, writing of the manuscript, and the decision were to submit the publication, were done by the authors of the study. This study was sponsored by Zoetis Belgium S.A. Rudiger Raue and Ellen Ons are employees of Zoetis.
Acknowledgments
The authors would like to thank all veterinarians who participated in this study and Equi Focus Point Belgium for the communication of this study to the Belgian veterinarians.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetmic.2015.01.011.
Contributor Information
Jelle Matthijnssens, Email: Jelle.matthijnssens@uz.kuleuven.ac.be.
Ellen Ons, Email: Ellen.ons@zoetis.com.
Sarah De Coster, Email: Sarah.decoster@uzleuven.be.
Nádia Conceição-Neto, Email: Nadia.daconceicaoneto@uzleuven.be.
Annick Gryspeerdt, Email: Annick.Gryspeerdt@dgz.be.
Marc Van Ranst, Email: Marc.vanranst@uzleuven.be.
Rudiger Raue, Email: Rudiger.raue@zoetis.com.
Appendix A. Supplementary data
References
- Bailey K.E., Gilkerson J.R., Browning G.F. Equine rotaviruses--current understanding and continuing challenges. Vet. Microbiol. 2013;167:135–144. doi: 10.1016/j.vetmic.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandeguy M., Parreño V., Lagos Marmol M., Pont Lezica F., Rivas C., Valle C., Fernandez F. Prevention of rotavirus diarrhoea in foals by parenteral vaccination of the mares: field trial. Dev. Biol. Stand. 1998;92:253–257. [PubMed] [Google Scholar]
- Browning G.F., Chalmers R.M., Fitzgerald T.A., Snodgrass D.R. Serological and genomic characterization of L338, a novel equine group A rotavirus G serotype. J. Gen. Virol. 1991;72:1059–1064. doi: 10.1099/0022-1317-72-5-1059. [DOI] [PubMed] [Google Scholar]
- Browning G.F., Chalmers R.M., Fitzgerald T.A., Snodgrass D.R. Evidence for two serotype G3 subtypes among equine rotaviruses. J. Clin. Microbiol. 1992;30:485–491. doi: 10.1128/jcm.30.2.485-491.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning G.F., Chalmers R.M., Snodgrass D.R., Batt R.M., Hart C.A., Ormarod S.E., Leadon D., Stoneham S.J., Rossdale P.D. The prevalence of enteric pathogens in diarrhoeic thoroughbred foals in Britain and Ireland. Equine Vet. J. 1991;23:405–409. doi: 10.1111/j.2042-3306.1991.tb03751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.J., Cullinane A., Martella V., O'Shea H. Molecular characterization of equine rotavirus in Ireland. J. Clin. Microbiol. 2008;46:3346–3354. doi: 10.1128/JCM.00995-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner M.E., Darlington R.W. Rotavirus infection in foals. Am. J. Vet. Res. 1980;41:1699–1703. [PubMed] [Google Scholar]
- Dhama K., Chauhan R.S., Mahendran M., Malik S.V. Rotavirus diarrhea in bovines and other domestic animals. Vet. Res. Commun. 2009;33:1–23. doi: 10.1007/s11259-008-9070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaicoechea L., Mino S., Ciarlet M., Fernandez F., Barrandeguy M., Parreno V. Molecular characterization of equine rotaviruses circulating in Argentinean foals during a 17-year surveillance period (1992-2008) Vet. Microbiol. 2011;148:150–160. doi: 10.1016/j.vetmic.2010.08.032. [DOI] [PubMed] [Google Scholar]
- Higgins W.P., Gillespei J.H., Schiff E.I. Equine rotavirus: three epizootics. Equine Practice. 1990;12:15–18. [Google Scholar]
- Imagawa H., Kato T., Tsunemitsu H., Tanaka H., Sato S., Higuchi T. Field Study of Inactivated Equine Rotavirus Vaccine. J. Equine Sci. 2005;16:35–44. [Google Scholar]
- Imagawa H., Sekiguchi K., Anzai T., Fukunaga Y., Kanemaru T., Ohishi H., Higuchi T., Kamada M. Epidemiology of equine rotavirus infection among foals in the breeding region. J. Vet. Med. Sci. 1991;53:1079–1080. doi: 10.1292/jvms.53.1079. [DOI] [PubMed] [Google Scholar]
- Imagawa H., Tsunemitsu H., Wada R., Fukunaga Y. Sero-Epidemiological Survey of Equine Rotavirus Infections in 1-year-old Thoroughbred Horses in the Hidaka Region of Hokkaido. J. Equine Sci. 2002;13:71–74. [Google Scholar]
- Matthijnssens J., Ciarlet M., McDonald S.M., Attoui H., Banyai K., Brister J.R., Buesa J., Esona M.D., Estes M.K., Gentsch J.R., Iturriza-Gomara M., Johne R., Kirkwood C.D., Martella V., Mertens P.P., Nakagomi O., Parreno V., Rahman M., Ruggeri F.M., Saif L.J., Santos N., Steyer A., Taniguchi K., Patton J.T., Desselberger U., Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch. Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., Mino S., Papp H., Potgieter C., Novo L., Heylen E., Zeller M., Garaicoechea L., Badaracco A., Lengyel G., Kisfali P., Cullinane A., Collins P.J., Ciarlet M., O'Shea H., Parreno V., Banyai K., Barrandeguy M., Van Ranst M. Complete molecular genome analyses of equine rotavirus A strains from different continents reveal several novel genotypes and a largely conserved genotype constellation. J. Gen. Virol. 2012;93:866–875. doi: 10.1099/vir.0.039255-0. [DOI] [PubMed] [Google Scholar]
- Monini M., Biasin A., Valentini S., Cattoli G., Ruggeri F.M. Recurrent rotavirus diarrhoea outbreaks in a stud farm, in Italy. Vet. Microbiol. 2011;149:248–253. doi: 10.1016/j.vetmic.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Nemoto M., Tsunemitsu H., Murase H., Nambo Y., Sato S., Orita Y., Imagawa H., Bannai H., Tsujimura K., Yamanaka T., Matsumura T., Kondo T. Antibody response in vaccinated pregnant mares to recent G3BP[12] and G14P[12] equine rotaviruses. Acta Vet. Scand. 2012;54:63. doi: 10.1186/1751-0147-54-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherwood T., Wood J.L., Townsend H.G., Mumford J.A., Chanter N. Foal diarrhoea between 1991 and 1994 in the United Kingdom associated with Clostridium perfringens, rotavirus, Strongyloides westeri and Cryptosporidium spp. Epidemiol. Infect. 1996;117:375–383. doi: 10.1017/s0950268800001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntafis V., Fragkiadaki E., Xylouri E., Omirou A., Lavazza A., Martella V. Rotavirus-associated diarrhoea in foals in Greece. Vet. Microbiol. 2010;144:461–465. doi: 10.1016/j.vetmic.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Ohta C., Hoshi A., Goto H., Tsunoda N., Tagami M., Akita H. Epizootiological and virological studies of foal diarrhea associated with serotype 3 rotavirus. Nippon Juigaku Zasshi. 1990;52:1049–1056. doi: 10.1292/jvms1939.52.1049. [DOI] [PubMed] [Google Scholar]
- Papp H., Matthijnssens J., Martella V., Ciarlet M., Banyai K. Global distribution of group A rotavirus strains in horses: a systematic review. Vaccine. 2013;31:5627–5633. doi: 10.1016/j.vaccine.2013.08.045. [DOI] [PubMed] [Google Scholar]
- Powell D.G., Dwyer R.M., Traub-Dargatz J.L., Fulker R.H., Whalen J.W., Jr., Srinivasappa J., Acree W.M., Chu H.J. Field study of the safety, immunogenicity, and efficacy of an inactivated equine rotavirus vaccine. J. Am. Vet. Med. Assoc. 1997;211:193–198. [PubMed] [Google Scholar]
- Ramos F., Daly S., Bouley S., Gueneau E., Asdrubal P., Nicollet P., Maingourd C., Mathevet P., Sellal E. European Buiatrics Forums; Marseille: 2009. Development of a New Real Time RT-PCR System for Detection of Rotavirus and Coronavirus in Faeces from Calf Diarrhea - Comparison with ELISAs; p. 103. [Google Scholar]
- Sellon D.C., Long M. 2nd Ed. Elsevier; Saunders: 2014. Equine Infectious Diseases. [Google Scholar]
- Trojnar E., Sachsenroder J., Twardziok S., Reetz J., Otto P.H., Johne R. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. J. Gen. Virol. 2013;94:136–142. doi: 10.1099/vir.0.047381-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


